Abstract
Coronary heart disease (CHD) is a major cause of death in Western countries. We used genome-wide association scanning to identify a 58-kilobase interval on chromosome 9p21 that was consistently associated with CHD in six independent samples (more than 23,000 participants) from four Caucasian populations. This interval, which is located near the CDKN2A and CDKN2B genes, contains no annotated genes and is not associated with established CHD risk factors such as plasma lipoproteins, hypertension, or diabetes. Homozygotes for the risk allele make up 20 to 25% of Caucasians and have a ~30 to 40% increased risk of CHD.
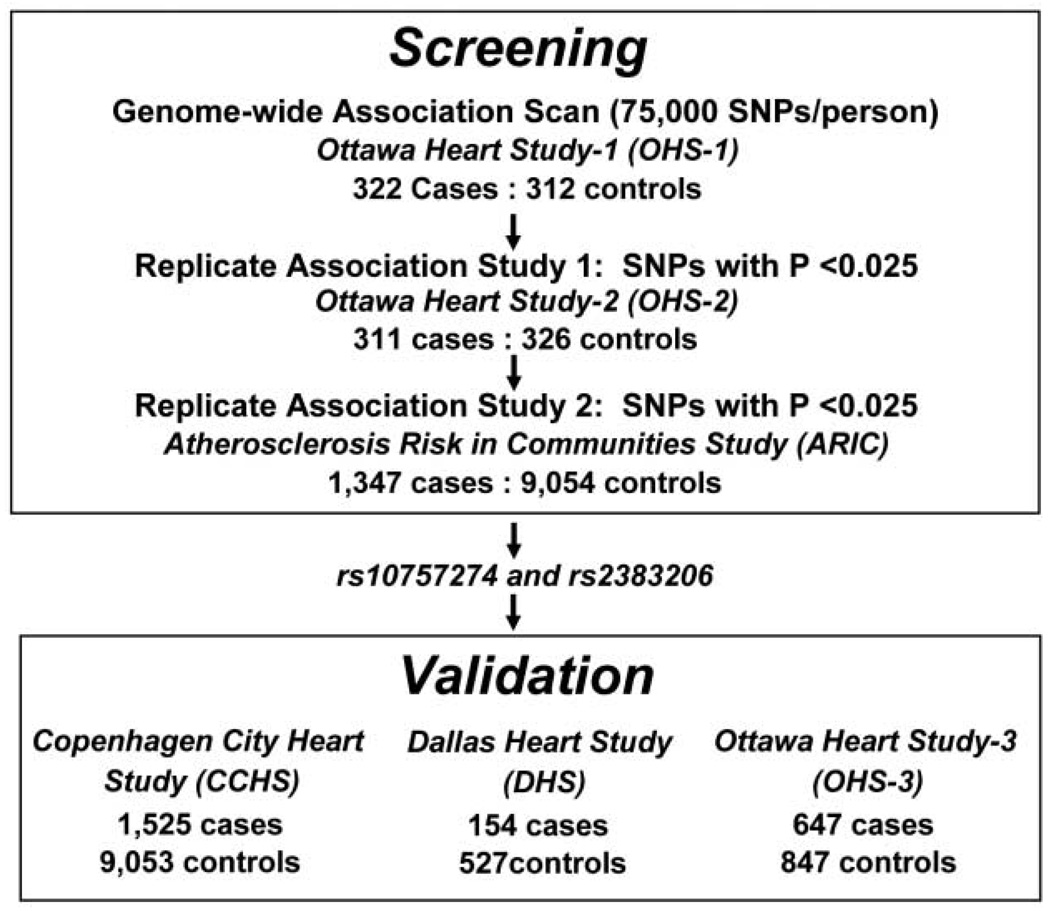
Coronary heart disease (CHD) is the single greatest cause of death worldwide (1, 2). Although CHD is highly heritable, the DNA sequence variations that confer cardiovascular risk remain largely unknown. To identify sequence variants associated with CHD, we undertook a genome-wide association study using 100,000 single-nucleotide polymorphisms (SNPs). To minimize false positive associations without unduly sacrificing statistical power, we designed the study to comprise three sequential case-control comparisons performed at a nominal significance threshold of P < 0.025 (Fig. 1). For the initial genome-wide scan, cases and controls were Caucasian men and women from Ottawa, Canada who participated in the Ottawa Heart Study (OHS). Cases had severe, premature CHD with a documented onset before the age of 60 years and culminating in coronary artery revascularization (table S1). To limit confounding by factors that strongly predispose to premature CHD, we excluded individuals with diabetes or plasma cholesterol levels consistent with monogenic hypercholesterolemia (>280 mg/dL). Controls were healthy Caucasian men (>65 years) and women (>70 years) from Ottawa who had no symptoms or history of CHD (table S1).
