Abstract
Recent studies have identified a conserved WG/GW-containing motif, known as the Argonaute (AGO) hook, which is involved in the recruitment of AGOs to distinct components of the eukaryotic RNA silencing pathways. By using this motif as a model to detect new components in plant RNA silencing pathways, we identified SPT5-like, a plant-specific AGO4-interacting member of the nuclear SPT5 (Suppressor of Ty insertion 5) RNA polymerase (RNAP) elongation factor family that is characterized by the presence of a carboxy-terminal extension with more than 40 WG/GW motifs. Knockout SPT5-like mutants show a decrease in the accumulation of several 24-nt RNAs and hypomethylation at different loci revealing an implication in RNA-directed DNA methylation (RdDM). Here, we propose that SPT5-like emerged in plants as a facultative RNAP elongation factor. Its plant-specific origin and role in RdDM might reflect functional interactions with plant-specific RNA Pols required for RdDM.
Keywords: Arabidopsis, AGO4, SPT5, RdDM, silencing
Introduction
The discovery of small non-coding RNAs with specific regulatory roles has changed our views on the regulation of eukaryotic gene expression. Small RNAs (sRNAs), 20–25 nt in length, are essential effectors of RNA silencing, a process with crucial roles in many cellular processes. Although their mode of action might vary, plant and animal sRNAs usually act as sequence-specific guides to target endogenous messenger RNAs (mRNAs) for cleavage or translational repression, a process known as post-transcriptional gene silencing (PTGS; Jones-Rhoades et al, 2006). They can also guide DNA and histone methylation, allowing heterochromatin assembly at homologous DNA loci, a process referred to as transcriptional gene silencing (TGS) in plants and fission yeast (Zaratiegui et al, 2007).
To achieve their regulatory functions, sRNAs recruit Argonaute (AGO)-containing RNA-induced silencing complexes (RISCs) that carry out effector functions, leading to gene silencing at the transcriptional or post-transcriptional level. MicroRNAs/sRNAs associate with AGOs to form RISCs that direct PTGS (Peters & Meister, 2007), heterochromatic sRNAs interact with AGOs to form RNA-induced initiation of transcriptional silencing complexes (RITSs) in Schizosaccharomyces pombe (Verdel et al, 2004), whereas a hypothetical RITS-like complex probably guides RNA-directed DNA methylation (RdDM) in plants (Qi et al, 2006), leading to TGS. RISC- and RITS-associated AGO components have ‘slicing' activity thought to initiate silencing through cleavage of mRNA/nascent transcripts bearing sRNA-complementary sites (Qi et al, 2006; Peters & Meister, 2007).
Several studies have recently provided new insights into the function and composition of RISC/RITS by identifying proteins that are likely to act with, or downstream from, AGOs in RNA-mediated interference (RNAi)-based silencing pathways. These are human GW182 (Homo sapiens glycine/tryptophane-rich protein.182; HsGW182) for the miRNA pathway (Behm-Ansmant et al, 2006), S. pombe TAS3 (SpTAS3) for the yeast TGS pathway (Partridge et al, 2007) and Arabidopsis thaliana nuclear RNA polymerase V largest subunit (AtNRPE1), the largest subunit of RNA polymerase V (PolV), for the plant RdDM pathway (Li et al, 2006). It has recently been shown that the HsGW182, SpTAS3 and AtNRPE1 proteins indeed show conserved AGO-binding capacity through a new WG/GW-containing peptide of similar amino-acid composition, also known as the ‘AGO hook' (El-Shami et al, 2007; Till et al, 2007). We have shown that the homologous WG/GW-rich regions of AtNRPE1 and HsGW182 are functionally interchangeable, interacting with AGOs in a tryptophan-dependent manner (El-Shami et al, 2007). These observations suggest that the AGO hook is an evolutionarily and functionally conserved peptide motif that recruits AGO proteins to distinct components of the RISC/RITS effector complexes.
