Abstract
Inflammatory cytokines, particularly the neutrophil chemoattractant IL-8, are elevated in the cystic fibrosis (CF) airway, even in the absence of detectable infection. The transcriptional regulation of many inflammatory genes, including IL8 (CXCL8), involves chromatin remodeling through histone acetylation. NF-κB is known to facilitate histone acetylation of IL8 and other proinflammatory gene promoters, but we find that increased NF-κB activation cannot explain the elevated IL8 expression and promoter acetylation seen in CFTR-deficient cells. Recognized components of the NF-κB-coactivator complex, acetyltransferase CBP, p300, and the histone deacetylase HDAC1, are unchanged by CFTR activity. However, we find that the histone acetyltransferase (HAT)/HDAC balance is sensitive to CFTR function, as cells with reduced or absent CFTR function have decreased HDAC2 protein, resulting in hyperacetylation of the IL8 promoter and increased IL8 transcription. Reduced HDAC2 and HDAC2 activity, but not HDAC2 mRNA, is observed in cells deficient in CFTR. Suppressing HDAC2 expression with HDAC2 short hairpin RNA (shRNA) results in increased IL8 expression and promoter acetylation comparable with CFTR-deficient cells. Treating CFTR-deficient cells with N-acetyl-cysteine (NAC) increases HDAC2 expression to near control levels. Our data suggest that there is an intrinsic alteration in the HAT/HDAC balance in cells lacking CFTR function in vitro and in native CF tissue and that oxidative stress is likely contributing to this alteration. This mechanism, found in other inflammatory airway diseases, provides an explanation for the apparent dysregulation of inflammatory mediators seen in the CF airway, as reduced histone deacetylation would potentially influence many genes.
Keywords: cystic fibrosis, acetylation, interleukin-8, oxidative stress
cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encodes a cAMP-regulated chloride channel. Inflammation is believed to be a primary cause of CF pulmonary pathology and leads to tissue destruction and ultimately respiratory failure. The link between loss of functional CFTR and CF lung disease is still not fully understood. Inflammatory cytokine responses, particularly that of the chemoattractant IL-8, are much greater in bronchoalveolar lavage fluid (BALF) obtained from CF patients compared with controls (2, 3, 7, 9, 15, 18, 27, 37, 51, 56). Although the production of inflammatory mediators is increased from CF lung epithelial cells, it is still unclear how a mutation in a chloride channel results in an exaggerated innate inflammatory response.
Recently, chromatin structure has been recognized as an important component in the regulation of inflammatory genes. The activation of many inflammatory genes involves the remodeling of chromatin structure through changes in the level of acetylation. Acetylation of lysine residues in the histone tails can be directly related to the level of gene transcription (35), and acetylation of a particular combination of residues may influence transcription factor access to a distinct set of genes. Histone deacetylases (HDACs) are thought to have a key role in the regulation of inflammatory gene expression because HDAC inhibitors enhance NF-κB-dependent inflammatory gene transcription with various stimuli (1). Small alterations in the histone acetyltransferase (HAT)/HDAC balance could affect transcription of many inflammatory genes, potentially having a profound effect on the initiation, duration, and resolution of an inflammatory response. Loss of tight regulation of inflammatory genes through hyperacetylation could explain some of the dysregulation of inflammatory mediators seen in the CF lung. This dysregulation includes elevated cytokine levels early in life (37, 39), exaggerated levels relative to the bacterial burden present (27), and a prolonged increase in inflammatory mediators after an infection (37). We (6) have previously reported that three cell line pairs lacking functional CFTR showed consistent changes in IL8 acetylation and expression.
There is evidence that this delicate balance between acetylation and deacetylation is altered in other inflammatory lung diseases. Chronic obstructive pulmonary disease (COPD) and asthma are also characterized by localized chronic inflammation, increased expression of multiple inflammatory genes, and high levels of oxidative stress. Lung biopsies from asthmatic patients showed reduced HDAC activity, reduced HDAC1 and HDAC2 protein expression, and increased HAT activity (20). The major cause of COPD is smoking. Lung biopsies from smokers also show reduced HDAC activity and protein expression of HDAC2 (23). To date, there are no reported studies on HAT/HDAC levels in the CF lung or in cells lacking functional CFTR. Global HAT or HDAC levels have been evaluated in five CF lung samples, and no difference was observed compared with controls (22). Different HDACs likely target different patterns of acetylation and regulate a specific set of genes (42), and an alteration in one HDAC may not be detected in global activity levels. An alteration in the balance of specific HAT or HDAC proteins could in part explain the dysregulation of inflammatory genes and IL8 hyperacetylation seen in cells lacking CFTR (6).
Oxidative stress is known to increase histone acetylation and can activate inflammatory gene transcription (43, 52). The airways of CF patients are exposed to increased levels of oxidative stress (19). High levels of oxidative stress are generated from the massive infiltration of neutrophils, which dominate the inflammatory response in the CF lung. CF patients may also have inadequate antioxidant defenses to cope with the elevated oxidative stress that they regularly experience. Oxidative stress contributes to the decline in pulmonary function in CF patients (12), and CF patients have increased levels of oxidative stress markers in their plasma (10, 11). Oxidative stress may lead to an imbalance in the levels of HATs and HDACs, and this may amplify lung inflammation. This imbalance could lead to a chronic and exaggerated inflammatory response, as seen in the CF lung.
It is not clear whether loss of CFTR directly affects the redox state in the CF lung. An alteration in lung glutathione levels, however, is a factor in several inflammatory lung diseases, including CF. Glutathione is a protective antioxidant in the lungs and plays a key role in regulating oxidant-induced lung injury. A reduction in glutathione levels would result in loss of antioxidant protection and increased oxidative burden to the lung. Bronchoalveolar lavage levels of glutathione are diminished in CF patients (47), and CFTR-null mice lack a significant increase in epithelial lining fluid glutathione when challenged with Pseudomonas aeruginosa (14). We (6) previously reported that cell lines with impaired CFTR function show consistent changes in IL8 acetylation and expression that could be modulated to a non-CF state by an agent known to reduce the redox state of the cells. Conversely, the lines expressing functional CFTR could be induced to a CF state of IL8 acetylation by treatment with an agent that increases oxidative load (6). These studies indicate an intrinsic change that contributes to a high level of oxidative stress in the CF lung. The CF airways may represent an environment that results in hyperacetylation of inflammatory loci and a primed situation for an exaggerated immune response.
