Abstract
Endurance exercise is known to provide cardioprotection against ischemia-reperfusion-induced myocardial injury, and mitochondrial adaptations may play a critical role in this protection. To investigate exercise-induced changes in mitochondrial proteins, we compared the proteome of subsarcolemmal and intermyofibrillar mitochondria isolated from the myocardium of sedentary (control) and exercise-trained Sprague-Dawley rats. To achieve this goal, we utilized isobaric tags for relative and absolute quantitation, which allows simultaneous identification and quantification of proteins between multiple samples. This approach identified a total of 222 cardiac mitochondrial proteins. Importantly, repeated bouts of endurance exercise resulted in significant alterations in 11 proteins within intermyofibrillar mitochondria (seven increased; four decreased) compared with sedentary control animals. Furthermore, exercise training resulted in significant changes in two proteins within subsarcolemmal mitochondria (one increased; one decreased) compared with sedentary control animals. Differentially expressed proteins could be classified into seven functional groups, and several novel and potentially important cardioprotective mediators were identified. We conclude that endurance exercise induces alterations in mitochondrial proteome that may contribute to cardioprotective phenotype. Importantly, based on our findings, pharmacological or other interventions could be used to develop a strategy of protecting the myocardium during an ischemic attack.
Keywords: cardioprotection, mitochondrial proteome
one of the major causes of morbidity and mortality around the world is coronary artery disease, which can lead to angina and/or an ischemia-reperfusion (IR) injury (43). Unfortunately, the incidence of IR injury in the heart is very high, and, each year, thousands of people die from this form of myocardial injury. Therefore, developing a strategy that results in a cardioprotective phenotype is important. In this regard, several approaches to achieve cardioprotection have been investigated. Importantly, regular bouts of endurance exercise have been shown to provide cardioprotection (2, 19, 39). Although it is clear that exercise promotes a cardioprotective phenotype, a detailed understanding of the mechanisms responsible for cardioprotection remains incomplete. Therefore, we performed this investigation to identify exercise-induced changes in potentially cardioprotective mitochondrial proteins, as recognition of putative cardioprotective mediators is the first step in unveiling the mechanism(s) responsible for exercise-induced cardioprotection.
Over the years, investigators have proposed several proteins and organelles as mechanistic candidates to explain exercise-induced cardioprotection. However, the specific proteins that are responsible for exercise-induced cardioprotection remain unclear. Nonetheless, several researchers agree that mitochondrial adaptations are likely to be important in exercise-induced cardioprotection (4, 50). Indeed, mitochondria are vital organelles that can serve as the final arbitrators of life or death during an IR insult, as they not only are required to produce ATP, but can also trigger both necrosis and apoptosis (16, 17). Moreover, studies show that endurance exercise results in a reduction in mitochondrial oxidant production (21, 45) and enhanced mitochondrial antioxidant enzyme activity (20, 21, 45). Furthermore, our laboratory has recently reported that exercise training promotes important adaptations to cardiac mitochondria, conferring an apoptotic resistant phenotype (23). Nonetheless, to date, no published reports have identified cardiac mitochondrial proteome changes following exercise training.
Modern advances in the area of proteomics [e.g., automated two-dimensional gel analysis, tandem mass spectrometry (MS/MS), etc.], have enhanced the importance of proteomics as a nonbiased tool for protein discovery (32, 38, 48). In this regard, one of the most powerful and innovative proteomic techniques is the newly developed isobaric tags for relative and absolute quantitation (iTRAQ) of proteins. Because iTRAQ technique allows free amines to be labeled on peptides with a stable isotope containing an isobaric tag, all of the proteins can be labeled without any specificity, and quantitative data will be representative of several peptides for each protein. Furthermore, the iTRAQ reagent strategy enables multiplexed quantitative analysis of separate protein mixtures in one MS/MS experiment. As a result, the iTRAQ methodology provides increased flexibility in the design of experiments. Additionally, the utilization of two-dimensional liquid chromatography of labeled peptides before MS/MS analysis allows for extensive separation of the peptide fragments (35). Thus we employed this innovative technique (iTRAQ) to investigate changes within the cardiac mitochondrial proteome that occur in response to exercise training.
Specifically, we used a robust experimental design, utilized state-of-the-art iTRAQ techniques, and analyzed protein changes in each of the two mitochondrial subfractions [i.e., subsarcolemmal (SS) and intermyofibrillar (IMF) mitochondria] from hearts of both sedentary control and exercised-trained rats. We hypothesized that exercise training would promote significant changes in protein abundance in cardiac mitochondrial proteins involved in energy metabolism, redox regulation, and apoptosis. Our results support this hypothesis and reveal changes in several exercise-induced alterations in mitochondrial proteins, many of which have the potential to be cardioprotective.
MATERIALS AND METHODS
Experimental design.
These experiments were approved by the Institutional Animal Care and Use Committee and followed guidelines established by the American Physiological Society for the use of animals in research. Adult Sprague-Dawley (male) rats (4–6 mo old) were randomly assigned to one of two experimental groups: sedentary controls (n = 11) or endurance exercise training (n = 8). Animals were further distributed for cardiac functional analysis (sedentary controls: n = 7, and endurance exercise trained: n = 4) and proteomic analysis (sedentary controls: n = 4, and endurance exercise trained: n = 4). Throughout the experimental period, all animals were housed on a 12:12-h light-dark cycle and provided rat chow and water ad libitum.
Exercise training protocol.
Animals assigned to the endurance exercise training group were habituated to treadmill exercise for 5 consecutive days. This habituation period involved a gradual increase in running time, beginning with 10 min/day and ending with 50 min/day. After 2 days of rest, the animals then performed 5 consecutive days of treadmill exercise for 60 min/day at 30 m/min, 0% grade (estimated work rate of 70% maximum O2 consumption) (12). All hearts were excised and used 24 h after the final exercise bout.
In vitro working heart protocol, cardiac function measurements, and mitochondrial isolation.