Fig. 1.
Study design for identification and validation of sequence variants associated with CHD. Assuming independence, the probability of any single SNP achieving a nominal significance level of 0.025 in all three studies with the associations being in the same direction was 3.9 × 10−6 (0.0253 × 0.52); thus, none of the 100,000 SNPs would be expected by chance to replicate consistently in all three comparisons.
Custom oligonucleotide arrays (3) were used to assay 100,000 SNPs arranged at ~30-kb intervals throughout the genome in 322 cases and 312 controls (data set designated as OHS-1). Of these, 9636 SNPs deviated strongly from Hardy-Weinberg equilibrium (P < 0.001) or did not meet quality-control criteria (3), and 17,500 were subpolymorphic (minor allele frequency < 1%) in the sample. The remaining 72,864 SNPs were entered into the analysis, and 2586 were associated with CHD at a nominal significance threshold of 0.025 (table S2). These 2586 SNPs were genotyped in an independent sample of 311 cases and 326 controls from Ottawa (OHS-2) using the same criteria as OHS-1 (table S1). Of these, 50 were associated with CHD at a nominal significance threshold of 0.025, with the same direction of effect (table S2).
To limit attrition of true positive associations due to inadequate statistical power, we performed the third case-control comparison in a much larger prospective study of CHD risk, the Atherosclerosis Risk in Communities (ARIC) study, which enrolled and followed 11,478 Caucasians (4). Only 2 of the 50 SNPs identified in the Ottawa cohorts were significantly associated with incident CHD in the ARIC population (table S2). These two SNPs, rs10757274 and rs2383206, were located within 20 kb of each other on chromosome 9p21 and were in strong linkage disequilibrium (r2 = 0.89).
To validate the association between rs10757274 and rs2383206 and CHD, we assayed both SNPs in three additional independent cohorts: the Copenhagen City Heart Study (CCHS), a prospective study of ischemic heart disease in 10,578 Danish men and women (5); the Dallas Heart Study (DHS), a population-based probability sample of Dallas County residents (6); and a third sample of 647 cases and 847 controls from the Ottawa Heart Study population (OHS-3). In the CCHS, cases were participants who experienced an ischemic cardiovascular event during the 20-year follow-up period, whereas controls were those who did not develop CHD over the same time interval. In the DHS, cases and controls were defined by using electron-beam computer tomography to measure coronary artery calcium, an index of coronary atherosclerosis (7). In OHS-3, cases were participants that had documented CHD before the age of 55 (men) or 65 (women) years, whereas controls were men aged > 65 years and women aged > 70 years who did not have symptoms of CHD (table S1). In all three validation studies, both SNPs were significantly associated with CHD (Table 1).
Table 1.
