Abstract
Sepiapterin reductase (SPR) catalyzes the final step of tetrahydrobiopterin (H4B) biosynthesis and the first step of H4B regeneration from an exogenous precursor sepiapterin. Despite the potential significance of SPR in regulating H4B-dependent nitric oxide (NO•) production, the endothelium-specific sequence and functions of SPR remain elusive. We first cloned endothelial SPR cDNA from bovine aortic endothelial cells (Genebank: DQ978331). In cells transiently transfected with SPR gene, SPR activity (HPLC) was dramatically increased by 19-fold, corresponding to a significant increase in endothelial H4B content (HPLC) and NO• production (electron spin resonance). In vivo delivery of SPR gene significantly increased vascular SPR protein expression (mouse vs. bovine antibodies to differentiate endogenous vs. exogenous), activity, H4B content, and NO• production, as well as NO•-dependent vasorelaxation. In endothelial cells transfected with small interfering RNA specific for SPR, ∼87% of mRNA were attenuated (real-time quantitative RT-PCR), corresponding to a significant reduction in SPR protein expression and activity, which was associated with decreases in both intracellular H4B content and NO• level. Exogenous administration of sepiapterin to endothelial cells significantly upregulated H4B and NO• levels, which were attenuated by SPR RNA interference (RNAi). H4B-stimulated increase in NO• production, however, was SPR RNAi independent. GTP cyclohydrolase 1 expression and activity, as well as dihydrofolate reductase expression, were not affected by SPR RNAi, whereas dihydrofolate reductase activity was significantly downregulated. These data represent the first to study endothelial SPR functionally and clearly demonstrate an important role of endothelial SPR in modulating H4B and NO• bioavailability.
Keywords: endothelial nitric oxide synthase, aortic endothelial cells, guanosine 5′-triphosphate cyclohydrolase 1, dihydrofolate reductase
tetrahydrobiopterin (h4b) is required for many enzymatic activities in cells and thus essential for various biological processes. For example, the nitric oxide radical (NO•)-producing activity of endothelial NO synthase (eNOS) is regulated by intracellular H4B levels (32). When H4B content is adequate, eNOS favors NO• production; when H4B level is limiting, eNOS becomes uncoupled and starts to generate superoxide (4, 16, 33, 39, 40, 42). This uncoupling of eNOS may contribute to endothelial dysfunction via two mechanisms: 1) NO• production is diminished; and 2) eNOS turns into a peroxynitrite generator (4, 29). From this point of view, NO• bioavailability could be decidedly affected by H4B-regulated eNOS function.
H4B is rapidly degradable by oxidation. Under most circumstances, H4B levels are determined by a balance of its generation, oxidation/degradation, and salvation (34). For the de novo biosynthesis, H4B is synthesized from GTP via the three-step pathway of GTP cyclohydrolase 1 (GTPCH1; rate limiting enzyme), 6-pyruvoyl-tetrahydropterin synthase, and sepiapterin reductase (SPR) (41). Alternatively, H4B can be synthesized from the exogenous pterin precursor sepiapterin, which is metabolized to dihydrobiopterin (H2B) by SPR first, and further to H4B by dihydrofolate reductase (DHFR). When H4B is endogenously oxidized to H2B, i.e., by peroxynitrite (18, 25), it can also be reduced back to H4B by DHFR (28, 35, 41). Our laboratory has previously shown that a DHFR deficiency occurs in response to ANG II-mediated uncoupling of eNOS.
Different from other H4B metabolic enzymes, SPR is uniquely involved in both synthetic and salvage pathways of H4B. The expression and activity of SPR in endothelial cells, particularly relevant to H4B level and NO• bioavailability, have not yet been explored. In the present study, we initiated such an effort by cloning endothelium-specific SPR from bovine aortic endothelial cells (BAECs), followed by monitoring endothelial H4B and NO• levels in SPR overexpressed or suppressed cells and mice. Importantly, we found that, whereas overexpression of SPR was associated with increased production of NO• and H4B content in vitro and in vivo, RNA interference (RNAi) silencing of SPR (not affecting GTPCH1 activity) attenuated H4B and NO• bioavailability. Sepiapterin or H4B supplementation increased endothelial cell production of NO• and H4B, but only the effect of sepiapterin was attenuated by SPR RNAi. These data clearly demonstrate an important role of SPR in modulating endothelial H4B and NO• bioavailability, which is more prominent when sepiapterin is administrated.
METHODS
Materials and animals.
Reagents and kits used were as follows: customized oligonucleotides from Integrated DNA Technologies (Coralville, IA); Platinum Taq polymerase, dNTP mix, and 1 kb plus ladder from GIBCO BRL (Carlsbad, CA), pGEMT-easy vector from Promega (Madison, WI); RNeasy Mini Kit, QIAprep Miniprep Kit, and QIAEX II Gel Extraction Kit from Qiagen (Hilden, Germany); SMART RACE cDNA Amplification Kit and BD Advantage 2 Polymerase Mix from Clontech Laboratories (Palo Alto, CA); ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (version 3.0) from PE Applied Biosystems (Foster City, CA); Luria Bertani broth and agar from Sigma (St Louis, MO); and Superscript II reverse transcriptase from Invitrogen (Carlsbad, CA). The tissue culture reagents used were as follows: FBS (Sigma, St Louis, MO), medium 199, l-glutamine, penicillin/streptomycin, and MEM vitamin (Mediatech, Herndon, VA). Other chemicals were purchased from Sigma in the highest purity available. C57BL6 mice were purchased from Harlan (Indianapolis, IN). The procedure was carried out based on protocols approved by the University of California Los Angeles Institutional Animal Care and Use Committee.
Rabbit antiserum for SPR that cross reacts with both mouse and bovine SPR (common domain selected as antigen) was customized from YenZym (San Francisco, CA). Mouse-specific rabbit antiserum was a generous gift of Dr. Y. S. Park from Inje University, Republic of Korea.
Cell culture and immunoblots.