To show that our exercise training protocol used in this study provided cardioprotection, sedentary (n = 7) and endurance exercise-trained (n = 4) animals were subjected to an IR insult using an isolated working heart preparation, as previously described by our group (26, 27, 40), with an ischemia duration of 25 min and reperfusion duration of 35 min. Measurements collected included aortic flow, coronary flow, peak systolic pressure, diastolic pressure, and heart rate.
Mitochondrial isolation.
Cardiac muscle contains two morphologically and biochemically distinct subfractions (SS and IMF) of mitochondria located in different regions of the cardiomyocyte (36, 42). Differential centrifugation was used to fractionate SS and IMF mitochondria, as described previously (sedentary controls: n = 4, and endurance exercise trained: n = 4) (23).
Trypsin digestion and iTRAQ labeling.
The protocol followed for trypsin digestion and iTRAQ labeling of peptides is fully described in Ref. 29. Briefly 60 μg of protein from each sample were aliquoted into individual tubes. Proteins were reduced with 50 mM tris-(2-carboxyethyl)phosphine, and thiol groups of cysteine were blocked with 200 mM methyl methanethiosulfonate in isopropanol before adding 10 μl of a 0.8 μg/μl trypsin solution (Promega, Madison, WI) to each tube. After digestion, each of the four labels (114, 115, 116, and 117) were added to each sample (114 for IMF mitochondria samples isolated from sedentary animals; 115 for IMF mitochondria samples isolated from endurance exercised animals; 116 for SS mitochondria samples isolated from sedentary animals; 117 for SS mitochondria samples isolated from endurance exercised animals) and incubated at 22°C for 2 h. The labeling procedure was independently performed for each mitochondrial sample isolated from each animal (four samples were labeled with 114, four samples were labeled with 115, four samples were labeled with 116, and four samples were labeled with 117). After labeling, each set of samples (i.e., 114, 115, 116, 117) was pooled in a new tube and dried, thus resulting in four pooled samples.
Two-dimensional liquid chromatography MS/MS.
The four pooled samples were desalted using a macrospin Vyadac Silica C18 column (The Nest Group, Southboro, MA). The eluted, labeled peptides were dried and resuspended in 95 μl buffer A [75% 0.01M ammonium formate, 25% acetonitrile (ACN)] for use in the offline strong cation exchange fractionation. Ninety microliters were injected onto a polysulfoethyl A column with a 5-μm particle size and a column dimension of 100 × 2.1 mm inner diameter, 200 Å pore size (PolyLC, Columbia, MD). Peptides were eluted during a linear ramp of 0–20% buffer B (75% 0.5M ammonium formate, 25% ACN) over 40 min and 20–100% buffer for 5 min. The peptide absorbance at 280 nm was used to indicate in which fractions the peptides eluted. Eight fractions for each replicate were collected, dried, and resuspended in 3% ACN, 0.1% acetic acid, and 0.01% trifluoroacetate. NanoLC-MS/MS analysis was carried out the same way as described by Martyniuk et al. (29) using LC Packing C18 Pep Map HPLC column (DIONEX, Sunnyvale, CA) for the peptide separation on line with a hybrid quadrupole-TOF QSTAR mass spectrometer (Applied Biosystems) for peptide detection.
Protein search database and statistical analysis.
Tandem mass spectra were extracted by Analyst version 1.1 (Applied Biosystems). Searches were performed using MS/MS data interpretation algorithms within ProteinPilot (Paragon algorithm, version 2.0, Applied Biosystems). The database search engine was set up to do a “thorough” search against a concatenated database of the forward and randomized sequences of the IPI_rat (version 3.32) database (41,240 sequences). The database allowed for iTRAQ reagent labels at NH2-terminal residues, internal K and Y residues, and methyl methanethiosulfonate-labeled cysteine as fixed modifications, deamidation, O-phosphorylation (STY), and oxidation (M) as variable modifications and one missed cleavage. The confidence level for protein identification was set to 1.3 (95%). However, proteins with less than two peptides above 80% confidence were manually filtered out. A measure for false-positive discovery rate was also calculated using twice the total identification spectra above 80% confidence from the decoy database, as outlined by Elias and Gygi (14). Relative quantification of proteins using ProteinPilot is performed on the MS/MS scans and is calculated based on the ratio of the areas of the 114-, 115-, 116-, and 117-Da iTRAQ reporter ions. ProteinPilot calculates average protein ratios using only ratios from the spectra that are distinct to each protein, excluding the shared peptides of isoforms. The peptides without iTRAQ modification of free amine in the lysine were excluded from calculation of the protein ratios. Furthermore, to exclude low spectral counts present in a subset of spectra from the calculation of averages, the intensity threshold for the sum of the signal-to-noise ratio for all the peak pairs was >9. All of the quantitative ratios were then corrected for bias to create the ProGroup algorithm results. The median average protein ratio was calculated and applied to the quantitation results, and statistically significant changes (P < 0.05) were weighted by the error factor (EF < 2). For a protein to be identified as significantly different from one sample to another, the protein had to be quantified with a P < 0.05, EF ≤ 2, in each biological replicate where the protein was detected.
Western blotting.
Isolated IMF and SS mitochondrial protein extracts were separated by performing sodium dodecyl sulfate polyacrylamide gel electrophoresis and subsequently electroblotted onto nitrocellulose membranes. The resulting membranes were then stained with Ponceau S and analyzed to verify equal loading and transfer. Membranes were blocked (2 h) with 5% skim milk in phosphate-buffered saline solution containing 0.05% Tween 20 (PBST). Blots were then incubated in blocking buffer with antibody directed against monoamine oxidase A (MAO-A; 1:1,000 dilution; ab40835; Abcam, Cambridge, MA) and peroxiredoxin III (PRDXIII; 1:1,000 dilution; sc-23973; Santa Cruz Biotechnology, Santa Cruz, CA). After being washed with PBST, blots were incubated at room temperature for 1 h with the appropriate secondary antibody coupled to horseradish peroxidase and washed again with PBST. The membranes were then treated with chemiluminescent reagents (luminol and enhancer; Amersham Biosciences, Pittsburgh, PA) and exposed to light-sensitive film. Images of these films were captured and analyzed using the 440CF Kodak Imaging System (Kodak, New Haven, CT).