By using the AGO hook as a model to functionally characterize new components in the plant RNA silencing pathways, we identified several putative Arabidopsis proteins with a large WG/GW platform having no known AGO-related function (El-Shami et al, 2007). Among these, Suppressor of Ty insertion 5 (SPT5)-like is a plant-specific member of the SPT5 family of transcription elongation factors (Ponting, 2002). It differs from known multicellular SPT5 homologues by the presence of a large carboxy-terminal extension with more than 40 WG/GW repeats, ranking SPT5-like as the largest WG/GW platform in Arabidopsis. We show that SPT5-like is indeed expressed and that its C-terminal extension is able to interact specifically with AGO4. Mutant alleles of SPT5-like show a significant reduction of cytosine methylation and accumulation of several siRNAs, indicating a requirement for SPT5-like in RdDM. On the basis of our results, we propose that SPT5-like has emerged in plants as a facultative RNA polymerase (RNAP) elongation factor that is required for RdDM.
Results And Discussion
Identification of an AGO4-interacting STP5-like factor
Previously, we used an empirical approach of classical sequence comparison to identify proteins containing AGO hook motifs in various organisms, including Arabidopsis (El-Shami et al, 2007). One of these, encoded by gene At5g04290, shows considerable similarity to SPT5 proteins, which are conserved throughout eukaryotes and form an SPT4–SPT5 complex that is an essential nuclear RNAPII elongation factor (Hartzog et al, 1998). There are in fact three genes in the Arabidopsis genome encoding SPT5 family proteins, two of which are highly similar (At4g08350 and At2g34210), whereas the third, encoded by gene At5g04290, has an additional long C-terminal extension, containing 44 WG/GW motifs, encoded by two large exons (Fig 1A and B; supplementary Fig S3A online). Reverse transcription–PCR analysis indicated that all three SPT5-type genes are ubiquitously expressed in Arabidopsis organs (supplementary Fig S1A online and data not shown).
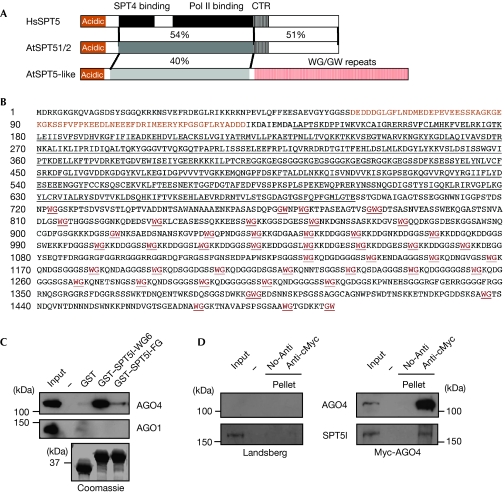
Figure 1.
SPT5-like proteins are plant-specific members of the SPT5 family. (A) Structures of human SPT5, its Arabidopsis homologues and the SPT5-like protein. Conserved acidic, SPT4-binding, RNAPII-binding and carboxy-terminal repeats (CTRs) are indicated as the conservation percentages. (B) Primary sequence of the Arabidopsis SPT5-like sequence. The amino-terminal acidic domain is shown in orange and the conserved binding region is underlined. The GW/WG residues in the SPT5-like amino-terminal repeats are shown in orange and underlined. (C) Preferential binding of AGO4 on the SPT5-like WG repeats. Myc-AGO4 or Flag-AGO1 extracts were applied to equimolar amounts of GST and GST-based fusion protein beads, and the bound proteins were detected by immunoblotting with Myc or M2 antibodies. The GST protein was used as a control. (D) Physical interaction between AGO4 and SPT5-like was detected by coimmunoprecipitation. Proteins from both control and Myc-AGO4 transgenic lines were immunoprecipitated using cMyc antibodies, and analysed to detect the Myc epitope (AGO4) and SPT5-like (SPT5l). AGO, Argonaute; GST, glutathione S-transferase; RNAP, RNA polymerase; SPT5, Suppressor of Ty insertion 5.
Fig 1A shows the domain organization of human and Arabidopsis SPT5s and the new SPT5-like protein. All have a conserved amino-terminal acidic domain, and central SPT4 and RNAPII-binding domains (Guo et al, 2008; Yamaguchi et al, 1999). There is no significant similarity in the C-terminal region, which is composed exclusively of WG/GW motifs in the SPT5-like protein. In contrast to the considerable divergence of AGO hooks in NRPE1 (El-Shami et al, 2007), WG/GW motifs in SPT5-like are highly similar, containing two slightly different conserved motifs (supplementary Fig S1B online). We identified putative homologues of SPT5 and SPT5-like in rice by sequence comparison, correcting the predicted structure of the latter after identification of additional 3′ exons (supplementary Fig S2 online). Phylogenetic analysis of representative SPT5 sequences from plants and other eukaryotes groups the classical SPT5 proteins from animals and plants, with distinct branches that correspond to the yeast- and the plant-specific SPT5-like proteins (supplementary Fig S1C online).