BAECs (Cell Systems, Kirkland, WA), between the second and sixth passage, were cultured in medium 199 supplemented with 10% FBS, 1% l-glutamine, 1% penicillin-streptomycin, and 1% MEM vitamin to confluence. For immunoblotting, cells were lysed, and 20 to 40 μg of protein were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes, and probed with DHFR (1:250) or β-actin (1:5,000) antibodies.
Cloning of SPR by 5′- and 3′-RACE.
The 5′- and 3′-rapid amplification of cDNA ends (RACE) cDNA libraries were constructed using the SMART RACE cDNA Amplification Kit, according to the manufacturer's manual. For bovine SPR cDNA amplification, the 5′-RACE reaction mix consisted of 2.5 μl 5′-RACE-Ready cDNA, 5 μl universal primer mix containing long primer 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ and short primer 5′-CTAATACGACTCACTATAGGGC-3′, and the final concentration of 1× Advantage 2 Polymerase Mix, 1× PCR buffer, 0.2 mmol/l dNTP mix, and 0.2 μmol/l SPR-GR2 (antisense primer) 5′-GGGCTCCAGACTCGAACTTGTC- 3′, in a total volume of 50 μl. The three-step thermal cycling profile was as follows: five cycles of denaturation at 94°C/20 s, followed by an extension at 72°C/2 min; 30 cycles of denaturation at 94°C/20 s, annealing at 68°C/30 s, extension at 72°C/2 min, followed by a final extension of 72°C/10 min. The 3′-RACE reaction mix, which yielded the full-length ∼1.4-kb cDNA, consisted of 2.5 μl 3′-RACE-Ready cDNA, 5 μl universal primer mix, and the final concentration of 1× Advantage 2 Polymerase Mix, 1× PCR buffer, 0.2 mmol/l dNTP mix, and 0.2 μmol/l SPR-GSP1F (sense) 5′-ATGGTAGCGGCTCCAGCTAGCAGGTAGGAGGCATG-3′ in a total volume of 50 μl. The three-step thermal cycling profile was as follows: one cycle of hot start at 94°C/20 s; 35 cycles of denaturation at 94°C/20 s, annealing at 68°C/30 s, extension at 72°C/2 min, followed by a final extension of 72°C/10 min. The PCR products were fractionated by 1% agarose gel electrophoresis. The most intense bands from both the 5′- and 3′-RACE amplifications were excised, purified, and ligated into TA vector (Qiagen), and at least 8–10 clones from both the 5′- and 3′-RACE libraries were fully sequenced using T7, SP6, and various internal primers. To express in BAEC, the SPR gene was further cloned into pEGFPN1 vector (Invitrogen). GFP region in the vector was removed during the cloning process to avoid possible intervention of fluorescence with HPLC measurement of SPR activity and H4B content.
Transient transfection experiments.
BAEC were plated at a density of 70–80% confluence 24 h before transfection. Transfection of cells was performed according to the standard TransPass V protocol (BioLab). The cells were processed 48–72 h after the transfection for analysis of H4B and NO• levels. Plasmid pcDNA 3.0 (Invitrogen) was used as a control.
Real-time PCR.
Real-time PCR was performed on a Bio-Rad iCYCLER iQ real-time PCR machine. The PCR reaction consisted of 2× iQ SYBR Green Supermix (Bio-Rad), 25 pmol of each primer, and 5 μl of cDNA as template in a final reaction volume of 25 μl. The PCR conditions were 95°C for 3 min and 40 cycles of 95°C/15 s, 52°C/15 s, and 60°C/15 s. BSPR primers were as follows: (forward) 5′-AGCGAGTGCTGCTCATCAACAATG-3′, (reverse) 5′-TCACTTCAGTTGGGTCAGTCAGGT-3′. β-actin primers were as follows: (forward) 5′-GGCACCCAGCAC AATGAAGATCAA-3′, (reverse) 5′-AGCTAACAGTCCGCCTAGAAGCAT-3′. GTPCH1 primers were as follows: (forward) 5′-AACAAGCAAGTTCTTGGCCTCA-3′, (reverse) 5′-AAGGCCTCTATGATTGCTACGG-3′.
Determination of SPR activity.
The SPR activity assay was performed according to Ferre and Naylor (8) with modification. In brief, the reaction was carried out in phosphate buffer (0.1 mol/l, pH 7.5) containing the final concentration of 0.125 mmol/l sepiapterin, 0.25 mmol/l NADPH, and 5 μg cell lysate. The enzymatic reaction was allowed at 37°C for 2 h in the dark, and the biopterin conversion rate was used as an index of SPR activity. The reaction was stopped by incubation with 25 μl of iodine solution for 10 min in the dark. The precipitated proteins were removed by centrifugation at 15,000 g for 3 min at room temperature. The excessive iodine was then quenched by addition of 25 μl of 2% ascorbic acid. The resulting biopterin content was measured by HPLC system (SCL-10AVP) (Shimadzu, Columbia, MD). The separation was carried out in a reverse-phase column (C18, 5 μm, 4.6 mm × 250 mm) (Alltech, Nicholasville, KY) at isocratic gradient using 5% methanol as mobile phase. Fluorescence was monitored with a fluorescent detector (RF-10AXL) (Shimadzu) at the wavelengths of 362 nm for excitation and 435 nm for emission. The flow rate was kept at 1 ml/min. Chromatographic profile was analyzed with the aid of EZstart chromatography software (Shimadzu).
Determination of H4B contents.
Cells were lysed using trichloroacetic acid containing 10 mmol/l dithiothreitol. Lysates were subjected to differential oxidation in acidic (0.2 mol/l trichloroacetic acid) or alkalytic (0.1 mol/l NaOH) solutions containing 2.5% I2/10% KI or 0.9% I2/1.8% KI, as shown previously. After centrifugation, 20 μl supernatant were injected into a HPLC system with a fluorescent detector. Excitation and emission wavelengths of 350 and 450 nm, respectively, were used to detect fluorescent H4B and its oxidized species, as previously shown (4, 30).
Detection of NO• with electron spin resonance.