Statistical analysis for cardiac function measurements and Western blotting.
Statistical significance between groups for dependent variables for cardiac function and Western blotting measurements was determined by a one-way analysis of variance followed with Tukey's multiple-comparison test, where appropriate. Significance was established at P < 0.05. Results are presented as means ± SD.
RESULTS
Myocardial performance during IR.
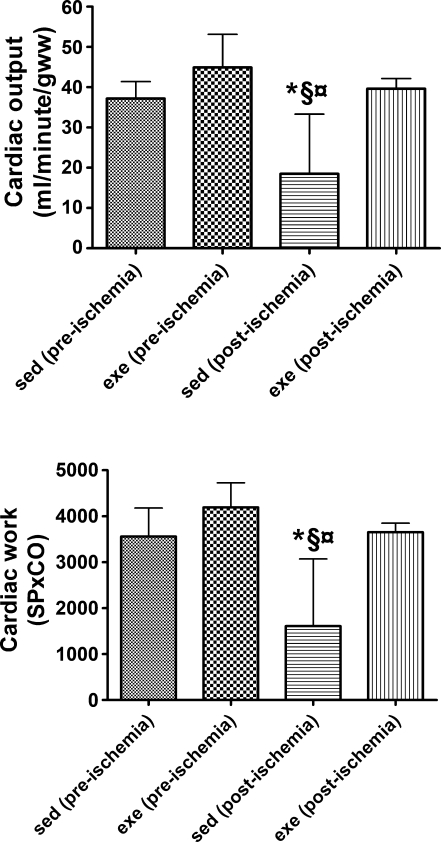
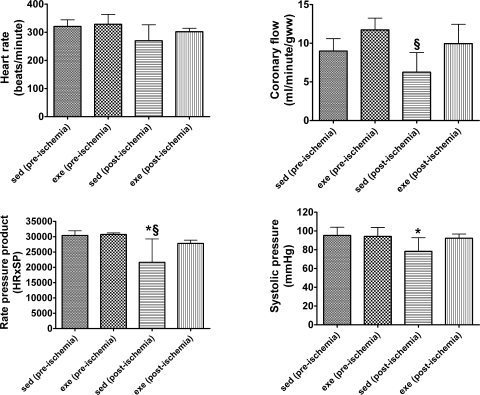
The effects of endurance exercise training on pre- and postischemic cardiac function are shown in Figs. 1 and 2. Our results show that the exercise training program used in this study was sufficient to provide cardioprotection against an IR insult. Before ischemia, no differences existed between experimental groups in cardiac output or cardiac work (Fig. 1). Furthermore, before ischemia, heart rate, coronary flow, systolic pressure, and rate pressure product were not different between sedentary and exercise-trained animals (Fig. 2). However, 25 min of global ischemia resulted in a significant ventricular dysfunction in sedentary control animals. For example, compared with preischemic values, percent recovery of cardiac work was depressed by 57% in sedentary control animals at 35 min of reperfusion. In contrast to sedentary control animals, exercise-trained animals maintained significantly higher cardiac outputs and cardiac work at 35 min of reperfusion.
Fig. 1.
Functional characteristics [cardiac output (CO; top) and cardiac work (bottom)] of hearts isolated from sedentary (sed) and exercise-trained (exe) animals before an ischemic insult (preischemia) and following 25 min of ischemia (postischemia). Values are means ± SD. *P < 0.05 between sed (preischemia) and sed (postischemia); §P < 0.05 between exe (preischemia) and sed (postischemia); ¤P < 0.05 between sed (postischemia) and exe (postischemia). SP, systolic pressure.
Fig. 2.
Functional characteristics [heart rate (HR), coronary flow, rate pressure product, and SP] of hearts isolated from sed and exe animals preischemia and postischemia. Values are means ± SD. *P < 0.05 between sed (preischemia) and sed (postischemia); §P < 0.05 between exe (preischemia) and sed (postischemia).
Identification and quantification of protein expression in cardiac SS and IMF mitochondria.
Four biological replicates of iTRAQ labeling were performed to specifically identify the differential protein expression in IMF and SS mitochondria isolated from the myocardium of both sedentary and exercise-trained rats. Between 158 and 185 proteins were identified in each replicate, with a 95% confidence level and at least two unique peptides (Table 1). The false positive discovery rate calculated from the number of spectra matching positively randomized sequences vs. the total number of spectra matched was extremely low and comprised between 0 and 0.16% (Table 1). Ninety-six percent of the proteins identified from the cardiac SS and IMF mitochondrial subfractions are reported in previous studies on human and murine heart mitochondrial proteomes (6, 15, 46). However, seven new proteins, including membrane proteins, NADH dehydrogenase subunits 3 and 12, cytochrome-c oxidase subunit 1, transmembrane protein 126A, and apolipoprotein-O were identified in our study.
Table 1.
Total number of proteins identified and quantified using Paragon and Pro Group Algorithms from ProteinPilot for the 4 replicates of iTRAQ experiments
| Replicate No. | Proteins Identified, no. | False Positive, % | Proteins Quantified, no. |
|---|---|---|---|
| 1 | 185 | 0.05 | 154 |
| 2 | 185 | 0.11 | 150 |
| 3 | 158 | 0.00 | 126 |
| 4 | 164 | 0.16 | 143 |
The false positive discovery rate of protein identification for each replicate is also indicated. iTRAQ, isobaric tags for relative and absolute quantitation.