To investigate the importance of the SPT5-like WG/GW-rich domain in AGO binding, we generated fusion proteins containing either six wild-type (glutathione S-transferase–SPT5l–WG6; GST–SPT5l–WG6) or mutant (GST–SPT5l–FG6) versions of the SPT5-like repeats fused with GST and monitored the ability of these constructs to interact either with AGO1 or with AGO4 compared with the GST control protein. Pull-down assays indicated that GST–SPT5l–WG6, but not GST–SPT5l–FG6, interacts specifically with AGO4 with high affinity (Fig 1C), showing that SPT5-like is indeed an AGO hook-containing protein. Interestingly, under the same experimental conditions, AGO1 failed to bind to the GST–SPT5l–WG6 fusion protein, indicating that the SPT5-like WG-containing platform shows distinct binding specificity for AGO proteins. As a single AGO hook motif is sufficient for binding AGO (El-Shami et al, 2007) and as SPT5-like contains the largest WG/GW platform in Arabidopsis, we infer that SPT5-like should have considerable AGO4-binding capacity.
To assess further the interaction between SPT5-like and AGO4 in vivo, whole-cell extracts generated from the previously reported Myc-AGO4 line (Li et al, 2006) were immunoprecipitated with Myc antibodies and analysed using a specific SPT5-like antibody (Ab72; see Fig 2A; supplementary Fig S3 online). Fig 1D shows that SPT5-like coimmunoprecipitates with Myc-AGO4, therefore supporting the interaction between these proteins. Taken together, our results indicate that SPT5-like is an authentic AGO hook-containing protein that interacts with AGO4 in Arabidopsis.
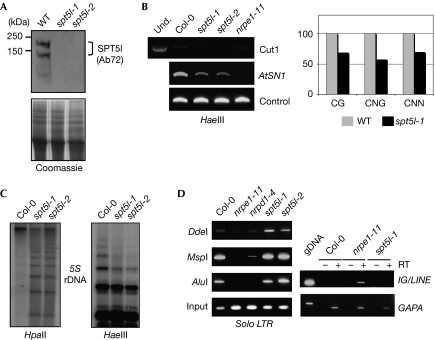
Figure 2.
DNA methylation defects in spt5-like knockout mutants. (A) Western blot and Coomassie blue staining on wild-type (WT) and mutant flower protein extracts (∼20 μg) using an SPT5-like specific antibody. (B) DNA methylation at the AtSN1 locus in spt5-like and control genotypes Col-0 and nrpe1-11 (polV). Left panel, HaeIII-digested DNA was used as a template for PCR reactions using AtSN1 primers (412 and 413) and control primers. Cut 1 primers (400 and 402) should not allow DNA amplification if the digestion is complete. Control primers (410 and 411) allow equilibration of DNA quantities used as templates. Right panel, DNA methylation was analysed by bisulphite sequencing. Histograms represent the percentage of CG, CNG and CNN methylation in the indicated genotypes. (C) DNA methylation at the 5S ribosomal RNA locus in spt5-like and Col-0. Genomic DNA was digested with HpaII or HaeIII and blots were hybridized with a 5S ribosomal RNA repeat probe. (D) DNA methylation at the solo LTR locus and IG/LINE expression in spt5-like and control genotypes Col-0, nrpd1-4 (polIV) nrpe1-11. Left panel, genomic DNA was digested by various methylation-sensitive enzymes before being used for PCR amplification using solo LTR-specific primers (769 and 770). Input indicates amplification from equal quantities of undigested DNA. Right panel, RT-PCR performed on Col-0, nrpe1-11 and spt5-like-1 messenger RNA. gDNA correspond to amplification from genomic Col-0 DNA. +/− RT indicates whether or not transcriptase treatment was carried out. GAPAs (glyceraldehyde 3-dehydrogenase A) have been used to check equilibration. All primers are described in supplementary Table 1 online. Ab, antibody; AtSN1, Arabidopsis thaliana short interspersed element 1; IG/LINE, intergenic/long interspersed element; LTR, long terminal repeat; RT–PCR, reverse transcription–PCR; SPT5, Suppressor of Ty insertion 5; Und., undigested DNA.