Association between SNPs rs10757274 and rs2383206 and CHD. Values are numbers of individuals in each genotype group. P values were calculated by χ2 tests on allele counts. HW, Hardy-Weinberg equilibrium.
| rs10757274 | rs2383206 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||||||||||||
| AA | AG | GG | AA | AG | GG | χ2P | HW P |
AA | AG | GG | AA | AG | GG | χ2P | HW P |
|
| OHS-1 | 85 | 149 | 78 | 49 | 148 | 125 | 3.7 × 10−6 | 0.08 | 77 | 147 | 88 | 45 | 140 | 137 | 6.7 × 10−6 | 0.19 |
| OHS-2 | 85 | 161 | 80 | 56 | 140 | 108 | 0.0009 | 0.4 | 80 | 160 | 86 | 50 | 141 | 113 | 0.0008 | 0.34 |
| ARIC | 2063 | 3822 | 1858 | 230 | 525 | 282 | 0.004 | 0.11 | 2140 | 4161 | 2231 | 230 | 600 | 324 | 0.0007 | 0.21 |
| CCHS | 2752 | 4543 | 1758 | 393 | 792 | 340 | 0.0004 | 0.56 | 2489 | 4583 | 1981 | 372 | 782 | 371 | 0.016 | 0.58 |
| DHS | 147 | 258 | 122 | 27 | 85 | 42 | 0.025 | 0.99 | 131 | 258 | 138 | 24 | 84 | 46 | 0.045 | 0.95 |
| OHS-3 | 228 | 418 | 201 | 121 | 333 | 193 | 0.0003 | 0.96 | 197 | 416 | 229 | 115 | 327 | 209 | 0.011 | 0.98 |
The magnitude of CHD risk associated with the risk allele was determined by Cox proportional-hazards modeling in the ARIC and CCHS cohorts. The hazard ratios associated with the risk alleles were comparable in the two populations and indicated a graded increase in risk from noncarriers to heterozygotes to homozygotes (Table 2). The two SNPs (rs10757274 and rs2383206) define an allele that was associated with an ~15 to 20% increase in risk in the 50% of individuals who were heterozygous for the allele and an ~30 to 40% increase in CHD in the 25% of Caucasians who were homozygous for the allele. The population attributable risk associated with the risk allele was 12.5 to 15% in the ARIC population and 10 to 13% in the CCHS cohort.
Table 2.
Risk of CHD as a function of rs10757274 and rs2383206 in the ARIC study and the CCHS. ARIC expected event values are based on the log-rank test. ARIC incidence rates are measured in number of events per 10,000 person years of followup. Ranges in parentheses indicate 95% confidence interval.
| ARIC study | CCHS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of events | Number of events | |||||||||
| n (%) | Observed | Expected | Incidence | Hazard ratio | n (%) | Observed | Expected | Incidence | Hazard ratio | |
| rs10757274 | ||||||||||
| AA | 2293 (26) | 255* | 295 | 79 (70–89) | 1 | 3145 (30) | 393‡ | 473 | 61 (55–68) | 1 |
| AG | 4347 (50) | 564 | 553 | 93 (86–101) | 1.18 (1.02–1.37) | 5335 (50) | 792 | 755 | 73 (68–79) | 1.26 (1.12–1.42) |
| GG | 2140 (24) | 298 | 269 | 101 (90–114) | 1.29 (1.09–1.52) | 2098 (20) | 340 | 296 | 80 (72–89) | 1.38 (1.19–1.60) |
| rs2383206 | ||||||||||
| AA | 2370 (25) | 259† | 310 | 78 (69–88) | 1 | 2,861 (27) | 372§ | 425 | 64 (58–71) | 1 |
| AG | 4761 (49) | 643 | 610 | 97 (90–105) | 1.26 (1.09–1.46) | 5,365 (51) | 782 | 772 | 72 (67–77) | 1.16 (1.02–1.31) |
| GG | 2555 (26) | 345 | 327 | 97 (88–108) | 1.26 (1.07–1.48) | 2,352 (22) | 371 | 327 | 78 (71–87) | 1.29 (1.12–1.50) |
P < 0.0111.
P < 0.0041.
P < 0.00001.
P < 0.005.