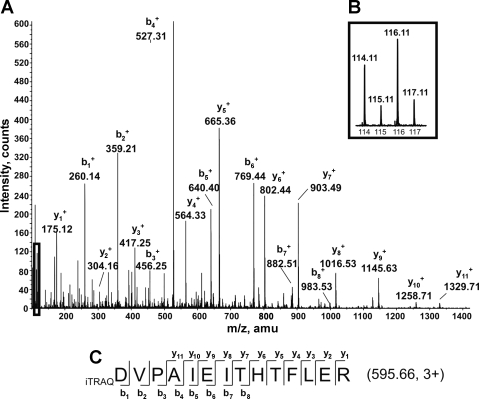
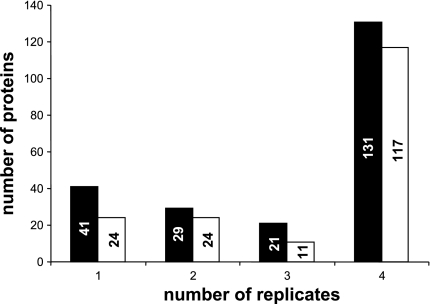
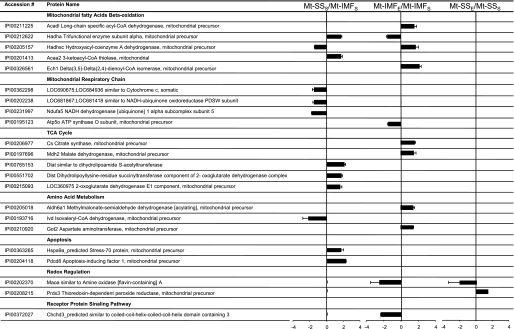
An average of 82% of the proteins identified was quantifiable, which means that at least three MS/MS spectra with iTRAQ reporter peaks (from the same peptide or different peptide) were monitored. Figure 3 illustrates an example of MS/MS spectrum obtained, confirming the identification of one peptide (i.e., MAO-A) and the intensity changes from the iTRAQ labels between samples. The reproducibility in protein identification and quantification through the four replicates was manually inspected and validated. Figure 4 illustrates the number of proteins identified in one, two, three, or four replicates. One hundred and thirty-one proteins were confidently and reproducibly identified in all of the replicate runs (4), with 117 proteins with quantification data. The difference between the number of identified proteins and quantified proteins is explained by unlabeled spectra, spectra quality not meeting standards for quantitation, or peptides shared by different proteins. This represent 60% of the total protein identified (222 proteins) from the four replicates (Supplemental Table 1). (The online version of this article contains supplemental data.) Data analysis of differential protein expression was performed using, as first threshold, the following criteria: EF ≤ 2, P < 0.05 (Supplemental Table 2). For a quantitative difference between samples to be significant, it also had to be identified and quantified in at least two of the four replicates, with an average ratio greater than or equal to +1.4 or −1.4. Twenty-two proteins were significantly differentially expressed between the groups (Fig. 5). Following identification, we grouped these 22 differential expressed proteins into functional categories. Seven types of cellular proteins, including proteins involved in mitochondrial β-oxidation of fatty acids, mitochondrial respiratory chain, the TCA cycle, amino acid metabolism, apoptosis, redox regulation, and receptor protein signaling pathway displayed changes (Fig. 5).
Fig. 3.
Example of tandem mass spectrometry (MS/MS) spectrum of an isobaric tag for relative and absolute quantitation (iTRAQ)-labeled peptide matching to the monoamine oxidase A (MAO-A). A: MS/MS fragmentation spectrum from the tryptic peptide 595.6 mass-to-charge ratio (m/z) (3+) of MAO-A. The y and b series of ions detected by the mass spectrometer are indicated. B: the enlarge region corresponds to the lower m/z range of the spectrum showing the peaks intensity from the iTRAQ reagents 114 [intermyofibrillar (IMF) mitochondria isolated from sed animals], 115 (IMF mitochondria isolated from exe animals), 116 [subsarcolemmal (SS) mitochondria isolated from sed animals], and 117 (SS mitochondria isolated from exe animals). C: y and b ion coverage of the peptide sequence observed on the MS/MS spectrum.
Fig. 4.
Reproducibility of proteins identified (solid bars) and quantified (open bars) from the four biological replicates of mitochondrial proteins from the myocardium of sed and exe rats (see text for detailed explanation).
Fig. 5.
Cardiac mitochondrial (Mt) proteins identified showing a significant difference between groups: Mt-SS and Mt-IMF [isolated from sed (S) and exe (E) animals]. The proteins were classified into 7 categories according to their protein function. The histogram shows the average and the SD of iTRAQ ratio calculated using the replicates, showing only a ratio with P < 0.05.
Protein abundance is altered in cardiac IMF mitochondria following exercise training.
In IMF mitochondria, seven proteins were upregulated, and four proteins were downregulated after endurance exercise (Fig. 5). Importantly, IMF mitochondria isolated from hearts of exercised animals had lower (−2.52) levels of MAO-A protein compared with IMF mitochondria isolated from sedentary animals. This finding is of great interest, since this mitochondrial enzyme is an important source of reactive oxygen species in the heart. Importantly, oxidative stress induced by MAO-A leads to apoptosis during postischemic myocardial injury (5, 37). In addition to decreased MAO-A protein levels, repeated bouts of endurance exercise resulted in altered expression of proteins involved in fatty acid β-oxidation, the TCA cycle, and amino acid metabolism.
Protein abundance is altered in cardiac SS mitochondria following exercise training.
In SS mitochondria, one protein increased and one protein decreased in abundance following endurance exercise (Fig. 5). These two SS mitochondrial proteins were MAO-A and PRDXIII. Notably, following exercise training, MAO-A abundance was reduced by a factor of 2. Furthermore, the mitochondrial-specific H2O2-scavenging enzyme PRDXIII was upregulated (+1.40) in SS mitochondria after endurance exercise training. The upregulation of PRDXIII is an important finding, because published data suggest that depletion of PRDXIII results in increased intracellular levels of H2O2 and sensitizes cells to induction of apoptosis (10).