Bioavailable NO• in BAECs were detected using electron spin resonance (ESR), as described previously (1, 2). In brief, endothelial cells were rinsed with modified Krebs/HEPES buffer and incubated with freshly prepared NO•-specific spin trap Fe2+(DETC)2 colloid (0.5 mmol/l) for 60 min. Gently collected cell suspensions were snap-frozen in liquid nitrogen and loaded into a finger Dewar for analysis with a Bio-Spin ESR spectrophotometer (Bruker) at the following settings: center field, 3485; sweep width, 100 G; frequency, 9.74 GHz; microwave power, 13.26 mW; modulation amplitude, 9.82 G; 512 points resolution; receiver gain 356.
In vivo SPR gene delivery.
To enhance the in vivo transfection efficiency, SPR construct (pEGFPN1) was mixed with a cocktail of OptiMEM (Invitrogen) and Lipofectamine (Invitrogen), according to the manufacturer's instruction. Tail vein injection was used to deliver SPR construct, and 48 h later, mice aorta were harvested and collected for NO• and SPR activity measurements.
Assessment of vasorelaxation.
The aorta were cut into 2- to 3-mm rings and mounted under 100-mmHg tension in 5-ml organ bath in Kreb's bicarbonate solution and gassed with 5% CO2 in O2 at 37°C. Isometric tension was recorded via two stainless steel wires through the lumen, one of which was fixed and the other attached to a transducer (Fort 10, WPI). Each preparation was allowed to equilibrate for 120 min before exposure to potassium physiological saline solution (122 mM). In aortic rings precontracted with 1 μmol/l phenylephrine to reach 70% maximal contraction, relaxant responses to acetylcholine (ACh) (10−9-10−6 mol/l, 1 log unit increment) were then assessed between groups. Vasodilator responses were calculated as percent recovery of phenylephrine response.
RNAi.
Based on the cloned sequence, short interfering RNAs (siRNAs) against SPR of bovine species were designed in house and synthesized from Qiagen (Hilden, Germany). The target sequence is 5′- CCCAACTGAAGTGAACAACTA-3′. Proliferating endothelial cells at 95% confluency were incubated with 50 nmol/l siRNA-oligofectamine mixtures in the presence of 10% growth media. To determine effects of RNAi on SPR expression, real-time PCR analysis was performed. β-Actin mRNA levels were used as references.
H4B, sepiapterin supplementation.
In additional experiments, siRNA or SPR construct (pEGFPN1) transfected cells were supplemented in serum-free media with H4B (5 μmol/l) or sepiapterin (5 μmol/l) for 2 h before NO• measurement and HPLC detection of H4B.
Detection of GTPCH1 activity.
To access GTPCH1 activity, cell lysates were incubated with GTP (0.75 mmol/l final concentration) at 37°C for 90 min in the dark. The reaction product was oxidized to neopterin triphosphate by acidic iodine (1% I2 and 2% KI in 2 mol/l TCA). After reduction of excessive iodine by ascorbic acid (2%) and adjustment of pH to 7.0, reaction products were monitored by HPLC with fluorescent detection (365/446 nm). The detailed chromatographic procedure has been described elsewhere (13).
Detection of DHFR activity.
The enzymatic assay was performed at 37°C. The assay buffer consisted of 0.1 mol/l potassium phosphate buffer (pH 7.4), 1 mmol/l DTT, 0.5 mol/l KCl, 1 mmol/l EDTA, and 20 mmol/l sodium ascorbate. NADPH (8 μl, 200 μmol/l final) was mixed with 132 μl assay buffer containing recombinant DHFR or cell/tissue lysate and then preincubated at 37°C for 3 min. Final protein concentration in sample was 6 μg/ml, unless another amount is specified. The reaction was initiated by the addition of 20-μl assay buffer with convenient concentration of dihydrolate. After 20 min of incubation, the reaction was terminated by 20 μl of trichloroacetic acid (final 0.2 mol/l), followed by the addition of 20 μl of stabilization solution (200 mg of sodium ascorbate and 30 mg of DTT in 1 ml of water). We found that, under these conditions, tetrahydrofolic acid (THF, specific product) is stable, and that samples can be analyzed by autosampler at 4°C. The THF content was monitored by the Shimadzu HPLC system, consisting of C-18 column (250 4.6-mm C18 column, Alltech) and fluorescent detector with wavelengths of 295 nm for excitation and 365 nm for emission and calculated against a standard curve prepared using known concentrations of THF solutions in assay buffer. Data are expressed as nanomole production of THF.
RESULTS
Cloning and characterization of endothelial SPR.
A full-length cDNA clone encoding endothelial SPR was isolated by screening the constructed bovine cDNA libraries. PCR products were cloned into pEGFPN1 vectors and sequenced. The cDNA sequence of SPR was deposited into Genbank under the accession number DQ978331 (date of submission: Sept 1, 2007). The SPR cDNA clone is 1,463 bp long. The cDNA predicatively encodes a 267-amino acid protein and showed 80% similarity with its human isoform (encodes 261-amino acid protein, Genebank ID NM_003124) using clustal W program (17). Additionally, 77% homology was found between bovine and mouse sequences (the mouse cDNA encodes 262-amino acid protein, Genebank ID NM_011467). Alignment with the only other bovine sequence (BC118299, neuronal) indicates that this endothelial isoform is 99% identical to SPR from neuronal tissues (Fig. 1), implicating ubiquitous expression and high tissue homology. The one amino acid difference (E236Q) between these two sequences needs to be further confirmed and studied, although artificial effect cannot be excluded.
Fig. 1.
Sepiapterin reductase (SPR) cDNA clone. The SPR amino acid sequences from endothelial (DQ978331) and neuronal (BC118299) SPR were aligned. The asterisks (“*”) represent the conservative part of the two sequences. The colon (“:”) represents the distinctive part of the two sequences. Underlining indicates from where the RNA interference (RNAi) was designed.
Effects of SPR overexpression on H4B and NO• bioavailability in vitro.
Since SPR is enzymatically involved in H4B synthesis, we speculate that increased SPR expression would increase intracellular H4B levels and NO production. We first investigated whether transfection of the cloned SPR gene could lead to increased SPR mRNA expression and activity. Aortic endothelial cells were transfected with plasmid pEGFPN1, containing SPR, and the transfection efficiency was determined by GFP expression by transfection of an empty GFP expression pEGFN1 vector in parallel. The transfection rate was ∼40% (Fig. 2A). Even with this efficiency, SPR mRNA and protein expression were markedly increased (Fig. 2, B and C). As shown in Fig. 2, D–F, augmented SPR expression was translated into significantly higher SPR activity, H4B level, and NO• production.