Here, we report that increased NF-κB activation alone cannot explain the elevation in IL8 expression and promoter acetylation seen in cells deficient in CFTR. No difference was found in the expression or activity of acetyltransferases CBP or p300, which are other components of the NF-κB-coactivator complex. No difference was found in the level of HDAC1. Cells lacking CFTR show a reduction in HDAC2 protein level. A corresponding reduction in HDAC2 activity level was also seen. Reducing HDAC2 levels with HDAC2 short hairpin RNA (shRNA) results in exaggerated IL8 expression and promoter acetylation as seen in CFTR-deficient cells. Treating CFTR-deficient cells with N-acetyl-cysteine (NAC) brings HDAC2 expression closer to control levels. Our data suggest that there is an intrinsic alteration in the HAT/HDAC balance in cells lacking CFTR function in cell culture models and in CF primary tissue and that oxidative stress is likely contributing to this imbalance.
MATERIALS AND METHODS
9/HTEo− pCEP and pCEP-R cell lines.
Human tracheal epithelial cell lines were created with 9/HTEo− cells overexpressing the CFTR regulatory (R) domain (pCEP-R; CF phenotype) and mock-transfected 9/HTEo− cells (pCEP; wild-type phenotype). Those cells in which CFTR function has been impaired (pCEP-R) are referred to here as CFTR−, and pCEP cells are referred to as CFTR+. These cell lines were provided by Dr. Pamela B. Davis (Case Western Reserve University, Cleveland, OH). Cells were grown at 37°C in 95% O2-5% CO2 on Falcon 10-cm diameter tissue culture dishes cultured as previously described (40).
16HBEo− antisense and sense cell lines.
Human bronchial epithelial cells (16HBEo−) were stably transfected with plasmids containing the first 131 nucleotides of human CFTR in the sense (16HBE-S) or antisense (16HBE-AS) orientations. The 16HBE-AS cells have a CF-like physiology. 16HBE-AS cells in which CFTR function has been impaired are referred to as CFTR−, and 16HBE-S cells are referred to as CFTR+. Cells were grown at 37°C in 95% O2-5% CO2 on Falcon 10-cm diameter tissue culture dishes and were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 200 μg/ml G418 as previously described (31). 16HBE-S and 16HBE-AS cells were provided by Dr. Pamela B. Davis (Case Western Reserve University, Cleveland, OH).
IB3-1 and S9 cells.
IB3 bronchial epithelial cells (ΔF508/W1282X) and S9 cells (IB3-1 cells stably transfected with the full-length wild-type CFTR as controls) were developed by Dr. Pamela L. Zeitlin (Johns Hopkins University, Baltimore, MD). IB3 cells are referred to as CFTR− and S9 cells, transduced to express wild-type CFTR, are referred to as CFTR+. These cells were grown at 37°C in 95% O2-5% CO2 on Falcon 10-cm diameter tissue culture dishes in LHC-8 Basal Medium (Biosource International, Camarillo, CA) with 5% fetal bovine serum.
Human nasal scrapings.
Nasal epithelia from CF patients and control individuals were obtained as previously described (29). Four scrapes from each nasal passage were taken from three CF subjects and three non-CF volunteers and immediately placed in ice-cold HBSS. Cells were pelleted at 4°C at 1,000 rpm for 5 min, and nuclear lysate was prepared from cells using the Active Motif Nuclear Extract Kit (Carlsbad, CA) according to manufacturer's instructions. Extracts were stored at −80°C until Western blotting analysis was performed. The protocol was approved by the Institutional Review Board. Human patients and volunteers gave informed consent under protocol approved by the Institutional Review Board (University Hospitals, Cleveland, OH).
Cell treatments.
Experimental and control cells were placed in serum-free media 24 h before any treatment. For inflammatory stimulation, cells were treated with a combination of 1 ng/ml TNFα and 0.5 ng/ml IL-1β (R&D Systems, Minneapolis, MN) for 24 h in serum-free media. For antioxidant treatment, cells were exposed to 5 mM NAC (Sigma, St. Louis, MO) in serum-free medium for 2 h before isolation of nuclear extract.
Preparing nuclear extracts.
Nuclear lysate was prepared from cells using the Active Motif Nuclear Extract Kit according to manufacturer's instructions. The total protein concentrations of nuclear extracts were determined with a Bio-Rad (Hercules, CA) protein assay. Nuclear extracts were stored at −80°C.
NF-κB (p65) activation assay.
An EZ-Detect Transcription Factor Kit for NF-κB p65 (Pierce, Rockford, IL) was used to assay the amount of active NF-κB p65. The kit was used according to manufacturer's instructions. Ten micrograms of nuclear lysate was added to wells coated with NF-κB consensus sequence. Bound, activated NF-κB was then detected with a p65 antibody. Chemiluminescence was measured with a luminometer.
Real-time quantitative PCR.
Total RNA was extracted from cells using Qiagen (Valencia, CA) RNeasy columns with DNase treatment. cDNA was synthesized using 1 μg of RNA and random primers. RT-PCR samples excluding Moloney murine leukemia virus RT (M-MLV-RT) served as negative controls in quantitative PCR (QPCR). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used to normalize all QPCR mRNA data. Real-time QPCR was performed using the DyNAmo HS SYBR Green qPCR Kit and the DNA Engine Opticon 2 System (MJ Research, Waltham, MA). PCR conditions were as follows: 95°C for 15 min, 95°C for 10 s, 60°C for 30 s, 72°C for 30 s repeated 50 cycles, 72°C for 10 min. For quantification, 8 10-fold dilutions of purified PCR product were used to make a standard curve. QPCR primers: GAPDH 5′-ACAACTTTGGTATCGTGGAAGGAC-3′, 5′-AGGCAGGGATGATGTTCTGGAG-3′; IL8 5′-TCTCTTGGCAGCCTTCCTGATTTC-3′, 5′-TCCAGACAGAGCTCTCTTCCATCA-3′.
H4 chromatin immunoprecipitation (ChIP).
Cells were lysed, and chromatin was sheared using an Active Motif Enzymatic Shearing Kit. Enzymatic shearing for 4 min resulted in chromatin fragments of 100–1,000 bp in size. Chromatin immunoprecipitation (ChIP) was performed using the Upstate Biotechnology (Billerica, MA) ChIP kit with an acetyl-histone H4 antibody (17-295). ChIP samples and input DNA were purified using a phenol/chloroform extraction followed by ethanol precipitation. QPCR as described above was used with promoter-specific primers to quantify ChIP samples and input DNA. For quantification, 8 10-fold dilutions of purified PCR product was used to make a standard curve. Initial experiments were performed using an IgG control antibody (ChIP-IT Control Kit; Active Motif) to confirm specificity of H4 ChIP. Immunoprecipitations (IP) using the control IgG antibody did not yield detectable enrichment of IL-8 promoter sequence (data not shown). QPCR primers: IL8 5′-GTTGTAGTATGCCCCTAAGAG-3′, 5′-CTCAGGGCAAACCTGAGTCATC-3′.
Western immunoblotting.