Differential protein expression in SS and IMF mitochondria isolated from hearts of sedentary animals.
In sedentary animals, 12 proteins were significantly different between IMF and SS mitochondria (Fig. 5). Specifically, the expression of seven proteins was higher in SS mitochondria compared with IMF mitochondria. Also, the expression of five proteins was lower in SS mitochondria compared with IMF mitochondria. Importantly, proteins present in lower abundance in SS mitochondria are involved in the mitochondrial respiratory chain. This agrees with previously published data indicating that SS mitochondria exhibit lower rates of oxidative phosphorylation compared with IMF mitochondria (36, 42).
Confirmation of iTRAQ by Western blotting study.
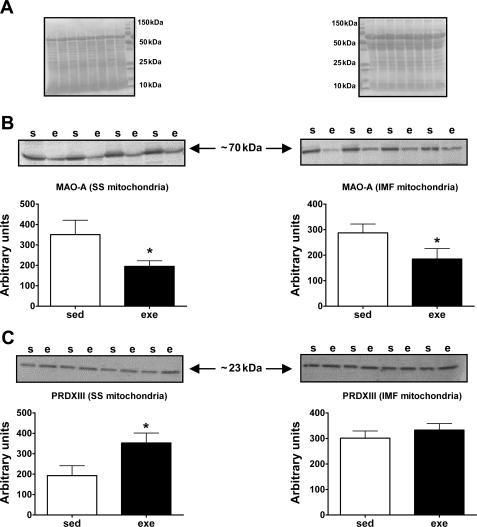
We performed Western blot analysis of selected proteins detected by the iTRAQ study to provide confirmation of differentially expressed proteins. Specifically, Western blot analysis was carried out for both MAO-A and PRDXIII. These proteins were chosen to represent different fold and directional changes (i.e., increased, decreased, and no change). Figure 6 shows the Western blot images and quantification for MAO-A (Fig. 6B) and PRDXIII (Fig. 6C). In agreement with iTRAQ data, the Western blot analysis revealed that, following exercise training, MAO-A protein expression was significantly decreased in both SS and IMF mitochondria. Additionally, PRDXIII protein expression was significantly increased in SS mitochondria, but it was not altered in IMF mitochondria following repeated bouts of endurance exercise. Therefore, the validation by Western blotting provides evidence that iTRAQ is a reliable method for large-scale protein quantification.
Fig. 6.
MAO-A and peroxiredoxin III (PRDXIII) protein levels detected by Western blotting. A: membranes used for MAO-A and PRDXIII protein analysis were stained with Ponceau S and visually inspected to ensure equal protein loading and transfer. B: Western blot images for MAO-A obtained from SS mitochondria (left) and IMF mitochondria (right). Arbitrary optical density values for MAO-A between sed and exe animals are shown below the Western blot images. C: Western blot images for PRDXIII obtained from SS mitochondria (left) and IMF mitochondria (right). Arbitrary optical density values for PRDXIII between sed and exe animals are shown below the Western blot images. s, sed animals; e, exe animals. Data are means ± SD. *P < 0.05.
DISCUSSION
Overview of principle findings.
This study reinforces the notion that endurance exercise training provides cardioprotection against IR-induced myocardial damage. Indeed, our present findings agree with previous work by our laboratory and others that indicate that short-term endurance exercise results in enhanced postischemic myocardial recovery. Although several investigators have shown that exercise promotes a cardioprotective phenotype, a detailed understanding of the mechanisms responsible for cardioprotection remains incomplete. Therefore, we tested the hypothesis that exercise training induces alterations in the abundance of several potentially cardioprotective proteins in cardiac SS and IMF mitochondria. Our results support this postulate. Importantly, we identified several novel mitochondrial proteins (e.g., MAO-A and PRDXIII) as potential candidates for exercise-induced cardioprotection. A brief discussion of these and related issues follows.
Changes in myocardial SS and IMF mitochondria protein abundance following exercise training.
IR-induced myocardial injury is manifested due to the complex interaction of numerous factors. However, increased production of reactive oxygen species, during both ischemia and reperfusion, appears to play a major role in this type of myocardial injury (39). In this regard, IR-induced oxidative stress appears to occur due to the activation of several oxidant production systems. One of these enzymatic systems is MAOs (25, 31). MAOs are mitochondrial enzymes involved in the oxidative deamination of biogenic amines. These oxidases have been subdivided into two major forms, A and B, based on genetic criteria, substrate specificity, and inhibition by synthetic compounds. However, in cardiac tissue, MAO-A is the principal enzyme involved in the deamination of endogenous or exogenous amines (37). Specifically, MAO-A catalyzes the oxidative deamination of several monoamines (i.e., serotonin, noradrenaline, dopamine), resulting in reactive oxygen species (e.g., H2O2) production (37).
Recent work indicates that oxidative stress induced by MAO-A is responsible for apoptosis during postischemic myocardial injury (5, 37). For example, an in vivo study using MAO-A knockout (KO) mice showed that the MAO-A KO animals were protected from IR-induced cardiac damage (37). Notably, the protection of MAO-A KO was related to significantly lower reactive oxygen species generation following IR injury (37). Furthermore, gene expression profiling by microarray revealed an upregulation of MAO-A gene in hypertrophy and cardiac failure (24). Also, several investigators have reported an increase in cardiac MAO-A activity with age (8, 33). Importantly, MAO-A protein levels and MAO-A-dependent H2O2 production strongly increase in the senescent heart, and the authors proposed that MAO-A was a key factor involved in cardiac oxidative stress during aging (31). In the present study, MAO-A protein levels were significantly reduced in both SS and IMF cardiac mitochondria following exercise training. In view of the results discussed above, it is possible that downregulation of MAO-A protein expression following endurance exercise is one of the mechanisms for exercise-induced cardioprotection. Therefore, it is conceivable that the downregulation of MAO by endurance exercise represents a physiological and practical approach to prevent cardiac oxidative stress and subsequent cell death and apoptosis, especially in situations in which the production of reactive oxygen species increases (i.e., IR).