Fig. 2.
Effects of SPR gene transfection on tetrahydrobiopterin (H4B) and nitric oxide radical (NO•) bioavailability in vitro. A: transfection efficiency demonstrated by green fluorescence due to green fluorescent protein expression. B: quantification of SPR mRNA expression from cells [control (Ctrl) vs. SPR gene transfection] using real-time PCR. C: SPR protein expression after SPR gene transfection. D: effects of SPR gene transfection on SPR activity monitored by HPLC. E: effects of SPR gene transfection on intracellular H4B level. F: effects of SPR gene transfection on NO• production. Data are shown as means ± SE from 4–10 individual experiments. *P < 0.05 compared with Ctrl.
Effects of SPR overexpression on SPR activity, H4B content, NO• production, and vasorelaxation in vivo.
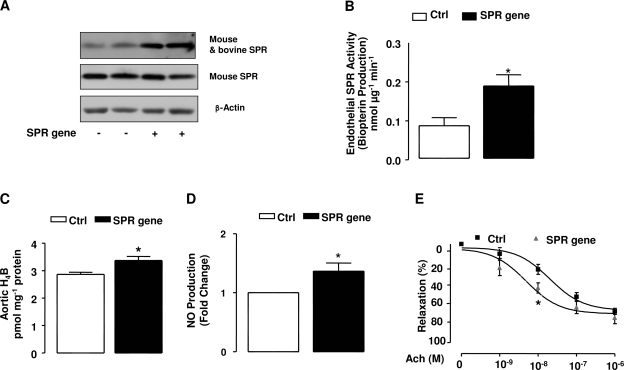
Recent evidence has established success of liposome reagents in gene deliveries in vivo in various disease models (19, 24). By tail vein, plasmid-containing SPR gene (pEGFPN1) was delivered every 12 h to mice for 48 h. Mouse-specific antiserum was used to detect basal expression of SPR. After detection, the mouse-specific antiserum was stripped from the membrane, and another customerized antibody, cross-reacting with both bovine and mouse SPR, was used to blot the membrane. As shown in Fig. 3A, overexpression of bovine SPR resulted in a marked increase in SPR abundance in mouse aortas. The aortas were then harvested for analysis of SPR activity, H4B content, and NO• production, and showed significant increases in all (Fig. 3, B–D), implicating that overexpression of SPR is effective in improving SPR-dependent NO• bioavailability in vivo. In additional experiments, ACh-induced vasorelaxation was also improved in wild-type C57BL6 mice after in vivo SPR gene delivery (Fig. 3E, significant improvement in response to ACh at 10−8 M).
Fig. 3.
Effects of SPR gene transfection on SPR activity and NO• production in vivo. Lipofectamine complexes containing empty vector or SPR gene were transferred to mice circulation via mouse tail vein injection twice a day for 2 days. Mouse aorta was harvested, as indicated in methods, and examined for SPR activity and NO• production. A: SPR protein expression after SPR gene tail vein injection (original mouse SPR and transfected bovine SPR level are both examined). B: effects of SPR gene transfection on SPR activity of mouse aorta. C: effect of SPR gene transfection on H4B content from aorta. D: effect of SPR gene transfection on NO• production from aorta. E: effect of SPR gene transfection on aortic relaxation in response to acetylcholine (ACh). Data are shown as means ± SE from 4–6 individual experiments. *P < 0.05 compared with Ctrl.
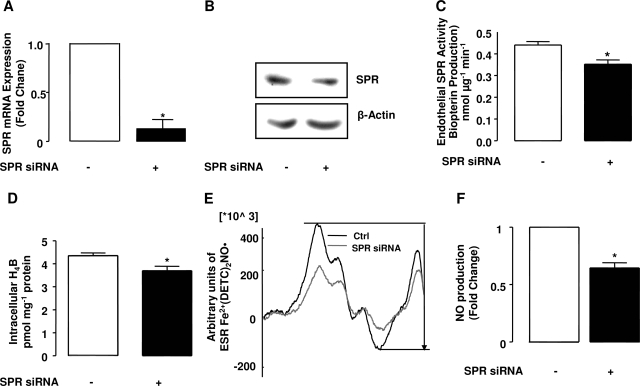
Effects of SPR RNAi on SPR mRNA expression, protein expression, and activity.
To further examine an endogenous role of SPR in modulating H4B and NO• availability, a RNAi tool was employed. The abundance of SPR mRNA in SPR or control RNAi-transfected endothelial cells was determined using competitive real-time PCR with endogenous control of β-actin (for details, see methods). As is obvious in Fig. 4A, SPR mRNA expression, normalized by steady β-actin mRNA, was significantly reduced by ∼87% upon transfection with SPR RNAi, which corresponded to a significant decrease in SPR protein expression and activity (Fig. 4, B and C). It is worth noting that, allegedly, SPR activity assay is not specific, as carbonyl reductase can catalyze similar reaction (2). However, the marked inhibition of SPR activity by SPR RNAi suggested that carbonyl reductase had limited interference.
Fig. 4.
Effects of SPR RNAi on SPR mRNA gene expression, SPR activity, H4B, and NO• bioavailability. Oligofectamine/RNAi complexes were transfected to endothelial cells, and the effects of SPR RNAi on SPR gene expression, SPR activity, H4B, and NO• level were examined 48 h after the transfection. A: real-time PCR results of SPR mRNA expression after RNAi transfection. SPR gene mRNA expression levels were standardized with β-actin, and the relative gene expression was calculated according to Livak and Schmittgen (21). B: effects of SPR RNAi on SPR protein expression. C: effects of SPR RNAi on SPR activity represented by SPR-driven pterin formation. D: summary of HPLC measurements of H4B contents after RNAi transfection. E: representative electron spin resonance (ESR) spectrum of Fe2+(DETC)2-NO•. F: fold changes of NO• production in endothelial cells transfected with SPR RNAi over control. Results are shown as means ± SE from 4 individual experiments. *P < 0.05 compared with Ctrl. siRNA, short interfering RNA.