spt5-like mutants show specific DNA methylation defects
To investigate the functional role of SPT5-like, two independent homozygous mutant lines, spt5l-1 and spt5l-2, were characterized. None of the mutant alleles showed a readily discernible phenotype. The T-DNA insertion position for each mutant is indicated in supplementary Fig S3A online. No full-length SPT5-LIKE transcript was detected by reverse transcription–PCR in either of the mutant lines (supplementary Fig S3B online). Furthermore, no SPT5-like protein was detected using either Ab71 or Ab72 SPT5-like antibodies, indicating that spt5l-1 and spt5l-2 are probably null mutants (Fig 2A; supplementary Fig S3A,C online). Both SPT5-like antibodies revealed a prominent 150–180 kDa protein doublet with the apparent mass of the largest protein being slightly greater than the predicted molecular size of SPT5-like (∼150 kDa; Fig 2A; supplementary Fig S3C online). Whatever the explanation for the apparent increase in the molecular weight of the detected protein, the fact that these two bands are absent in the knockout insertion lines shows their specificity and confirms the accumulation of SPT5-like protein in wild-type extracts.
As AGO4 is known to be involved in RdDM and SPT5-like is a nuclear protein (data not shown), we tested the impact of spt5-like mutation on DNA methylation at repeated endogenous loci. The retrotransposon Arabidopsis thaliana short interspersed element 1 (AtSN1) locus is protected against cleavage by the methylation-sensitive enzyme HaeIII in wild-type plants. A similar reduction in protection, indicating decreased methylation, is seen in both spt5-like mutants compared with the wild type (Fig 2B, left panel). The reduction is, however, not as strong as that seen in the nrpe1-11 mutant (Fig 2B, left panel). Decreased methylation at the AtSN1 locus was confirmed by bisulphite sequencing, which indicated a reduction by about 43% of CNG (C, cytosine; N, adenine, thymine or cytosine; G, guanine) and 32% of both CG and CNN methylation in the spt5l-1 mutant compared with the wild type (Fig 2B, right panel; supplementary Fig S4 online). DNA methylation defects in both spt5-like mutants were also observed for the 5S rDNA cluster for which digestions with HpaII and HaeIII allow the monitoring of both CG and CNN methylation, respectively (Fig 2C). Interestingly, DNA methylation assays of the solo LTR (long terminal repeat) locus, a well-known target of the RdDM pathway (Huettel et al, 2006), indicate that the two spt5-like mutants, in contrast to the nrpd1-4 and nrpe1-11 mutants, do not lose DNA methylation (Fig 2D, left panel). As expected, the adjacent IG/LINE (intergenic/long interspersed element) transcript is activated in the nrpe1-11 mutant, owing to hypomethylation of the solo LTR locus, but not in the spt5l-1 mutant (Fig 2D, right panel). Taken together, our results suggest a target-specific requirement for the AGO-interacting protein SPT5-like in RdDM.
Altered levels of 24-nt sRNA in spt5-like mutants
Cytosine methylation is guided to RdDM targets by 24-nt sRNAs produced by components of the RNAi machinery (Xie et al, 2004), and the plant-specific PolIV and PolV (Herr et al, 2005; Kanno et al, 2005; Onodera et al, 2005; Pontier et al, 2005; Zhang et al, 2007). According to these observations and to extend our analysis, we assessed the impact of spt5-like null mutations on the accumulation of a set of 24-nt sRNAs that are produced either from PolIV-dependent (TR2558 and siRNA02) or PolIV/PolV-dependent (AtSN1, SimpleHAT2, 45S rDNA and 5S rDNA) loci (Pontier et al, 2005; Mosher et al, 2008). As expected, all 24-nt sRNAs were absent or greatly reduced in samples from the nrpd1-4 mutant lacking PolIV, whereas only a subset of the sRNAs, corresponding to PolIV/PolV-dependent loci, was also affected in the nrpe1-11 mutant (Fig 3). Our analysis showed that although the siRNA02 and TR2558 24-nt sRNAs remain unaffected in both spt5-like mutants, the level of 24-nt sRNAs corresponding to AtSN1 was considerably reduced in these mutants (Fig 3), an observation that is consistent with the methylation defect observed previously at this locus. Similarly, the accumulation of SimpleHAT2, 45S and 5S 24-nt sRNAs was also decreased in the spt5-like mutants to a level similar to that detected in nrpe1-11 (Fig 3). Taken together, these results indicate that the methylation defects noted in spt5-like mutants are linked to a defect in the production/accumulation of 24-nt sRNA and that the PolIV/PolV-dependent loci are preferentially sensitive to the spt5-like mutations.