The mechanistic basis for the association between the risk allele defined by rs10757274 and rs2383206 and CHD is not known. The allele may increase the development of atherosclerotic plaques, promote thrombogenesis, or increase the tendency of atherosclerotic plaques to rupture. The finding that the risk allele was associated with coronary artery calcification in the DHS and with severe premature atherosclerosis in OHS-1 suggests that it promotes CHD by increasing the atherosclerotic burden. The risk allele was not associated with any of the major risk factors for atherosclerosis in ARIC or CCHS (table S3 and table S4), and the association remained significant in models that considered multiple possible confounding covariates (including age, gender, plasma lipid levels, blood pressure, diabetes, and plasma C-reactive protein concentrations; table S3). These analyses suggest that the effect of the chromosome 9 risk allele on CHD was not mediated by any of the established risk factors for cardiovascular disease.
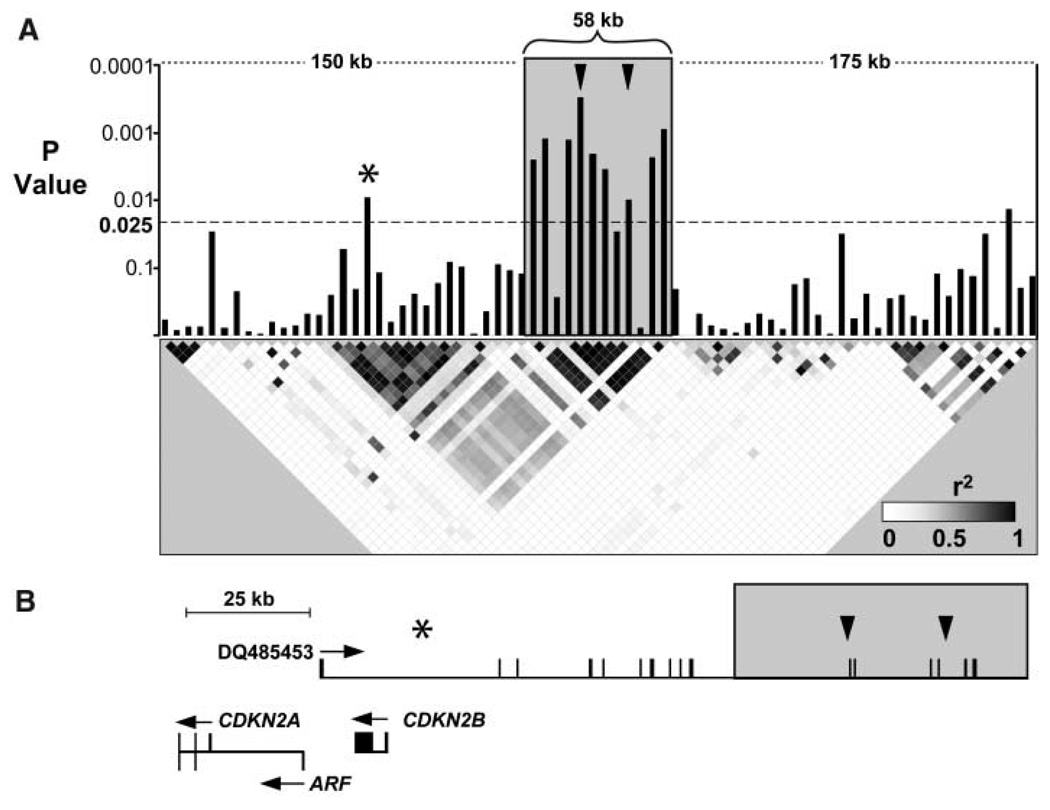
To fine-map the locus associated with CHD, we assayed SNPs spaced at ~5-kb intervals across the region extending 175 kb upstream and downstream of rs10757274 and rs2383206 in 500 cases and 500 controls from OHS-2 and OHS-3. Eight additional SNPs at the locus spanning a 58-kb region (extending from 22,062,301 to 22,120,389) were significantly associated with CHD (Fig. 2). All eight were in strong linkage disequilibrium with each other and with rs10757274 and rs2383206. The region demarcated by these SNPs was flanked on both sides by ~50-kb regions in which none of the 30 SNPs tested were associated with CHD. Two of 65 SNPs in the 350-kb region surrounding the 58-kb risk locus were associated with CHD at the nominal significance threshold, but neither was in strong linkage disequilibrium with rs10757274 and rs2383206. These data indicate that the risk allele comprises a single haplotype that spans ~58 kb.