Effects of SPR RNAi on H4B and NO• bioavailability.
Since SPR is responsible for both de novo synthetic and salvage pathways of H4B production, we next examined the effects of RNAi silencing of SPR on H4B and NO• bioavailability. As shown by Fig. 4D, intracellular H4B content was significantly decreased in SPR siRNA transfected cells. Endothelial NO• production, as shown by representative ESR spectra and quantitative data (Fig. 4, E and F), was also markedly diminished by RNAi silencing of SPR.
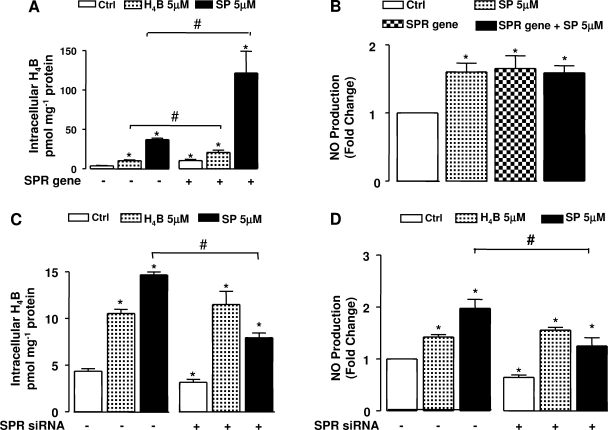
Influence of sepiapterin challenge on H4B and NO• bioavailability in SPR-overexpressed and SPR-silenced cells.
Exogenous H4B (5 μmol/l) and sepiapterin (5 μmol/l) were used to further examine whether SPR preferably improves H4B levels in the presence of its substrate sepiapterin vs. other biopterin precursors, such as H4B itself. Our study showed that both exogenous H4B and sepiapterin were significant in enhancing endothelial production of NO• and H4B bioavailability (Fig. 5). Moreover, there is an additive effect of exogenous H4B/sepipaterin supplementation over SPR gene transfection regarding elevated H4B levels, although the additive effect of H4B is much smaller (Fig. 5A). However, the additive effect of sepipaterin supplementation on intracellular H4B level (over SPR gene transfection) did not result in a proportional increase in NO• generation, indicating a possible saturation of H4B after SPR gene overexpression (Fig. 5B). In contrast, the beneficial effects of sepiapterin on intracellular H4B and NO• production were attenuated by SPR RNAi (Fig. 5, C and D), implicating that an enzymatic conversion of sepiapterin to H4B is involved in NO• production in resting endothelial cells. The effects of H4B, however, were unaffected by SPR RNAi.
Fig. 5.
Effects of SPR gene or SPR RNAi on endothelial NO• and H4B bioavailability in sepiapterin (SP) or H4B challenged endothelial cells. After the endothelial cells were transfected with SPR gene or SPR RNAi for 48 h, the cells were further incubated with H4B or sepiapterin (5 μmol/l, 2 h). A: effect of sepiapterin or H4B supplementation on intracellular H4B content in the presence or absence of SPR gene transfection. B: effect of sepiapterin supplementation on NO• production in the presence or absence of SPR gene transfection. C: effect of sepiapterin or H4B supplementation on intracellular H4B content in the presence or absence of SPR RNAi. D: effect of sepiapterin or H4B supplementation on NO• production in the presence or absence of SPR RNAi. Results are means ± SE from 3–6 individual experiments. *P < 0.05 compared with Ctrl. #P < 0.05, when comparisons were conducted between conditions indicated by brackets.
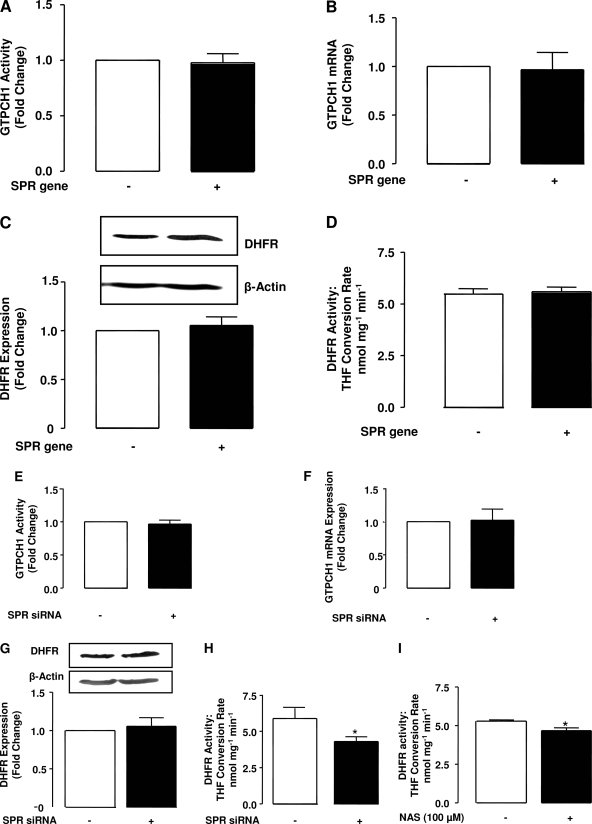
Effects of SPR overexpression or suppression on DHFR and GTPCH1 activity.
Although SPR seems important in regulating H4B bioavailability, other key enzymes involved in H4B metabolic pathways, such as DHFR and GTPCH1, might be affected by SPR overexpression/suppression. However, as shown in Fig. 6, A–D, both expression and activities of DHFR and GTPCH1 were not altered during SPR overexpression. Likewise, GTPCH1 mRNA expression and activity were both unaffected by SPR RNAi (Fig. 6, E and F). However, DHFR activity was significantly downregulated by SPR siRNA, although the mRNA expression of the enzyme was unchanged (Fig. 6, G and H). Of note, incubation of pharmacological inhibitor of SPR, N-acetyl-serotonin (100 μmol/l), with endothelial cell lysates also resulted in a modest but significant decrease in DHFR activity (Fig. 6I).