Antibodies against CBP (cat. no. 06-297), p300 (cat. no. 05-257), HDAC2 (cat. no. 05-814), and HDAC1 (cat. no. 06-720) were obtained from Upstate Biotechnology. Antibodies against nucleolin (cat. no. KAM-CP100) were obtained from Stressgen Bioreagents (Ann Arbor, MI). Nuclear extracts were prepared using a Nuclear Extract Kit (Active Motif) according to manufacturer's instructions. Proteins were separated using SDS-PAGE with a 10% acrylamide gel. Fifty micrograms of protein were loaded per sample. The samples were transferred to a nitrocellulose membrane (Schleicher & Schuell BioScience, Keene, NH) at 100 V for 60 min using a Bio-Rad wet-transfer apparatus. Blots were blocked with Tris-buffered saline with Tween 20 (TBST) containing 5% nonfat dehydrated milk for 2 h at room temperature or overnight at 4°C. Blots were incubated overnight with primary antibody (at 1:1,000 dilution) in TBST with 5% nonfat dehydrated milk at 4°C with rocking. Blots were washed three times in TBST and incubated with secondary antibody conjugated to horseradish peroxidase (1:10,000 dilution) in TBST 5% nonfat dehydrated milk for 2 h at room temperature with rocking. Blots were again washed three times in TBST. SuperSignal West Pico Chemiluminescent Substrate (Pierce), BioMax XAR film (Kodak, Rochester, NY), and densitometry software on the VersaDoc (Quantity One; Bio-Rad) were used to quantify protein expression. Blots were stripped with Restore Western Blot Stripping Buffer (Pierce) per manufacturer's instructions and reprobed with additional antibodies.
HDAC IP.
HDAC1 and HDAC2 were immunoprecipitated from nuclear extract using a Seize Classic (G) Immunoprecipitation Kit (Pierce) with HDAC1 (cat. no. 06-720) and HDAC2 (cat. no. 05-814) antibodies from Upstate Biotechnology. For a negative control, an IP was performed with no antibody present. Fifty micrograms of nuclear extract were diluted to 0.5 μg/μl in 1× PBS. Extract was incubated with 2 μg of antibody overnight at 4°C with rocking, to which 40 μl of immobilized protein G was added and incubated 4 h at 4°C with rocking. The mixture was centrifuged for 5 min at 13,000 rpm at 4°C, and the supernatant was removed. Immobilized protein G was washed 2× with 500 μl of Wash Buffer and then resuspended in 20 μl of lysis buffer (Active Motif Nuclear Extraction Kit). After 5-min centrifugation at 13,000 rpm, 4°C, the supernatant was removed for analysis.
HAT and HDAC activity assays.
HAT and HDAC activities were assayed from nuclear extracts using colorimetric enzymatic assay kits from BioVision Incorporated (Mountain View, CA), K332-100 and K331-100. Kits were used according to manufacturer's instructions to assay global HAT and HDAC activity. HDAC1 and HDAC2 activity were assayed after IP. The IP pellet was resuspended in 85 μl of water and 10 μl of 10× HDAC buffer. Five microliters of colorimetric substrate was added and incubated 1 h at 37°C with shaking. Ten microliters of lysine developer was added and incubated 30 min at 37°C with shaking, followed by a 5-min centrifugation at 13,000 rpm, 4°C, after which the supernatant was removed and transferred to a 96-well plate and read at 405 nm.
HDAC2 knockdown.
HDAC2 knockdown was performed using the HDAC2 RNAi Upstate Biotechnology AdenoSilence RNAi Virus HDAC2-v2 (cat. no. GAL10051-V2) per manufacturer's instructions. Cells were plated on 100-mm plates. On the day of transduction, media was removed from plates, and the cells were washed with PBS. Transduction was performed in serum-free medium at a multiplicity of infection (MOI) of 500. The transduction medium was left on the cells for 24 h and then replaced with normal medium. Cells were harvested after 5 days.
Data analysis.
All RNA data were normalized to GAPDH levels. ChIP samples were all normalized to the amount of input DNA for each sample. This normalization was performed after the QPCR and before any statistical analysis. All data are presented as the sample average with standard error bars. All data are presented with the sample average of the untreated CFTR+ set to 1 to aid in data interpretation. All the other averages were adjusted accordingly. Variances of sample groups were compared by F test, and no significantly different variances were detected. Subsequently, averages were compared using t-tests.
RESULTS
NF-κB activation in CFTR-deficient cells.
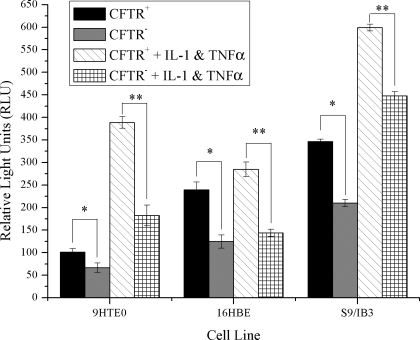
NF-κB is considered the master transcription factor for numerous genes involved in inflammation, immune responses, and cellular proliferation. NF-κB is also ubiquitously expressed (55). Enhanced NF-κB activation results in targeted increases in histone modifications, such as acetylation leading to inflammatory gene expression. We (6) have previously demonstrated that cell models deficient in CFTR have exaggerated IL8 message levels and H4 acetylation around the NF-κB site in the IL8 promoter. To determine whether this alteration in H4 acetylation can be explained by elevated NF-κB activation, we assayed the amount of active NF-κB p65 in these same three cell models. In the 9/HTEo and S9/IB3 cells, both CFTR− and CFTR+ cells responded appropriately to inflammatory stimulation with an increase in NF-κB activation. However, under all conditions tested, NF-κB was never greater in the CFTR− cells than the CFTR+ cells, as would be predicted if NF-κB were responsible for the increased histone acetylation state of the CFTR− cells. In fact, the amount of active NF-κB p65 observed in the three CFTR− cell lines under basal conditions, or after inflammatory stimulation (1 ng/ml TNFα and 0.5 mg/ml IL-1β; Fig. 1), was significantly lower in the CFTR− cells. Thus, in these cell models, it does not appear that increased NF-κB activation can explain the alteration in IL8 promoter acetylation or IL8 message levels. These results suggest that another component of the NF-κB-coactivator complex may be altered in cells deficient in CFTR.
Fig. 1.
There is no elevation in the amount of activated NF-κB in the nucleus of CFTR− cells. NF-κB p65 DNA binding activity was assayed in 9/HTEo cells and human bronchial epithelial (16HBE) cells with and without CFTR function. Cells were placed in serum-free medium 48 h before the preparation of nuclear extract. For inflammatory stimulation, cells were treated with a combination of 1 ng/ml TNFα and 0.5 ng/ml IL-1β for 24 h in serum-free media. A chemiluminescent ELISA-based assay was used to measure active NF-κB p65 transcription factors in 10 μg of nuclear protein. NF-κB p65 DNA binding activity is expressed as relative light units (RLU). Error bars represent SE. n = 6. *P < 0.02 for comparisons of CFTR+ with CFTR− cells for each cell pair. **P < 0.000007 for comparisons of CFTR+ cells treated with TNFα and IL-1β with CFTR− cells treated with TNFα and IL-1β for each cell pair.