Fig. 2.
Fine mapping of the genomic interval on chromosome 9 associated with CHD. (A) SNPs spaced ~5 kb apart in the interval extending 175 kb upstream and downstream of rs10757274 and rs2383206 were assayed in 500 cases and 500 controls from the OHS population with GeneChip Human Mapping 500K Array Sets (Affymetrix, Santa Clara, CA). Bars represent P values (determined with χ2 tests) for differences in allele frequency between cases and controls. Arrowheads indicate rs10757274 and rs2383206. The asterisk represents rs518394. The risk interval is indicated with a gray box. The linkage disequilibrium map indicates pairwise r2 values. Blocks are shaded on a continuous scale, where white represents an r2 of 0 and black represents an r2 of 1. (B) Physical map of the region showing the location of the risk interval (gray box) relative to the noncoding RNA DQ485453 and adjacent genes CDKN2A, ARF, and CDKN2B. Arrowheads indicate rs10757274 and rs2383206, and the asterisk represents rs518394 [see (A)].
Inspection of the UCSC Genome Browser (http://genome.ucsc.edu) and BLAST searches against the National Center for Biotechnology Information (NCBI) (www.ncbi.nih.gov/blast) nr (nonredundant) nucleotide sequence database revealed no annotated genes or microRNAs within the 58-kb interval. A number of spliced expressed sequence tags (ESTs) map within the interval, but none contained an open reading frame that extends more than a few amino acids. Resequencing of the 58-kb interval in two homozygotes for the risk allele and one homozygote for the reference allele revealed 66 polymorphisms (SNPs plus small insertions or deletions), of which 35 were specific to the risk allele (table S5). Only one of these variants, a copy number variation in a run of nine consecutive CAT repeats (extending from nucleotide 22110787 to 22110814, NCBI build 36.1) mapped to a spliced transcript (BI765545) that appears to be part of a large noncoding RNA of unknown function (8). Polymerase chain reaction (PCR) amplification of cDNAs confirmed expression of the transcript in placenta and transformed lymphocytes (fig. S1). It is possible that variation in the expression or function of this transcript may be associated with risk of CHD.
Alternatively the risk allele may alter a regulatory element that affects the expression of a gene (or genes) located outside of the 58-kb interval. Cross-species sequence alignments revealed several conserved segments within the 58-kb interval that may contain such regulatory elements (fig. S2). It is also possible that the risk allele extends beyond the 58-kb interval defined in this study and that the functional sequence variants that confer risk of CHD are located outside of the interval. Resequencing the coding regions of the two genes most proximal to the risk locus, CDKN2A and CDKN2B, revealed only a single variant (Ala158→Ser158 in CDKN2A) that was present in 6 of the 96 individuals examined and is thus unlikely to explain the CHD risk associated with the locus. The localization of the risk locus to a region devoid of known genes implicates a previously unrecognized gene or regulatory element that can substantially affect CHD independently of established risk factors. Further studies will be required to elucidate the mechanism by which the locus modulates CHD risk.
Comparison of the Yoruba and Centre d’ Etude du Polymorphisme Humain (CEPH) data from the International HapMap Project (www.hapmap.org) revealed notable ethnic differences in allele frequencies in the risk interval (table S6). Of the 10 alleles that were significantly associated with CHD in whites, 3 were virtually absent from the Yoruba population, and 6 others were much less common. Both rs10757274 and rs2383206 were present at appreciable frequencies among African-Americans in ARIC and DHS, but neither SNP was associated with CHD in either population (table S7). The apparent ethnic differences in association between these SNPs and CHD in ARIC may reflect differences in statistical power in ARIC but cannot explain the ethnic differences observed in DHS, where African-Americans are the largest group. Accordingly, it seems more likely that the functional sequence variants associated with the risk allele in whites are less common in African-Americans. This notion is consistent with our finding that the frequencies of several alleles associated with CHD risk factors differ widely among ethnic groups (9–11). Comprehensive analysis of the locus in African-Africans may allow further refinement of the risk interval.