Fig. 6.
Effects of SPR overexpression and knockdown on GTP cyclohydrolase 1 (GTPCH1) and dihydrofolate reductase (DHFR) expression and activity. A: effects of SPR gene transfection on GTPCH1 gene expression. B: effects of SPR gene transfection on GTPCH1 activity. C: effects of SPR gene transfection on DHFR protein expression. Top: representative immunoblot of DHFR expression from Ctrl vs. SPR gene transfected endothelial cells. Bottom: summary of desitometric analysis of immunoblots. D: effects of SPR gene transfection on DHFR activity, represented as tetrahydrofolate conversion rate. E: GTPCH1 mRNA expression after SPR RNAi. F: effect of SPR RNAi on GTPCH1 activity. G, top: representative immunoblot of DHFR expression from Ctrl vs. SPR RNAi-treated cells. Bottom: summary of desitometric analysis of immunoblots. H: effect of SPR RNAi on DHFR activity. I: effects of SPR inhibitor NAS (N-acetyl-serotonin, 100 μmol/l) on DHFR activity. Results are means ± SE from 3–6 individual experiments. *P < 0.05 compared with Ctrl.
DISCUSSION
In this study, we identified the SPR sequence in the endothelial cells to examine a potential role of SPR on H4B synthesis and NO• production. To our knowledge, this is the first time that SPR is identified and fully characterized in endothelial cells; with specific focus on the regulation of H4B level and NO• production. We established that SPR is important in the regulation of H4B and NO• bioavailability in vascular endothelial cells and mouse aortas, as convincingly demonstrated by both SPR gene overexpression and silencing data.
SPR was first discovered in chicken and rat liver by Matsubara et al. (22, 23). Purification and characterization of SPR from rat erythrocytes further demonstrated it to be an acidic protein composed of two 28-kDa subunits (37). SPR gene was then isolated and cloned from rat liver by Citron et al. (5). The complete amino acids sequence of SPR in the rat were further confirmed by Oyama and colleagues (31). Later on, the cDNA sequences of SPR from different species, human (11), drosphila (36), and mouse (20), were reported. However, endothelial SPR has never been reported or studied. In this study, full-length SPR containing the 5′ untranslated region was cloned from endothelial cells, which would be a good tool for future studies of SPR gene regulation in pathophysiological conditions.
It has been well accepted now that restoration of H4B is associated with restored eNOS function in vascular diseases (1, 3, 6, 10, 12). Many measures have been proposed to increase H4B and NO• bioavailability via eNOS recoupling (34). Examples include supplementation of H4B and enhancement of its de novo biosynthesis. However, the outcomes are not exactly satisfactory, which are likely consequent to the fact that 1) H4B decays very quickly (9); and 2) overexpression of GTPCH1 has been found to only partially restore endothelial function in vivo (7). While sepiapterin also recouples eNOS effectively (15, 43) in some studies, it failed to recouple eNOS in others (26, 38, 40), likely attributed to inhibited activity of salvage enzymes in vascular diseases. Although the rate-limiting salvage enzyme DHFR has been found to have a critical role in modulating eNOS coupling status and thus a therapeutic target (4), SPR, as an enzyme with dual actions in both H4B de novo synthesis and salvation, may also serve as a therapeutic approach, particularly in combination with sepiapterin supplementation. Interestingly, our study showed that overexpression of SPR gene in the endothelial cells increased H4B levels corresponding to an increase in NO• production, via improvement of eNOS function, both in vitro and in vivo. The importance of SPR in regulating H4B and NO availability was further confirmed with SPR RNAi experiments. Our results showed that, after inhibition of SPR gene expression with RNAi, the H4B level in the endothelial cells was significantly decreased, which was accompanied with a marked reduction in NO• generation. A similar effect of SPR RNAi was also observed in sepiapetrin-supplemented endothelial cells.
Sepiapterin-induced increase in H4B content is indirect, where SPR and DHFR play sequential roles. Acute sepiapterin administration over SPR gene overexpression exerted an additive effect on intracellular H4B level, but not NO• production. In BAECs, basal NO• production was corresponding to intracellular H4B content at 4.4 ± 0.5 pmol/mg protein (Fig. 5C). As shown in Fig. 5, A and B, no further increase of NO• production was observed when intracellular H4B production was higher than 50 pmol/mg, implicating a potential saturation of H4B in its ability to competitively bind eNOS. Although SPR is involved in both the final reduction of the de novo synthetic and the first step of salvage pathways of H4B production from sepiapterin, alterations of SPR alone does not necessarily affect H4B level, because interplay with other key enzymes, such as GTPCH1 and DHFR, are also involved in the modulation of H4B levels (4, 44). GTPCH1 is considered as the rate-limiting enzyme on H4B de novo synthesis. From the literature, it has been shown that overexpression of GTPCH1 gene augments H4B biosynthesis, which restores endothelium eNOS activity/NO• production from hyperglycaemic toxicity (3), and, in contrast, inhibition of GTPCH1 by 2,4-diamino-6-hydroxypyrimidine (GTPCH1 inhibitor) significantly decreased H4B synthesis in both aorta and porcine endothelial cells (14, 27).
Our results showed that both GTPCH1 expression and activity were not altered by either inhibition or overexpression of SPR, indicating that SPR regulates H4B level independently, and the upstream de novo synthetic pathway is not altered to compensate for changes in H4B levels. On the other hand, while SPR RNAi had no effects on DHFR expression, the activity of the enzyme was significantly reduced consequent to SPR inhibition, due to SPR RNAi transfection, or in the presence of SPR inhibitor N-acetyl-serotonin. Presumably, DHFR activity is dependent on its protein level in the total lysate during HPLC measurement. However, it cannot be excluded that, as an enzyme, DHFR activity could also be regulated by protein-protein interactions or posttranslational modifications, such as phosphorylation or oxidation. Moreover, whether or not this is consequent to a decreased level of oxidized H4B (the substrate for DHFR) when H4B level is also low remains to be fully investigated. Nonetheless, our study indicates that DHFR expression or activity is not compensatorily increased when SPR is down.