Levels of the acetyltransferases CBP or p300 in CFTR-deficient cells.
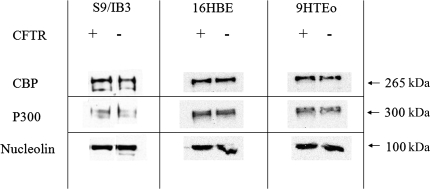
After translocation into the nucleus, NF-κB physically interacts with both CBP and p300 (16, 38, 49). Both of these enzymes contain an intrinsic HAT activity. The p65 component of NF-κB binds to the coactivator CBP and its structural homolog p300 (16, 41, 57). To determine whether increased expression of either of these proteins could account for the elevated acetylation at the IL-8 promoter in CFTR− cells, protein levels were evaluated in the three cell models. There was no significant difference in CBP or p300 protein levels between any of the cell models (Fig. 2). These results indicate that the expression of the most relevant acetyltransferases are not altered with loss of CFTR.
Fig. 2.
Loss of CFTR does not result in an elevation in the protein levels of the acetyltransferase CBP or p300. Protein levels were assayed in IB3 and S9 cells. The level of CBP and p300 expression in nuclear extract was determined by Western blotting. Nucleolin was used as a loading control.
HDAC2 protein level in CFTR-deficient cells.
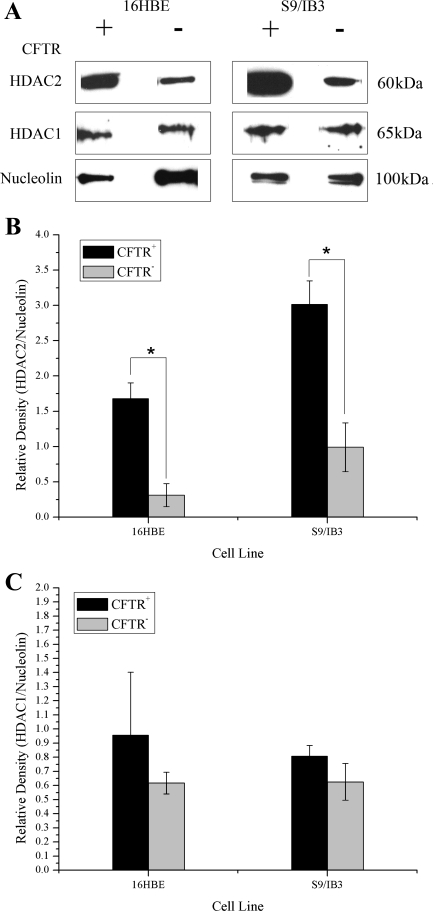
HDAC1 and HDAC2 target NF-κB through a direct association of HDAC1 with p65. HDAC2 does not interact with NF-κB directly but can regulate NF-κB activity through its association with HDAC1 (4). HDAC1 has been shown to be recruited to the IL8 promoter (25). In mammalian cells, HDAC1 and HDAC2 are typically found in a large complex together (28). Expression of HDAC2 protein has been shown to be lower in patients with increasing severity of COPD (22) and cigarette smokers (23). The 16HBE sense/antisense and the S9/IB3 cell pairs were compared for HDAC protein levels. Similar to findings in COPD and smokers, we also have found a decrease in HDAC2 protein levels in CFTR− cells compared with CFTR+ controls (Fig. 3, A and B). No significant alterations in HDAC1 protein levels were found in CFTR− cells (Fig. 3, A and C).
Fig. 3.
Loss of CFTR results in a reduction in the protein levels of the histone deacetylase HDAC2 but not HDAC1. The level of HDAC1 and HDAC2 expression in nuclear extract was determined by Western blotting. Nucleolin is used as a loading control. Protein levels were assayed in IB3/S9 and 16HBE cells. A: Western blot showing protein levels of HDAC1 and HDAC2. 16HBE, 9/HTEo, and S9/IB3 samples were run on different gels. B: densitometry analysis of HDAC2 protein levels relative to nucleolin protein level. n = 3. *P < 0.004 for comparisons of CFTR+ with CFTR− cells for each cell pair. C: densitometry analysis of HDAC1 protein levels relative to nucleolin protein level. n = 3.
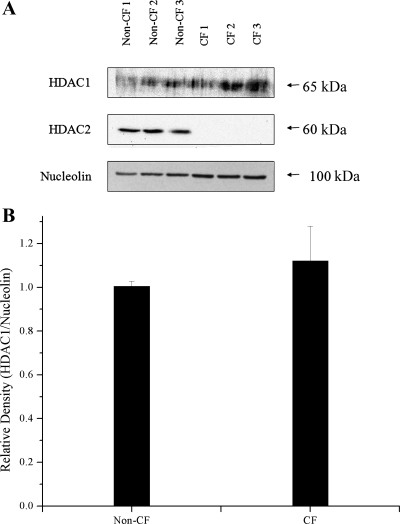
As all three of the CFTR-deficient cell models showed decreased HDAC2 protein compared with controls, we examined HDAC2 protein levels in tissue obtained by nasal scraping from three separate CF subjects and three non-CF controls. As shown in Fig. 4A, HDAC2 protein was not detectable in all three of the CF primary human tissue samples by Western blot analysis. The three non-CF control samples showed similar HDAC2 levels compared with each other. Although densitometry analysis for HDAC2 was not possible due to the undetectable levels of HDAC2 in the CF samples, it does appear that the loss of HDAC2 protein in CF primary tissue may be even more pronounced than in the CF cell line models. No significant difference in HDAC1 levels were found between CF and non-CF primary tissue samples by densitometry analysis (Fig. 4B). These data demonstrate in primary human nasal tissue and in cultured cell models of CF there is an intrinsic reduction in HDAC2 protein level in CF cells and tissues.
Fig. 4.
HDAC2 is not detectable in cystic fibrosis (CF) primary nasal tissue by Western blot analysis. HDAC1 levels are not significantly different between CF and non-CF primary tissue. A: Western blot showing HDAC1, HDAC2, and nucleolin expression in human nasal scrape tissue obtained from 3 non-CF and 3 CF subjects. B: densitometry analysis of HDAC1 protein expression normalized to nucleolin content. n = 3. P = 0.5 for the comparison non-CF with CF samples. Significance was determined by t-test. Densitometry analysis for HDAC2 was not possible due to the undetectable levels of HDAC2 in the CF samples.