The cardioprotective benefits of endurance exercise may also be due to increased myocardial antioxidant capacity. In this regard, most reactive oxygen species are generated as a result of the univalent reduction of molecular oxygen to superoxide anion by electrons that leak from the mitochondrial electron transport chain (7). Most of the superoxide anion is countered by manganese superoxide dismutase, an enzyme specifically localized in the mitochondrial matrix (9). However, this reaction only partially relieves oxidative stress in mitochondria, because the product from this reaction (i.e., H2O2) is also an oxidant. To counteract intracellular H2O2, animal cells have additional antioxidant enzymes that remove H2O2 [e.g., catalase, glutathione peroxidase, and the newly identified family of peroxidases (i.e., PRDX)] (10). Importantly, this family of peroxidases includes at least six isoforms in mammalian cells, and among them it is the mitochondria-specific isoform PRDXIII (41). It is postulated that the specific localization of PRDXIII within the mitochondria provides a primary line of defense against H2O2 produced by the mitochondrial respiratory chain (34, 41). Although several researchers have reported changes in antioxidant enzymes in cardiac tissue following endurance exercise, no report has specifically investigated the effects of endurance exercise on PRDXIII protein abundance. In the present experiments, we report that PRDXIII protein abundance was upregulated in SS mitochondria after endurance exercise training. This finding is important, because data indicate that intracellular accumulation of H2O2 caused by PRDXIII depletion or oxidation results in acceleration of apoptosis (10, 11). Furthermore, overexpression of PRDXIII inhibited left ventricular remodeling and heart failure following myocardial infarction. In addition, overexpression of PRDXIII reduced mitochondrial oxidative stress and mitochondrial dysfunction after myocardial infarction (30). Importantly, PRDXIII protein abundance was upregulated in the exercise preconditioned animals, albeit only in the SS mitochondrial subfraction. Therefore, it is possible that SS and IMF mitochondria adapt differently to exercise training, as the two mitochondrial subfractions possess different functional, compositional, and biochemical properties. Therefore, exercise-induced upregulation of myocardial PRDXIII may account, at least in part, for exercise-induced cardioprotection.
In the present study, the coiled-coil-helix coiled-coil-helix domain 3 (CHCHD3) protein was downregulated (−2.34) in IMF mitochondria after endurance exercise. The CHCHD3 protein has been identified before in mitochondria (44), but the functions of this protein are poorly understood (13). However, recently data suggest that CHCHD3 may play a role in metal binding similar to cytochrome-c oxidase 17 and 19 (18). In addition, new research suggests that CHCHD3 may be involved in protein import into the mitochondria (49). Also, recently, it has been proposed that CHCHD3 may be a candidate for immunogenic membrane antigens of pancreatic cancer. Despite the unknown function of this protein, it is important to further investigate its physiological and biochemical role in cardiac biology. Given the fact that endurance exercise induced a large downregulation of this protein in mitochondria, further research is warranted to determine its putative function in cardioprotection.
Furthermore, the quantity of other mitochondrial proteins was altered following repeated bouts of endurance exercise. Specifically, the abundance of several proteins involved in bioenergetics was changed following exercise in both SS and IMF mitochondria (Fig. 5). In this regard, mitochondria are highly dynamic organelles that continuously adjust ATP regeneration to match changing bioenergetic demands of cells. Therefore, regulation of proteins/enzymes involved in energy production may help to satisfy the increased energy demands during exercise and maintain and/or enhance cardiac function in the resting condition.
In all mammals under physiological conditions, the myocardium obtains much of its energy supply by the oxidation of fatty acids. However, during the development of heart disease, the myocardial energy source switches from fatty acid β-oxidation to glycolysis. Furthermore, genetic defects in mitochondrial fatty acid β-oxidation enzymes in humans suggest that reduced capacity for fatty acid utilization may lead to heart failure and cardiac rhythm disturbances. For example, acyl-CoA dehydrogenase is downregulated in a rodent model of heart failure, and samples obtained from cardiomyopathic hearts of humans have decreased levels of acyl-CoA dehydrogenase (3). In the present study, the protein levels of several proteins involved in β-oxidation of fatty acids [e.g., acyl-CoA dehydrogenase, hydroxyacyl-coenzyme A dehydrogenase, delta(3,5)-delta(2,4)-dienoyl-CoA isomerase] were increased following repeated bouts of endurance exercise. Moreover, the levels of two proteins (methylmalonate-semialdehyde dehydrogenase and aspartate aminotransferase) involved in amino acid metabolism were increased following endurance exercise. Specifically, aminotransferases can convert some amino acids into other amino acids, and amino acid transamination may be an important adaptive process in the immature heart, improving its resistance to ischemic damage (22).
Proteome differences between myocardial SS and IMF mitochondria.
Recent evidence indicates that SS and IMF mitochondria exhibit decreased susceptibility to apoptotic stimuli (1, 23). To investigate molecular differences between these two mitochondrial subpopulations, we compared the proteome of SS and IMF mitochondria isolated from hearts of sedentary animals. Our experiments reveal that, in sedentary animals, 12 proteins are differentially expressed between SS and IMF cardiac mitochondria. Importantly, three proteins involved in mitochondrial respiratory chain were found in lower abundance in SS mitochondria compared with IMF mitochondria, which is consistent with the observation that SS mitochondria exhibit lower rates of oxidative phosphorylation compared with IMF mitochondria (36, 42).