Figure 3.
Short interfering RNA accumulation defect in spt5-like mutants. Small RNA blot assays for U6 and various endogenous siRNAs in spt5-like, nrpd1-4 and nrpe1-11 mutants. Blots were stripped and reprobed multiple times with probes indicated on the right. siRNA accumulation dependency on polIV or polV is indicated on the left. U6 was used as an equilibration control. AtSN1, Arabidopsis thaliana short interspersed element 1; nrpd1, nuclear RNA polymerase IV largest subunit; nrpe1, nuclear RNA polymerase V largest subunit; siRNA, short interfering RNA; SPT5, Suppressor of Ty insertion 5.
Previous work has shown that the plant-specific PolV was probably derived by the modification of a duplicated PolIV gene (Luo & Hall, 2007). The WG motifs of NRPDE1 are encoded by two large exons inserted between the Pol and the DCL (defective chloroplast and leaves) domains of the ancestral gene (Pontier et al, 2005). It is intriguing to note that the AGO platform in Arabidopsis SPT5-like is also carried by two large exons at the 3′ end of the gene. This is therefore the second small gene family in which two forms exist: one resembling other non-plant proteins and the other that contains an AGO-binding platform. All members of the SPT5 family have been implicated in the control of RNAP processivity through direct binding to the core RNAP enzyme (Sims et al, 2004). As the conserved domain structure of SPT5-like suggests an interaction with RNAP, and as it is involved in RdDM, we propose that SPT5-like is a variant elongation factor needed for the activity of PolV (Fig 4). This idea is supported by the 24-nt sRNA analysis, which revealed that most of the loci that specifically require PolV for the accumulation of sRNA are affected in spt5-like mutants.
Figure 4.
Hypothetical model for the action of SPT5-like in RNA-directed DNA methylation. Two possible mechanisms for the mode of action of SPT5-like in RdDM are presented. Both involve the recruitment of AGO4 to PolV and SPT5-like (SPT5l) but differ in the subsequent role of the latter. (A) Recycling of AGO4 through SPT5-like. (B) Recruitment of SPT5l through the AGO hook domain. AGO, Argonaute; DCL3, dicer-like 3; DRM2, domain rearranged methylase 2; dsRNA, double-stranded RNA; meC, methylated cytosine; RdDM, RNA-directed DNA methylation; RDR2, RNA dependent RNA polymerase 2; siRNA, short interfering RNA; SPT5, Suppressor of Ty insertion 5; Spt5l, SPT5 like.
Our data also indicate that the AGO hook platform of SPT5-like shows specificity towards AGO proteins having a strong preference for AGO4 in our binding assays. As NRPE1 and SPT5-like contain the largest WG/GW platforms in Arabidopsis, we propose that a massive recruitment of AGO4 is likely to be essential in the RdDM pathway. In this context, it is possible that SPT5-like has evolved as a facultative elongation factor with a dual role in RdDM, modulating the processivity of PolV and ensuring the availability of AGO4 during the course of PolV transcription (Fig 4A). This claim is in agreement with the prevailing model, which suggests that base pairing between AGO4-bound siRNA and PolV transcripts is required for RdDM (Wierzbicki et al, 2008; Lahmy et al, 2009). This suggests that the complexity of the PolV-transcribed sequences will be significantly greater than that of the siRNA population that can be loaded simultaneously on the PolV CTD (carboxy-terminal domain; El-Shami et al, 2007; Mosher et al, 2008), requiring a dynamic supply of AGO4. Alternatively, SPT5-like could be acting merely as an SPT5-type transcription elongation factor, of which the post-initiation recruitment on the PolV/AGO4 complex takes place through its WG/GW domain (Fig 4B). Further experiments will be necessary to decipher the exact role of SPT5-like and its Pol partner in RdDM.