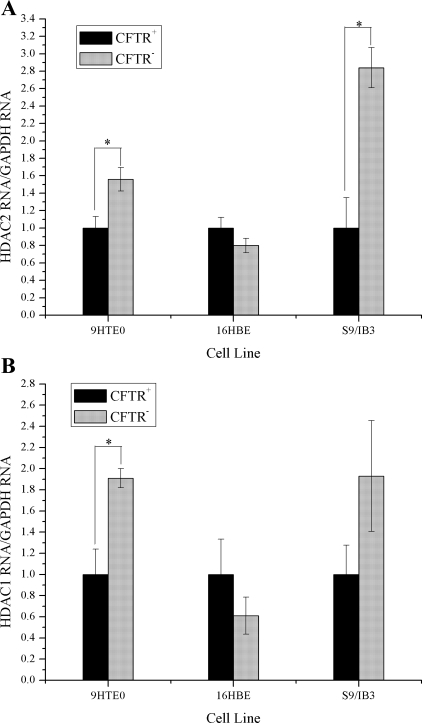
However, unlike the other mentioned studies, no corresponding reduction in HDAC2 RNA was observed in the CFTR− cells. In fact, HDAC2 message level is significantly increased in two of the CFTR− cell lines (Fig. 5A). There was no reduction in HDAC1 RNA levels in CFTR− cells either. HDAC1 message level is significantly increased in one of the CFTR− cell lines (Fig. 5B).
Fig. 5.
There is no reduction in the amount HDAC2 message in CFTR− cells. HDAC1 and HDAC2 RNA levels were evaluated in 3 CFTR− airway epithelial cell lines compared with CFTR+ controls. Cells were placed in serum-free medium 24 h before RNA isolation. All HDAC mRNA levels are normalized to GAPDH mRNA levels. The CFTR+ cell line averages have been set at 1 for each cell pair, and CFTR− averages are normalized to that average. Error bars represent SE. n = 3. A: HDAC2 message levels. *P < 0.02 for comparisons of CFTR+ with CFTR− cells for each cell pair. B: HDAC1 message levels. *P < 0.01 for comparisons of CFTR+ with CFTR− cells for each cell pair.
HAT and HDAC activity in CFTR-deficient cells.
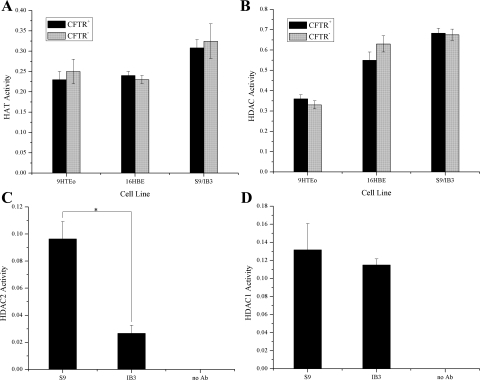
Total HAT and HDAC activity in nuclear extract samples was not altered in CFTR− cells compared with CFTR− controls (Fig. 6, A and B). No alteration in total HAT activity is consistent with the fact that no increase in CBP/p300 protein levels were observed in these cells. A reduction in total HDAC activity was not detectable with this assay even with the observed reduction in HDAC2 protein levels. However, when HDAC2 was immunoprecipitated from the nuclear extract and then assayed for HDAC activity, an ∼2.5-fold reduction in activity level was observed (Fig. 6C). No reduction in HDAC1 activity was seen in immunoprecipitated HDAC1 samples, consistent with the HDAC1 protein levels (Fig. 6D).
Fig. 6.
There is a reduction in HDAC2 activity with no global change in histone acetyltransferase (HAT) or HDAC activity levels in CFTR− cells. Enzymatic activity was assayed from nuclear extract using colorimetric activity assays. A: global HAT activity from nuclear extract samples. Cells were placed in serum-free medium 48 h before the preparation of nuclear extract. n = 6. B: global HDAC activity from nuclear extract samples. Cells were placed in serum-free medium 48 h before the preparation of nuclear extract. n = 6. C: deacetylase activity of immunoprecipitated HDAC2. No HDAC2 antibody (Ab) was used in 1 sample during the immunoprecipitation as a negative control. n = 3. *P < 0.004 for comparisons of CFTR+ with CFTR− cells. D: deacetylase activity of immunoprecipitated HDAC1. No HDAC1 antibody was used in 1 sample during the immunoprecipitation as a negative control. n = 3.
Reducing HDAC2 with RNAi results in exaggerated IL-8 message level and promoter acetylation.
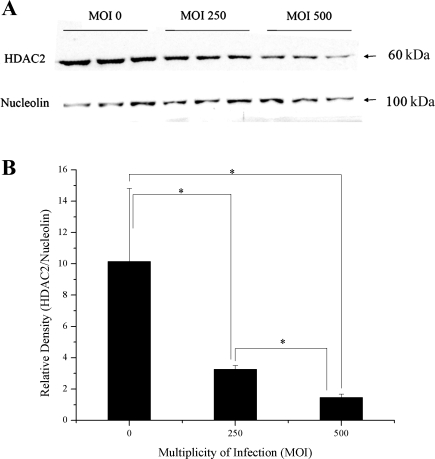
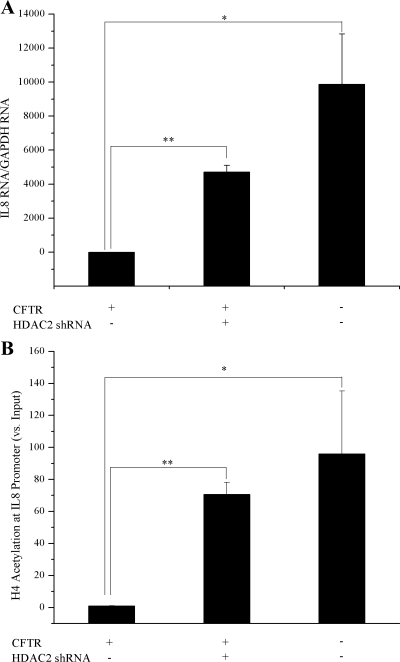
16HBE cells were used for HDAC2 knockdown experiments because they have endogenous CFTR expression (not CFTR corrected as the S9 cells) and showed a higher level of transduction than the 9/HTEo cells. By RNA interference (RNAi), we are able to achieve a 2- to 3-fold reduction in HDAC2 protein levels in 16HBE sense (CFTR+) cells. An MOI of 500 resulted in the greatest knockdown of HDAC2 and was chosen for use in future RNAi experiments (Fig. 7, A and B). Reducing HDAC2 protein levels in CFTR+ cells resulted in a significant increase in IL8 RNA levels. The level of IL8 RNA in the CFTR+ cells with HDAC2 RNAi was still less than the matched CFTR− cell line but only marginally (P = 0.08; Fig. 8A). Consistent with the IL8 RNA level, when HDAC2 protein level was reduced in CFTR+ cells, the level of H4 acetylation at the IL8 promoter was also increased (Fig. 8B). The level of IL8 RNA in the CFTR+ cells with HDAC2 shRNA was increased to a level similar to that seen in the matched CFTR− cell line (P = 0.3; Fig. 8A).