In summary, in the present study we fully characterized the functional significance of an endothelium-specific bovine SPR gene both in vitro and in vivo, with particular relevance to endothelial H4B and NO• bioavailability. We found that overexpression of SPR or RNAi silencing of SPR was highly effective in improving or reducing NO• and H4B levels, both in vitro and in vivo. Sepiapterin supplementation caused an augmented production of NO• that was associated with higher H4B content, and these responses were attenuated in SPR RNAi pretransfected cells. These data clearly demonstrate an endogenous role of SPR that is critical for endothelial H4B and NO• bioavailability. It also seems to suggest that regimes targeted at enhancing SPR function may prove to be novel therapeutic tools for vascular diseases where eNOS uncoupling occurs.
GRANTS
The authors' work has been supported by National Heart, Lung, and Blood Institute Grants HL077440 (H. Cai), HL081571 (H. Cai), and HL057422 (P. L. Li and H. Cai); American Heart Association Grant 0435189N (H. Cai); American Diabetes Association Award 7–04-RA-16 (H. Cai); a Laubisch Award; and a Start-up Fund from University of California Los Angeles (H. Cai).
REFERENCES
- 1.Alp NJ, McAteer MA, Khoo J, Choudhury RP, Channon KM. Increased endothelial tetrahydrobiopterin synthesis by targeted transgenic GTP-cyclohydrolase I overexpression reduces endothelial dysfunction and atherosclerosis in ApoE-knockout mice. Arterioscler Thromb Vasc Biol 24: 445–450, 2004. [DOI] [PubMed] [Google Scholar]
- 2.Blau N, Bonafe L, Thony B. Tetrahydrobiopterin deficiencies without hyperphenylalaninemia: diagnosis and genetics of dopa-responsive dystonia and sepiapterin reductase deficiency. Mol Genet Metab 74: 172–185, 2001. [DOI] [PubMed] [Google Scholar]
- 3.Cai S, Khoo J, Channon KM. Augmented BH4 by gene transfer restores nitric oxide synthase function in hyperglycemic human endothelial cells. Cardiovasc Res 65: 823–831, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Chalupsky K, Cai H. Endothelial dihydrofolate reductase: critical for nitric oxide bioavailability and role in angiotensin II uncoupling of endothelial nitric oxide synthase. Proc Natl Acad Sci USA 102: 9056–9061, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citron BA, Milstien S, Gutierrez JC, Levine RA, Yanak BL, Kaufman S. Isolation and expression of rat liver sepiapterin reductase cDNA. Proc Natl Acad Sci USA 87: 6436–6440, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crabtree MJ, Tatham AL, Al-Wakeel Y, Warrick N, Hale AB, Cai S, Channon KM, Alp NJ. Quantitative regulation of intracellular endothelial nitric-oxide synthase (eNOS) coupling by both tetrahydrobiopterin-eNOS stoichiometry and biopterin redox status: insights from cells with tet-regulated GTP cyclohydrolase I expression. J Biol Chem 284: 1136–1144, 2009. [DOI] [PubMed] [Google Scholar]
- 7.Du YH, Guan YY, Alp NJ, Channon KM, Chen AF. Endothelium-specific GTP cyclohydrolase I overexpression attenuates blood pressure progression in salt-sensitive low-renin hypertension. Circulation 117: 1045–1054, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Ferre J, Naylor EW. Sepiapterin reductase in human amniotic and skin fibroblasts, chorionic villi, and various blood fractions. Clin Chim Acta 174: 271–282, 1988. [DOI] [PubMed] [Google Scholar]
- 9.Fiege B, Ballhausen D, Kierat L, Leimbacher W, Goriounov D, Schircks B, Thony B, Blau N. Plasma tetrahydrobiopterin and its pharmacokinetic following oral administration. Mol Genet Metab 81: 45–51, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Heitzer T, Brockhoff C, Mayer B, Warnholtz A, Mollnau H, Henne S, Meinertz T, Munzel T. Tetrahydrobiopterin improves endothelium-dependent vasodilation in chronic smokers: evidence for a dysfunctional nitric oxide synthase. Circ Res 86: E36–E41, 2000. [DOI] [PubMed] [Google Scholar]
- 11.Ichinose H, Katoh S, Sueoka T, Titani K, Fujita K, Nagatsu T. Cloning and sequencing of cDNA encoding human sepiapterin reductase–an enzyme involved in tetrahydrobiopterin biosynthesis. Biochem Biophys Res Commun 179: 183–189, 1991. [DOI] [PubMed] [Google Scholar]
- 12.Ishii M, Shimizu S, Momose K, Yamamoto T. SIN-1-induced cytotoxicity in cultured endothelial cells involves reactive oxygen species and nitric oxide: protective effect of sepiapterin. J Cardiovasc Pharmacol 33: 295–300, 1999. [DOI] [PubMed] [Google Scholar]
- 13.Kerler F, Hultner L, Ziegler I, Katzenmaier G, Bacher A. Analysis of the tetrahydrobiopterin synthesizing system during maturation of murine reticulocytes. J Cell Physiol 142: 268–271, 1990. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita H, Milstien S, Wambi C, Katusic ZS. Inhibition of tetrahydrobiopterin biosynthesis impairs endothelium-dependent relaxations in canine basilar artery. Am J Physiol Heart Circ Physiol 273: H718–H724, 1997. [DOI] [PubMed] [Google Scholar]