The results of this study illustrate both the perils and the promise of whole-genome association. The initial scan and the first replicate screen both generated substantially more SNPs that achieved the prespecified significance threshold than would be predicted by chance alone, as indicated by permutation testing (table S2). Yet only two of these SNPs (comprising one allele) survived further replication, despite the use of a large sample (i.e., ARIC) with high statistical power. This finding highlights the necessity for adequate replication to protect against artifacts that may occur because of population stratification, multiple testing, or other factors to which whole-genome association studies are particularly susceptible. The consistent replication of the chromosome 9 risk allele in six independent study samples indicates that the approach can be productively applied to conditions as complex as CHD, which is known to be influenced by a variety of environmental and genetic factors (12). Furthermore, analysis of 50 randomly selected regions of 500 kb each indicated that the 72,864 informative SNPs used in the initial scan provided 30 to 40% of the power that would be obtained by assaying all phase II Hapmap SNPs. Therefore, scans with denser SNP panels and larger sample sizes may reveal further loci associated with CHD risk.
Footnotes
Information about obtaining reprints of this article or about obtaining permission to reproduce this article in whole or in part can be found at: http://www.sciencemag.org/about/permissions.dtl
Supporting Online Material
www.sciencemag.org/cgi/content/full/1142447/DC1
Materials and Methods
Figs. S1 and S2
Tables S1 to S7
References and Notes
- 1.Murray CJ, Lopez AD. Lancet. 1997;349:1436. doi: 10.1016/S0140-6736(96)07495-8. [DOI] [PubMed] [Google Scholar]
- 2.Mathers CD, Loncar D. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saccone SF, et al. Hum. Mol. Genet. 2007;16:36. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The ARIC Study Investigators. Am. J. Epidemiol. 1989;129:687. [PubMed] [Google Scholar]
- 5.Schnohr P, Jensen G, Scharling H, Appleyard M. Eur. Heart J. 2001;3(suppl H):H1. [Google Scholar]
- 6.Victor RG, et al. Am. J. Cardiol. 2004;93:1473. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 7.Agatston AS, et al. J. Am. Coll. Cardiol. 1990;15:827. doi: 10.1016/0735-1097(90)90282-t. [DOI] [PubMed] [Google Scholar]
- 8.Pasmant E, et al. Cancer Res. 2007;67:1. [Google Scholar]
- 9.Cohen JC, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1810. [Google Scholar]
- 10.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. N. Engl. J. Med. 2006;354:34. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 11.Romeo S, et al. Nat. Genet. 2007;39:513. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Circulation. 2002;106:3143. [PubMed] [Google Scholar]
- 13.We thank T. Hyatt, H. Doelle, T. Naing, L. Nie, K. Moller Rasmussen, M. Refstrup, W. S. Schackwitz, J. Martin, and A. Ustaszewska for excellent technical assistance and J. Schageman, C. Lee, K. Lawson, and K. Williams for statistical analyses. We are indebted to the staff and participants of the OHS, the DHS, the ARIC study, and the CCHS for their important contributions. Supported by grants from Foundation Le Ducq, the Donald W. Reynolds Foundation, NIH (HL-082896), the National Heart, Lung, and Blood Institute Program for Genomic Applications (no. HL-066681), the U.S. Department of Energy (contract no. DE-AC02-05CH11231), the Canadian Institutes of Health Research (no. 44360), the Canadian Foundation for Innovation, the Heart and Stroke Foundation of Ontario (no. NA-5413) the Danish Medical Research Council, the Danish Heart Foundation, and the Research Fund at Rigshospitalet, Copenhagen University Hospital. D.R.C. and D.A.H. have equity interest in Perlegen Sciences.