Fig. 7.
HDAC2 short hairpin RNA (shRNA) sequence does reduce the amount of HDAC2 protein in a multiplicity of infection (MOI)-dependent manner. HDAC2 protein levels were knocked down using an Adenoviral shRNA Expression Vector in 16HBE CFTR+ cells. A titration was performed using the Adenoviral shRNA Expression Vector at different MOIs. Knockdown was seen at a MOI of 250, however, greatest knockdown was seen at a MOI of 500. The level of HDAC2 protein from nuclear extract was determined by Western blotting. Nucleolin was used as a loading control. A: Western blot showing protein levels of HDAC2. Blot contains 3 replicate samples for each MOI, showing consistent HDAC2 knockdown. B: densitometry analysis of HDAC2 protein levels relative to nucleolin protein level. n = 3. *P < 0.05 for comparisons of MOI 0 with MOI 500, MOI 0 with MOI 250, and MOI 250 with MOI 500.
Fig. 8.
Knockdown of HDAC2 protein levels results in elevated IL-8 message levels and IL-8 promoter hyperacetylation. IL-8 RNA levels and promoter acetylation were evaluated in 16HBE CFTR+ cells after RNA interference (RNAi) using an HDAC2 Adenoviral shRNA Expression Vector. A: IL-8 message levels. All IL-8 mRNA levels are normalized to GAPDH mRNA levels. Error bars represent SE. n = 3. *P < 0.01 for comparisons of CFTR+ with CFTR− cells. **P < 0.0001 for comparisons of CFTR+ with CFTR+ treated with HDAC2 RNAi. B: IL-8 promoter acetylation was assayed using acetyl-H4 chromatin immunoprecipitation (ChIP). All IL-8 promoter H4 acetylation levels are normalized to amount of input DNA. Error bars represent SE. n = 3. *P < 0.04 for comparisons of CFTR+ with CFTR− cells. **P < 0.0004 for comparisons of CFTR+ with CFTR+ treated with HDAC2 RNAi.
Treatment of CFTR-deficient cells with NAC brings HDAC2 expression closer to control levels.
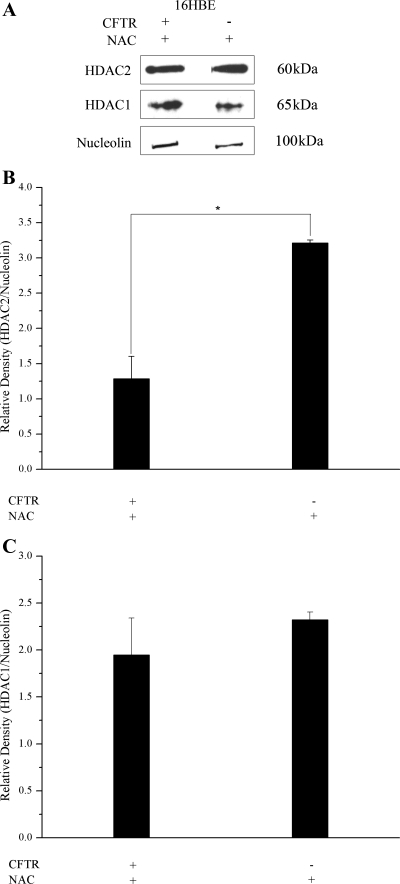
We (6) have shown previously that the elevated levels of IL8 RNA and promoter acetylation in cells deficient in CFTR is likely due to increased susceptibility to exogenous oxidative stress. The antioxidant NAC has been shown to attenuate the oxidant-mediated reduction in HDAC activity in human alveolar epithelial cells exposed to cigarette smoke condensate (36). To determine the effect of reducing exogenous oxidative stress on HDAC2 expression in CFTR− cells, we treated the cells with NAC. Treating 16HBE antisense (CFTR−) cells with NAC resulted in comparable levels of HDAC2 protein (relative to nucleolin) in CFTR+ and CFTR− cells. In some samples, the level of HDAC2 was even higher in the CFTR− cells than in the CFTR+ cells (Fig. 9, A and B). No significant difference was observed in HDAC1 protein levels between the CFTR+ and CFTR− cells (Fig. 9C). HDAC2 levels are reduced in CFTR− cells, but it appears that reducing oxidative stress through NAC treatment results in HDAC2 levels that are above that seen in CFTR+ cells.
Fig. 9.
Treatment with the antioxidant N-acetyl-cysteine (NAC) results in comparable HDAC2 protein levels in CFTR+ and CFTR− cells. The level of HDAC1 and HDAC2 expression in nuclear extract was determined by Western blotting. Nucleolin was used as a loading control. Protein levels were assayed in 16HBE cells. For antioxidant treatment, cells were exposed to 5 mM NAC in serum-free medium for 2 h before isolation of nuclear extract. A: Western blot showing protein levels of HDAC1 and HDAC2. B: densitometry analysis of HDAC2 protein levels relative to nucleolin protein level. n = 3. *P < 0.004 for comparisons of CFTR+ treated with NAC with CFTR− treated with NAC. C: densitometry analysis of HDAC1 protein levels relative to nucleolin protein level. n = 3. No significant difference was found between CFTR+ cells treated with NAC and CFTR− cells treated with NAC.
DISCUSSION
It is clear that lung inflammation leads to tissue destruction and ultimately respiratory failure in CF. The process begins early in life (37, 39), is prolonged (39), and may be exaggerated relative to the bacterial burden present (27). IL-8 levels are elevated in the sputum, BALF, and serum of CF patients (9, 15) and thought to be a response to infection. More recently, it has been suggested that CFTR dysfunction may itself lead to increased IL-8 levels in CF airways (3, 27, 37), an idea supported by the data presented here.
Because IL-8 is regulated primarily at the level of gene transcription (46), elucidating the factors involved in increasing transcription of the IL8 gene may be critical to understanding CF airway inflammation. H4 acetylation at the IL8 promoter is needed for maximal transcription and has been shown to be an important factor in IL8 transcriptional regulation (17, 45, 48, 52). A master regulator of inflammatory gene acetylation and expression, NF-κB activity does not correspond to the elevation of IL8 mRNA and promoter acetylation observed in CFTR-deficient cells. In fact, NF-κB activity was actually lower in the CFTR− cells compared with the CFTR+ cells. Other components of the NF-κB-coactivator complex, CBP, p300, and HDAC1, were not different between CFTR+ and CFTR− cells, but HDAC2 was found to be reduced in amount and activity. Furthermore, suppressing HDAC2 expression with HDAC2 shRNA resulted in increased IL8 expression and promoter acetylation similar to that observed in CFTR− cells. CFTR− cells exposed to NAC experience a reduction of HDAC2 protein to a level closer to CFTR+ cells. These data suggest that there is an intrinsic alteration in the HAT/HDAC balance in cells lacking CFTR function and that oxidative stress is likely contributing to this alteration.