Methods
Plant growth and genetic analysis. For plant growth, seeds were stratified at 4 °C for 2 days before growth at 23 °C with 16-h light on the soil. The spt5l-1 mutant corresponds to line SALK_001254C (Alonso et al, 2003). The spt5l-2 mutant corresponds to line SAIL SAIL_362_G07. nrpd1-4 and nrpe1-11 were described by Herr et al (2005) and El-Shami et al (2007).
DNA isolation and analysis. Genomic DNA was extracted from leaves using a Wizard Genomic kit (Promega, Charbonnières-les-Bains, France) before being used for PCR or Southern blot hybridizations. PCR was performed using 150 ng of digested or undigested genomic DNA. Primers are described in supplementary Table 1 online. All PCRs were performed with an annealing temperature of 55 °C and 30 amplifying cycles, except in some cases described in supplementary Table 1 online. For Southern blots, 5 μg of DNA was used and hybridized with a 5S probe (Pontier et al, 2005).
RNA isolation and analysis. Total RNA was isolated using the Trizol reagent (Invitrogen, Cergy-Pontoise, France). After DNAse treatment, cDNA was synthetized with an Affinity Multitemperature cDNA synthesis kit (Stratagene, Amsterdam, The Netherlands) using an oligodT primer with 1 μg of RNA according to the manufacturer's instructions. For sRNA analysis, total RNA extraction and northern blots were performed as described previously (Pontier et al, 2005), except that NX-Hybond membranes (Amersham Biosciences, Orsay, France) and EDC (Sigma, Saint-Quentin Fallavier, France)-mediated crosslinking were used. The probes used are described in supplementary Table 1 online.
Immunoblot. Total flower protein extracts were obtained following the method described by Pontier et al (2005). Protein samples from wild-type and transgenic lines were quantified and subjected to immunoblot analysis using a 1/1000 dilution of Spt5-like antibodies raised against peptides described in supplementary Table 1 online (Eurogentec, Angers, France).
Sodium bisulphite sequencing. Sodium bisulphite sequencing was performed as described previously (Fraga et al, 2008). Primer sequences are described in the supplementary information online. In total, 17 and 19 clones were sequenced for Col-0 and spt5l-1, respectively.
GST–CTD-binding experiments. The multimers were constructed following the strategy described by El-Shami et al (2007). Primers corresponding to each monomer (877/878 for SPT5l-WG1 and 919/920 for SPT5l-FG1) were annealed and subcloned into a BglII–BamHI-digested pBluescript vector. After three rounds of cloning, the multimers were introduced into a BamHI–HindIII-digested pET41a(+) vector and expressed in BL21. All the GST fusion proteins were purified on glutathione Sepharose 4B beads (Amersham Biosciences) and used in pull-down assays as described previously (El-Shami et al, 2007).
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Note added in proof. In a recent study, Huang et al (2009) identified peptides of SPT5-like in a PoIV-purified fraction. This finding is in agreement with the model of action reported here, that SPT5-like acts in RdDM as a PoIV-associated SPT5-type elongation factor in plants.
Supplementary Material
Supplementary Information
Acknowledgments
We thank J.-R. Pages for his help with artwork and Felipe K. Teixeira for his help with bisulphite sequencing. Cooke and Lagrange lab researches are supported by Centre National de la Recherche Scientifique and Agence Nationale de la Recherche ANR-08-BLAN-0206-01.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alonso JM et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T (2007) Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF et al. (2008) Epigenetic inactivation of the Groucho homologue gene TLE1 in hematologic malignancies. Cancer Res 68: 4116–4122 [DOI] [PubMed] [Google Scholar]
- Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L (2008) Core structure of the yeast spt4–spt5 complex: a conserved module for regulation of transcription elongation. Structure 16: 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F (1998) Evidence that SPT4, SPT5 and SPT6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe D (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC (2009) An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 16: 91–93 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and PolIV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T (2009) PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci USA 106: 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE (2006) An Argonaute4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Luo J, Hall B (2007) A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol 64: 101–112 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe D (2008) PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA 105: 3145–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ (2007) Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell 26: 593–602 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 8: 611–623 [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi M-A, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP (2002) Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res 30: 3643–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Sims RJ, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and the long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG (2007) A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase PolIVb/PolV mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H (1999) Structure and function of the human transcription elongation factors DSIF. J Biol Chem 274: 8055–8092 [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA (2007) Noncoding RNAs and gene silencing. Cell 128: 763–776 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE (2007) Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA 104: 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information