- 15.Korda M, Kubant R, Patton S, Malinski T. Leptin-induced endothelial dysfunction in obesity. Am J Physiol Heart Circ Physiol 295: H1514–H1521, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landmesser U, Dikalov S, Price SR, McCann L, Fukai T, Holland SM, Mitch WE, Harrison DG. Oxidation of tetrahydrobiopterin leads to uncoupling of endothelial cell nitric oxide synthase in hypertension. J Clin Invest 111: 1201–1209, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Laursen JB, Somers M, Kurz S, McCann L, Warnholtz A, Freeman BA, Tarpey M, Fukai T, Harrison DG. Endothelial regulation of vasomotion in apoE-deficient mice: implications for interactions between peroxynitrite and tetrahydrobiopterin. Circulation 103: 1282–1288, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Hargest R, Wasan H, Phillips RK. Liposome-mediated adenomatous polyposis coli gene therapy: a novel anti-adenoma strategy in multiple intestinal neoplasia mouse model. Dis Colon Rectum 47: 2105–2113, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Lee SW, Park IY, Hahn Y, Lee JE, Seong CS, Chung JH, Park YS. Cloning of mouse sepiapterin reductase gene and characterization of its promoter region. Biochim Biophys Acta 1445: 165–171, 1999. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 22.Matsubara M, Akino M. On the presence of sepiapterin reductase different from folate and dihydrofolate reductase in chicken liver. Experientia 20: 574–575, 1964. [DOI] [PubMed] [Google Scholar]
- 23.Matsubara M, Katoh S, Akino M, Kaufman S. Sepiapterin reductase. Biochim Biophys Acta 122: 202–212, 1966. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda N, Takano Y, Kageyama S, Hatakeyama N, Shakunaga K, Kitajima I, Yamazaki M, Hattori Y. Silencing of caspase-8 and caspase-3 by RNA interference prevents vascular endothelial cell injury in mice with endotoxic shock. Cardiovasc Res 76: 132–140, 2007. [DOI] [PubMed] [Google Scholar]
- 25.Milstien S, Katusic Z. Oxidation of tetrahydrobiopterin by peroxynitrite: implications for vascular endothelial function. Biochem Biophys Res Commun 263: 681–684, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell BM, Dorrance AM, Ergul A, Webb RC. Sepiapterin decreases vasorelaxation in nitric oxide synthase inhibition-induced hypertension. J Cardiovasc Pharmacol 43: 93–98, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Moat SJ, Ashfield-Watt PA, Powers HJ, Newcombe RG, McDowell IF. Effect of riboflavin status on the homocysteine-lowering effect of folate in relation to the MTHFR (C677T) genotype. Clin Chem 49: 295–302, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Moens AL, Kass DA. Therapeutic potential of tetrahydrobiopterin for treating vascular and cardiac disease. J Cardiovasc Pharmacol 50: 238–246, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Munzel T, Sinning C, Post F, Warnholtz A, Schulz E. Pathophysiology, diagnosis and prognostic implications of endothelial dysfunction. Ann Med 40: 180–196, 2008. [DOI] [PubMed] [Google Scholar]
- 30.Oak JH, Cai H. Attenuation of angiotensin II signaling recouples eNOS and inhibits nonendothelial NOX activity in diabetic mice. Diabetes 56: 118–126, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Oyama R, Katoh S, Sueoka T, Suzuki M, Ichinose H, Nagatsu T, Titani K. The complete amino acid sequence of the mature form of rat sepiapterin reductase. Biochem Biophys Res Commun 173: 627–631, 1990. [DOI] [PubMed] [Google Scholar]
- 32.Ozkor MA, Quyyumi AA. Tetrahydrobiopterin. Curr Hypertens Rep 10: 58–64, 2008. [DOI] [PubMed] [Google Scholar]
- 33.Pou S, Pou WS, Bredt DS, Snyder SH, Rosen GM. Generation of superoxide by purified brain nitric oxide synthase. J Biol Chem 267: 24173–24176, 1992. [PubMed] [Google Scholar]
- 34.Schmidt TS, Alp NJ. Mechanisms for the role of tetrahydrobiopterin in endothelial function and vascular disease. Clin Sci (Lond) 113: 47–63, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Schulz E, Jansen T, Wenzel P, Daiber A, Munzel T. Nitric oxide, tetrahydrobiopterin, oxidative stress, and endothelial dysfunction in hypertension. Antioxid Redox Signal 10: 1115–1126, 2008. [DOI] [PubMed] [Google Scholar]
- 36.Seong C, Kim YA, Chung HJ, Park D, Yim J, Baek K, Park YS, Han K, Yoon J. Isolation and characterization of the Drosophila melanogaster cDNA encoding the sepiapterin reductase. Biochim Biophys Acta 1443: 239–244, 1998. [DOI] [PubMed] [Google Scholar]
- 37.Sueoka T, Katoh S. Purification and characterization of sepiapterin reductase from rat erythrocytes. Biochim Biophys Acta 717: 265–271, 1982. [DOI] [PubMed] [Google Scholar]
- 38.Tarpey MM Sepiapterin treatment in atherosclerosis. Arterioscler Thromb Vasc Biol 22: 1519–1521, 2002. [DOI] [PubMed] [Google Scholar]
- 39.Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J 347: 1–16, 2000. [PMC free article] [PubMed] [Google Scholar]
- 40.Vasquez-Vivar J, Hogg N, Martasek P, Karoui H, Pritchard KA Jr, Kalyanaraman B. Tetrahydrobiopterin-dependent inhibition of superoxide generation from neuronal nitric oxide synthase. J Biol Chem 274: 26736–26742, 1999. [DOI] [PubMed] [Google Scholar]
- 41.Werner-Felmayer G, Golderer G, Werner ER. Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr Drug Metab 3: 159–173, 2002. [DOI] [PubMed] [Google Scholar]
- 42.Wever RM, van Dam T, van Rijn HJ, de Groot F, Rabelink TJ. Tetrahydrobiopterin regulates superoxide and nitric oxide generation by recombinant endothelial nitric oxide synthase. Biochem Biophys Res Commun 237: 340–344, 1997. [DOI] [PubMed] [Google Scholar]
- 43.Zerr-Fouineau M, Jourdain M, Boesch C, Hecker M, Bronner C, Schini-Kerth VB. Certain progestins prevent the enhancing effect of 17beta-estradiol on NO-mediated inhibition of platelet aggregation by endothelial cells. Arterioscler Thromb Vasc Biol 29: 586–593, 2009. [DOI] [PubMed] [Google Scholar]
- 44.Zheng JS, Yang XQ, Lookingland KJ, Fink GD, Hesslinger C, Kapatos G, Kovesdi I, Chen AF. Gene transfer of human guanosine 5′-triphosphate cyclohydrolase I restores vascular tetrahydrobiopterin level and endothelial function in low renin hypertension. Circulation 108: 1238–1245, 2003. [DOI] [PubMed] [Google Scholar]