The observations reported here are not specific to CF. HDAC2 has been shown to be reduced in smokers, individuals with COPD, and those with asthma (5, 20, 23, 50). We did not, however, find evidence for a reduction in global HDAC activity as is seen in asthma (20), consistent with other published data measuring total HDAC activity in five explanted lungs from patients with CF (22).
One paradoxical observation from the current work is the apparent inverse relationship between HDAC2 mRNA and protein levels. Many HDACs are regulated by posttranslational modifications, whereas very little is known about the transcriptional regulation of HDACs. Mouse studies suggest that HDAC1 can regulate its own expression as well as the expression of other class I HDACs (34). The class I HDAC inhibitor, valproic acid, reduces HDAC2 protein levels by inducing proteasomal degradation of HDAC2, with no reduction in HDAC2 mRNA (30). Proteasomal degradation is a major mechanism of regulation of HDAC function. Oxidative stress can decrease HDAC2 activity and is associated with the tyrosine nitration status of HDAC2 (21). Nitration of HDAC2 could target it for proteasomal degradation. Although we see a reduction in HDAC2 protein and activity levels in CFTR− cells, we do not see a corresponding reduction in HDAC2 mRNA. Reducing oxidative stress with treatment with NAC rescues the reduction in HDAC2 in CFTR− cells. These data would suggest that the oxidative stress caused by loss of CFTR is resulting in HDAC2 being degraded in the proteasome. Further work would be needed to investigate the nitration status of HDAC2 and the source of oxidative stress in cells without CFTR.
Using shRNA to reduce HDAC2 levels, we show that HDAC2 does play a role in the transcriptional regulation of IL8 in these cells. Reducing HDAC2 protein levels alone leads to a CF-like elevation in IL8 transcription and hyperacetylation. Again, it would be useful to determine genome-wide alterations in expression and histone acetylation after HDAC2 knockdown and compare that to data from CFTR− cells. It is very likely that a reduction in HDAC2 could affect the regulation of many genes in these cell models. The expression of many genes are altered in CF, including many genes regulated by NF-κB. A reduction in HDAC2 could explain the dysregulation of several inflammatory genes and could contribute to the persistent inflammation seen in CF. In all, the emerging picture is that multiple airway diseases are characterized by changes in histone acetylation processes and that reduction of HDAC2 is common to them. If IL8 is representative of other inflammatory genes, clinical intervention strategies that target acetylation directly or indirectly by suppressing oxidative stress may hold therapeutic promise.
GRANTS
This work was supported by National Institutes of Health Grants HL-68883, P30-DK-27651, and T32-GM-08613 and the Cystic Fibrosis Foundation.
REFERENCES
- 1.Adcock IM, Ford P, Barnes PJ, Ito K. Epigenetics and airways disease. Respir Res 7: 21, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutierrez JP, Hull J, Olinsky A, Phelan EM, Robertson CF, Phelan PD. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med 156: 1197–1204, 1997. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DS, Grimwood K, Carzino R, Carlin JB, Olinsky A, Phelan PD. Lower respiratory infection and inflammation in infants with newly diagnosed cystic fibrosis. BMJ 310: 1571–1572, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner BP, Westerheide SD, Baldwin AS Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol 21: 7065–7077, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes PJ, Ito K, Adcock IM. Corticosteroid resistance in chronic obstructive pulmonary disease: inactivation of histone deacetylase. Lancet 363: 731–733, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Bartling TR, Drumm ML. Oxidative stress causes IL8 promoter hyperacetylation in cystic fibrosis airway cell models. Am J Respir Cell Mol Biol 40: 58–65, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger M Lung inflammation early in cystic fibrosis: bugs are indicted, but the defense is guilty. Am J Respir Crit Care Med 165: 857–858, 2002. [DOI] [PubMed] [Google Scholar]
- 8.Boncoeur E, Roque T, Bonvin E, Saint-Criq V, Bonora M, Clement A, Tabary O, Henrion-Caude A, Jacquot J. Cystic fibrosis transmembrane conductance regulator controls lung proteasomal degradation and nuclear factor-kappaB activity in conditions of oxidative stress. Am J Pathol 172: 1184–1194, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, Berger M. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med 152: 2111–2118, 1995. [DOI] [PubMed] [Google Scholar]
- 10.Brown RK, Kelly FJ. Evidence for increased oxidative damage in patients with cystic fibrosis. Pediatr Res 36: 487–493, 1994. [DOI] [PubMed] [Google Scholar]
- 11.Brown RK, McBurney A, Lunec J, Kelly FJ. Oxidative damage to DNA in patients with cystic fibrosis. Free Radic Biol Med 18: 801–806, 1995. [DOI] [PubMed] [Google Scholar]
- 12.Brown RK, Wyatt H, Price JF, Kelly FJ. Pulmonary dysfunction in cystic fibrosis is associated with oxidative stress. Eur Respir J 9: 334–339, 1996. [DOI] [PubMed] [Google Scholar]
- 13.Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, Barnes PJ. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J Exp Med 200: 689–695, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day BJ, van Heeckeren AM, Min E, Velsor LW. Role for cystic fibrosis transmembrane conductance regulator protein in a glutathione response to bronchopulmonary pseudomonas infection. Infect Immun 72: 2045–2051, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean TP, Dai Y, Shute JK, Church MK, Warner JO. Interleukin-8 concentrations are elevated in bronchoalveolar lavage, sputum, and sera of children with cystic fibrosis. Pediatr Res 34: 159–161, 1993. [DOI] [PubMed] [Google Scholar]
- 16.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA 94: 2927–2932, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmour PS, Rahman I, Donaldson K, MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am J Physiol Lung Cell Mol Physiol 284: L533–L540, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Hubeau C, Puchelle E, Gaillard D. Distinct pattern of immune cell population in the lung of human fetuses with cystic fibrosis. J Allergy Clin Immunol 108: 524–529, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Hull J, Vervaart P, Grimwood K, Phelan P. Pulmonary oxidative stress response in young children with cystic fibrosis. Thorax 52: 557–560, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, Adcock IM. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med 166: 392–396, 2002. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Hanazawa T, Tomita K, Barnes PJ, Adcock IM. Oxidative stress reduces histone deacetylase 2 activity and enhances IL-8 gene expression: role of tyrosine nitration. Biochem Biophys Res Commun 315: 240–245, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, Barnes PJ. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med 352: 1967–1976, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Ito K, Lim S, Caramori G, Chung KF, Barnes PJ, Adcock IM. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J 15: 1110–1112, 2001. [PubMed] [Google Scholar]
