Abstract
Plasmodium falciparum thymidylate synthase-dihydrofolate reductase (TS-DHFR) is an essential enzyme in folate biosynthesis, and a major malarial drug target. Point mutations in P. falciparum TS-DHFR have caused widespread global antifolate resistance, and yet the most effective antifolate known to overcome drug-resistance, WR99210, has poor oral bioavailability. More specific, less toxic therapies are urgently needed. Antifolates target the conversion of methylene tetrahydrofolate to dihydrofolate by TS, and that of dihydrofolate to tetrahydrofolate by DHFR. In humans, TS and DHFR are two discrete enzymes. In P. falciparum, however, TS-DHFR is a bifunctional enzyme, with TS and DHFR encoded within a single protein, and tethered together with by a ‘linker’ region. This linker is not homologous to any other known TS or DHFR enzymes, and is essential for enzyme activity. This bifunctional enzyme thus presents different design approaches for developing novel inhibitors against drug-resistant mutants: developing active-site inhibitors equally effective against wildtype and drug-resistant parasites, or targeting unique non-active site regions for parasite-specific inhibitors. As a first step in identifying unique inhibitors, we performed a high-throughput in silico screen of a database of diverse, drug-like molecules against a non-active site pocket within the linker region of TS-DHFR. The top compounds from the virtual screen were evaluated by enzymatic and cellular assays. In vitro enzymatic studies and cell culture studies of wildtype and drug-resistant P. falciparum parasites identified three compounds active to 20 μM IC50s in both wildtype and antifolate-resistant enzymatic studies, as well as in P. falciparum cell culture. Moreover no inhibition of human DHFR enzyme was observed indicating the inhibitory effects appeared to be parasite-specific. Notably, all three compounds had a biguanide scaffold. Further computational analysis was utilized to determine the relative free energy of binding and these calculations suggested that the compounds might preferentially interact with the active site over the screened ‘linker’ region. To resolve the two possible modes of binding, co-crystallization studies of the compounds complexed with TS-DHFR enzyme were performed to determine the three-dimensional structures. Surprisingly, the structural analysis revealed that these novel, biguanide compounds, distinct from WR99210, do indeed bind at the active site of DHFR, and additionally revealed the molecular basis by which they overcome drug-resistance. To our knowledge, these are the first co-crystal structures of novel, biguanide, non-WR99210 compounds that are active against folate-resistant malaria parasites in cell culture. These studies reveal how serendipity coupled with computational and structural analysis can identify unique compounds as a promising starting point for rational drug design to combat drug-resistant malaria.
Keywords: Dihydrofolate reductase-thymidylate synthase (DHFR-TS), Plasmodium falciparum, antifolate, anti-malarial, biguanide, molecular docking
INTRODUCTION
Malaria is an infectious disease caused by Plasmodium spp parasites, and remains an epidemic of sweeping socioeconomic consequence in tropical countries (2). Between 1 and 3 million lives are lost annually, and over 40% of the world's population is at risk of contracting malaria, with some 350 million new infections each year (2). Notably, P. falciparum infections account for over 90% of malaria-related mortality (2). The last decade has seen a 25% increase in mortality from malaria in Africa alone, due in large part to a rise in drug-resistant parasites (2).
The history of malaria treatment is one of acquired drug resistance and toxic side effects. There is known, widespread resistance to chloroquine, mefloquine, atovaquone, proguanil and pyrimethamine (3-5). Artemisinin compounds, developed from ancient Chinese herbals, are the only antimalarials to which known resistance has not yet been identified (3). With the introduction of each new antimalarial drug, resistance has emerged more quickly than with the last (2, 6, 7). Novel, less toxic, more specific, nonartemisinin treatments are urgently needed to curb this global epidemic (2).
Antifolates like pyrimethamine and cycloguanil are active-site inhibitors of the malarial dihydrofolate reductase (DHFR) enzyme, and have been used successfully to treat falciparum malaria (3). They prevent the conversion of dihydrofolate (H2-folate) to tetrahydrofolate (H4-folate) by DHFR (3). Interestingly, unlike in humans where TS and DHFR are encoded as two discrete enzymes, the malarial DHFR is encoded on the same polypeptide chain as the thymidylate synthase (TS) enzyme (which catalyzes the upstream reaction of converting methylene tetrahydrofolate (CH2H4-folate to H2-folate). This bifunctional TS-DHFR enzyme is the target of antifolate drug design in P. falciparum (8).