Furthermore, in sedentary animals, proteins categorized in other functional groups were also differentially expressed between the two mitochondrial subpopulations. In this regard, an important mitochondrial protein is apoptosis-inducing factor (AIF), because AIF is associated with the induction of apoptosis during an IR insult (47). Our data indicate that, in sedentary animals, SS mitochondria contain higher AIF protein levels compared with IMF mitochondria. This finding may explain, at least in part, the differential susceptibility of the two mitochondrial subpopulations to apoptotic stimuli (1, 23). Specifically, several investigators have reported that AIF contains a DNA binding site responsible for chromatin condensation and DNA fragmentation. However, research suggests that AIF may also contain a redox-active region that could be anti-apoptotic (28, 47). Therefore, additional research is required to fully elucidate the cellular role of AIF.
Conclusions and recommendations for future studies.
These experiments provide novel and significant information about the proteome of the two subpopulations (i.e., SS and IMF) of cardiac mitochondria. Importantly, our results reveal that both SS and IMF mitochondrial protein expression is altered following exercise training. Specifically, MAO-A protein levels were downregulated following endurance exercise. Furthermore, PRDXIII expression was upregulated in cardiac mitochondria after exercise training. Although the beneficial effects of endurance exercise to the myocardium are evident, further work is needed to delineate the physiological impact of these protein alterations following exercise training. Thus this is an interesting area for future work that awaits further investigation.
GRANTS
This study was supported by an award from the American Heart Association awarded to A. N. Kavazis and National Heart, Lung, and Blood Institute grant R01HL067855 awarded to S. K. Powers.
Supplementary Material
Acknowledgments
We thank Marjory Chow and Scott McClung from the Proteomics Facility at the Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, for expertise in protein sample preparation and mass spectrometry analysis.
Present address of S. Alvarez: D. Danforth Plant Science Center, Proteomics and Mass Spectrometry Facility, St. Louis, MO, 63132.
REFERENCES
- 1.Adhihetty PJ, Ljubicic V, Menzies KJ, Hood DA. Differential susceptibility of subsarcolemmal and intermyofibrillar mitochondria to apoptotic stimuli. Am J Physiol Cell Physiol 289: C994–C1001, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Balady GJ Survival of the fittest–more evidence. N Engl J Med 346: 852–854, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Barger PM, Kelly DP. Fatty acid utilization in the hypertrophied and failing heart: molecular regulatory mechanisms. Am J Med Sci 318: 36–42, 1999. [DOI] [PubMed] [Google Scholar]
- 4.Bejma J, Ramires P, Ji LL. Free radical generation and oxidative stress with ageing and exercise: differential effects in the myocardium and liver. Acta Physiol Scand 169: 343–351, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi P, Kunduzova O, Masini E, Cambon C, Bani D, Raimondi L, Seguelas MH, Nistri S, Colucci W, Leducq N, Parini A. Oxidative stress by monoamine oxidase mediates receptor-independent cardiomyocyte apoptosis by serotonin and postischemic myocardial injury. Circulation 112: 3297–3305, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Bousette N, Kislinger T, Fong V, Isserlin R, Hewel J, Emili A, Gramolini A. Large scale characterization and analysis of the murine cardiac proteome. J Proteome Res. In press. [DOI] [PubMed]
- 7.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J 134: 707–716, 1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Danh H, Strolin Benedetti M, Dostert P, Mousset A. Age-related changes in benzylamine oxidase activity in rat tissues. J Pharm Pharmacol 36: 592–596, 1984. [DOI] [PubMed] [Google Scholar]
- 9.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev 59: 527–605, 1979. [DOI] [PubMed] [Google Scholar]
- 10.Chang TS, Cho CS, Park S, Yu S, Kang SW, Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J Biol Chem 279: 41975–41984, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Cox AG, Pullar JM, Hughes G, Ledgerwood EC, Hampton MB. Oxidation of mitochondrial peroxiredoxin 3 during the initiation of receptor-mediated apoptosis. Free Radic Biol Med 44: 1001–1009, 2008. [DOI] [PubMed] [Google Scholar]
- 12.Criswell D, Powers S, Dodd S, Lawler J, Edwards W, Renshler K, Grinton S. High intensity training-induced changes in skeletal muscle antioxidant enzyme activity. Med Sci Sports Exerc 25: 1135–1140, 1993. [PubMed] [Google Scholar]
- 13.Dreger M, Mika J, Bieller A, Jahnel R, Gillen C, Schaefer MK, Weihe E, Hucho F. Analysis of the dorsal spinal cord synaptic architecture by combined proteome analysis and in situ hybridization. J Proteome Res 4: 238–249, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 5: 608–619, 2006. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb RA Mitochondria and apoptosis. Biol Signals Recept 10: 147–161, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science 305: 626–629, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Heaton D, Nittis T, Srinivasan C, Winge DR. Mutational analysis of the mitochondrial copper metallochaperone Cox17. J Biol Chem 275: 37582–37587, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Ignarro LJ, Balestrieri ML, Napoli C. Nutrition, physical activity, and cardiovascular disease: an update. Cardiovasc Res 73: 326–340, 2007. [DOI] [PubMed] [Google Scholar]
- 20.Judge S, Jang YM, Smith A, Hagen T, Leeuwenburgh C. Age-associated increases in oxidative stress and antioxidant enzyme activities in cardiac interfibrillar mitochondria: implications for the mitochondrial theory of aging. FASEB J 19: 419–421, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Judge S, Jang YM, Smith A, Selman C, Phillips T, Speakman JR, Hagen T, Leeuwenburgh C. Exercise by lifelong voluntary wheel running reduces subsarcolemmal and interfibrillar mitochondrial hydrogen peroxide production in the heart. Am J Physiol Regul Integr Comp Physiol 289: R1564–R1572, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Julia P, Young HH, Buckberg GD, Kofsky ER, Bugyi HI. Studies of myocardial protection in the immature heart. II. Evidence for importance of amino acid metabolism in tolerance to ischemia. J Thorac Cardiovasc Surg 100: 888–895, 1990. [PubMed] [Google Scholar]