- 24.Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, Barnes PJ. A molecular mechanism of action of theophylline: induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci USA 99: 8921–8926, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamaluddin M, Choudhary S, Wang S, Casola A, Huda R, Garofalo RP, Ray S, Brasier AR. Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-kappaB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J Virol 79: 15302–15313, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph T, Look D, Ferkol T. NF-κB activation and sustained IL-8 gene expression in primary cultures of cystic fibrosis airway epithelial cells stimulated with Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol 288: L471–L479, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW. Early pulmonary inflammation in infants with cystic fibrosis. Am J Respir Crit Care Med 151: 1075–1082, 1995. [DOI] [PubMed] [Google Scholar]
- 28.Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell 99: 447–450, 1999. [DOI] [PubMed] [Google Scholar]
- 29.Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, Kowalczyk TH, Hyatt SL, Fink TL, Gedeon CR, Oette SM, Payne JM, Muhammad O, Ziady AG, Moen RC, Cooper MJ. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther 15: 1255–1269, 2004. [DOI] [PubMed] [Google Scholar]
- 30.Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Gottlicher M. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J 22: 3411–3420, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kube D, Sontich U, Fletcher D, Davis PB. Proinflammatory cytokine responses to P. aeruginosa infection in human airway epithelial cell lines. Am J Physiol Lung Cell Mol Physiol 280: L493–L502, 2001. [DOI] [PubMed] [Google Scholar]
- 32.Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-kappa B p65 (RelA) and NF-IL-6. J Immunol 153: 153–164, 1994. [PubMed] [Google Scholar]
- 33.Kunsch C, Rosen CA. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol 13: 6137–6146, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21: 2672–2681, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J Biol Chem 276: 14773–14783, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Moodie FM, Marwick JA, Anderson CS, Szulakowski P, Biswas SK, Bauter MR, Kilty I, Rahman I. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. FASEB J 18: 1897–1899, 2004. [DOI] [PubMed] [Google Scholar]
- 37.Muhlebach MS, Stewart PW, Leigh MW, Noah TL. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am J Respir Crit Care Med 160: 186–191, 1999. [DOI] [PubMed] [Google Scholar]
- 38.Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem 273: 10831–10834, 1998. [DOI] [PubMed] [Google Scholar]
- 39.Noah TL, Black HR, Cheng PW, Wood RE, Leigh MW. Nasal and bronchoalveolar lavage fluid cytokines in early cystic fibrosis. J Infect Dis 175: 638–647, 1997. [DOI] [PubMed] [Google Scholar]
- 40.Perez A, Risma KA, Eckman EA, Davis PB. Overexpression of R domain eliminates cAMP-stimulated Cl− secretion in 9/HTEo− cells in culture. Am J Physiol Lung Cell Mol Physiol 271: L85–L92, 1996. [DOI] [PubMed] [Google Scholar]
- 41.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275: 523–527, 1997. [DOI] [PubMed] [Google Scholar]
- 42.Peterson CL HDAC's at work: everyone doing their part. Mol Cell 9: 921–922, 2002. [DOI] [PubMed] [Google Scholar]
- 43.Rahman I Oxidative stress, chromatin remodeling and gene transcription in inflammation and chronic lung diseases. J Biochem Mol Biol 36: 95–109, 2003. [DOI] [PubMed] [Google Scholar]
- 44.Rahman I, Adcock IM. Oxidative stress and redox regulation of lung inflammation in COPD. Eur Respir J 28: 219–242, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Rahman I, Gilmour PS, Jimenez LA, MacNee W. Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol Cell Biochem 234–235: 239–248, 2002. [PubMed] [Google Scholar]
- 46.Roebuck KA Regulation of interleukin-8 gene expression. J Interferon Cytokine Res 19: 429–438, 1999. [DOI] [PubMed] [Google Scholar]
- 47.Roum JH, Buhl R, McElvaney NG, Borok Z, Crystal RG. Systemic deficiency of glutathione in cystic fibrosis. J Appl Physiol 75: 2419–2424, 1993. [DOI] [PubMed] [Google Scholar]
- 48.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol 3: 69–75, 2002. [DOI] [PubMed] [Google Scholar]
- 49.Sheppard KA, Rose DW, Haque ZK, Kurokawa R, McInerney E, Westin S, Thanos D, Rosenfeld MG, Glass CK, Collins T. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol Cell Biol 19: 6367–6378, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szulakowski P, Crowther AJ, Jimenez LA, Donaldson K, Mayer R, Leonard TB, MacNee W, Drost EM. The effect of smoking on the transcriptional regulation of lung inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 174: 41–50, 2006. [DOI] [PubMed] [Google Scholar]
- 51.Tirouvanziam R, de Bentzmann S, Hubeau C, Hinnrasky J, Jacquot J, Peault B, Puchelle E. Inflammation and infection in naive human cystic fibrosis airway grafts. Am J Respir Cell Mol Biol 23: 121–127, 2000. [DOI] [PubMed] [Google Scholar]
- 52.Tomita K, Barnes PJ, Adcock IM. The effect of oxidative stress on histone acetylation and IL-8 release. Biochem Biophys Res Commun 301: 572–577, 2003. [DOI] [PubMed] [Google Scholar]
- 53.Venkatakrishnan A, Stecenko AA, King G, Blackwell TR, Brigham KL, Christman JW, Blackwell TS. Exaggerated activation of nuclear factor-kappaB and altered IkappaB-beta processing in cystic fibrosis bronchial epithelial cells. Am J Respir Cell Mol Biol 23: 396–403, 2000. [DOI] [PubMed] [Google Scholar]
- 54.Weber AJ, Soong G, Bryan R, Saba S, Prince A. Activation of NF-κB in airway epithelial cells is dependent on CFTR trafficking and Cl− channel function. Am J Physiol Lung Cell Mol Physiol 281: L71–L78, 2001. [DOI] [PubMed] [Google Scholar]
- 55.Xiao W Advances in NF-kappaB signaling transduction and transcription. Cell Mol Immunol 1: 425–435, 2004. [PubMed] [Google Scholar]
- 56.Zahm JM, Gaillard D, Dupuit F, Hinnrasky J, Porteous D, Dorin JR, Puchelle E. Early alterations in airway mucociliary clearance and inflammation of the lamina propria in CF mice. Am J Physiol Cell Physiol 272: C853–C859, 1997. [DOI] [PubMed] [Google Scholar]
- 57.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell 1: 661–671, 1998. [DOI] [PubMed] [Google Scholar]