- 23.Kavazis AN, McClung JM, Hood DA, Powers SK. Exercise induces a cardiac mitochondrial phenotype that resists apoptotic stimuli. Am J Physiol Heart Circ Physiol 294: H928–H935, 2008. [DOI] [PubMed] [Google Scholar]
- 24.Kong SW, Bodyak N, Yue P, Liu Z, Brown J, Izumo S, Kang PM. Genetic expression profiles during physiological and pathological cardiac hypertrophy and heart failure in rats. Physiol Genomics 21: 34–42, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Kunduzova OR, Bianchi P, Parini A, Cambon C. Hydrogen peroxide production by monoamine oxidase during ischemia/reperfusion. Eur J Pharmacol 448: 225–230, 2002. [DOI] [PubMed] [Google Scholar]
- 26.Lennon SL, Quindry J, Hamilton KL, French J, Staib J, Mehta JL, Powers SK. Loss of exercise-induced cardioprotection after cessation of exercise. J Appl Physiol 96: 1299–1305, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Lennon SL, Quindry JC, French JP, Kim S, Mehta JL, Powers SK. Exercise and myocardial tolerance to ischaemia-reperfusion. Acta Physiol Scand 182: 161–169, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Lipton SA, Bossy-Wetzel E. Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell 111: 147–150, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Martyniuk C, Alvarez S, McClung S, Villeneuve D, Ankley G, Denslow N. Quantitative proteomic profiles of androgen receptor signaling in the liver of fathead minnows (Pimephales promelas). J Proteome Res 8: 2186–2200, 2009. [DOI] [PubMed] [Google Scholar]
- 30.Matsushima S, Ide T, Yamato M, Matsusaka H, Hattori F, Ikeuchi M, Kubota T, Sunagawa K, Hasegawa Y, Kurihara T, Oikawa S, Kinugawa S, Tsutsui H. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation 113: 1779–1786, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Maurel A, Hernandez C, Kunduzova O, Bompart G, Cambon C, Parini A, Frances B. Age-dependent increase in hydrogen peroxide production by cardiac monoamine oxidase A in rats. Am J Physiol Heart Circ Physiol 284: H1460–H1467, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Mayr M, Zhang J, Greene AS, Gutterman D, Perloff J, Ping P. Proteomics-based development of biomarkers in cardiovascular disease: mechanistic, clinical, and therapeutic insights. Mol Cell Proteomics 5: 1853–1864, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Meco M, Bonifati V, Collier WL, Ramacci MT, Amenta F. Enzyme histochemistry of monoamine oxidase in the heart of aged rats. Mech Ageing Dev 38: 145–155, 1987. [DOI] [PubMed] [Google Scholar]
- 34.Miranda-Vizuete A, Damdimopoulos AE, Spyrou G. The mitochondrial thioredoxin system. Antioxid Redox Signal 2: 801–810, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Mitulovic G, Stingl C, Smoluch M, Swart R, Chervet JP, Steinmacher I, Gerner C, Mechtler K. Automated, on-line two-dimensional nano liquid chromatography tandem mass spectrometry for rapid analysis of complex protein digests. Proteomics 4: 2545–2557, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977. [PubMed] [Google Scholar]
- 37.Pchejetski D, Kunduzova O, Dayon A, Calise D, Seguelas MH, Leducq N, Seif I, Parini A, Cuvillier O. Oxidative stress-dependent sphingosine kinase-1 inhibition mediates monoamine oxidase A-associated cardiac cell apoptosis. Circ Res 100: 41–49, 2007. [DOI] [PubMed] [Google Scholar]
- 38.Ping P Identification of novel signaling complexes by functional proteomics. Circ Res 93: 595–603, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Powers SK, Quindry JC, Kavazis AN. Exercise-induced cardioprotection against myocardial ischemia-reperfusion injury. Free Radic Biol Med 44: 193–201, 2008. [DOI] [PubMed] [Google Scholar]
- 40.Quindry J, French J, Hamilton K, Lee Y, Mehta JL, Powers S. Exercise training provides cardioprotection against ischemia-reperfusion induced apoptosis in young and old animals. Exp Gerontol 40: 416–425, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52: 35–41, 2001. [DOI] [PubMed] [Google Scholar]
- 42.Riva A, Tandler B, Loffredo F, Vazquez E, Hoppel C. Structural differences in two biochemically defined populations of cardiac mitochondria. Am J Physiol Heart Circ Physiol 289: H868–H872, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Rosamond W, Flegal K, Friday G, Furie K, Go A, Greenlund K, Haase N, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell CJ, Roger V, Rumsfeld J, Sorlie P, Steinberger J, Thom T, Wasserthiel-Smoller S, Hong Y. Heart disease and stroke statistics–2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 115: e69–e171, 2007. [DOI] [PubMed] [Google Scholar]
- 44.Schauble S, King CC, Darshi M, Koller A, Shah K, Taylor SS. Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J Biol Chem 282: 14952–14959, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Starnes JW, Barnes BD, Olsen ME. Exercise training decreases rat heart mitochondria free radical generation but does not prevent Ca2+-induced dysfunction. J Appl Physiol 102: 1793–1798, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nature Biotechnol 21: 281–286, 2003. [DOI] [PubMed] [Google Scholar]
- 47.van Empel VP, Bertrand AT, van der Nagel R, Kostin S, Doevendans PA, Crijns HJ, de Wit E, Sluiter W, Ackerman SL, De Windt LJ. Downregulation of apoptosis-inducing factor in harlequin mutant mice sensitizes the myocardium to oxidative stress-related cell death and pressure overload-induced decompensation. Circ Res 96: e92–e101, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Vondriska TM, Ping P. Multiprotein signaling complexes and regulation of cardiac phenotype. J Mol Cell Cardiol 35: 1027–1033, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett 581: 3545–3549, 2007. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X, Zuo L, Cardounel AJ, Zweier JL, He G. Characterization of in vivo tissue redox status, oxygenation, and formation of reactive oxygen species in postischemic myocardium. Antioxid Redox Signal 9: 447–455, 2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.