Although resistance to antifolates in P. falciparum emerged soon after their introduction, pyrimethamine continues to be used today, in combination therapy with sulfadoxine (sulfadoxine-pyrimethamine or SP, trade name Fansidar™) for malaria prophylaxis in pregnant women (9). In addition, SP combined with amiodaquine or artesenuate remains the first-line therapy for uncomplicated P. falciparum malaria in many parts of sub-Saharan Africa (5). It should be noted that the competitive inhibitors of P. falciparum DHFR like pyrimethamine are routinely used in combination therapy (5).
Antifolate resistance in P. falciparum TS-DHFR is caused by point mutations in the DHFR active site (10). The first mutation to occur is S108N, followed by C59R, then N51I, and finally I164L; each subsequent mutation progressively decreases the binding of both H2-folate (the natural substrate) and pyrimethamine, due to structural changes in the DHFR active site (8). The Ki's for pyrimethamine for the double mutant C59R/S108N and N51I/C59R/S108N/I164L DHFR are 50-fold and >500-fold, respectively, less inhibitory than WT (1.5 nM) (11). Note that these Ki's are only for the monofunctional P. falciparum DHFR enzyme and reaction. Pyrimethamine-resistant DHFR mutations are found throughout West and Central Africa and Asia (5).
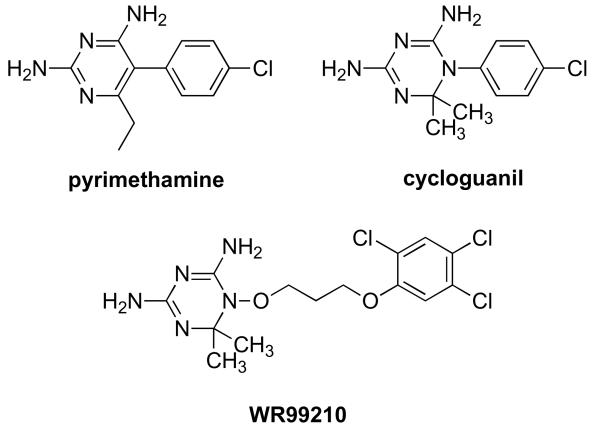
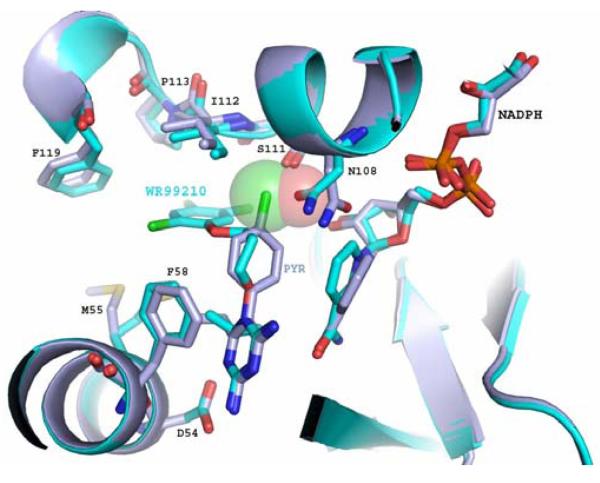
Several attempts have been made to develop novel antifolates which bind to the active site of the clinically important, quadruple mutant of P. falciparum TS-DHFR. One of these, the dihydrotriazene WR99210 (Figure 1a), has a sub-nanomolar Ki value for the WT, double mutant and quadruple mutant DHFRs (8). Structural studies have demonstrated that WR99210 is highly effective against the quadruple mutant because it lacks the p-chlorophenyl moiety of antifolates like pyrimethamine, and has a flexible linker, both of which enable it to avoid steric clash with Asn 108 (8). WR99210 is also bound within the ‘substrate space’ – the essential space within the active site which cannot undergo major conformational rearrangements if H2-folate binding (and thus catalytic activity) are to be preserved (8) (Figure 1b). Unfortunately, the poor oral bioavailability of WR99210 prevents its clinical use (12). To develop novel P. falciparum DHFR active-site inhibitors, molecular docking studies using derivatives of WR99210 (13) or pyrimethamine (14) are underway in some laboratories (15). There have also been reports of molecular docking studies using homology models of P. falciparum DHFR (16).
Figure 1.
Figure 1a Structure of WR99210 compared to those of clinically used antifolates, pyrimethamine and cycloguanil.
Figure 1b WR99210 (WR, cyan) binds successfully to the active site of the clinically important, drug-resistant, N51I/C59R/S108N/I164L “quadruple” mutant of P. falciparum DHFR. Unlike Pyrimethamine (Pyr, purple), WR99210 avoids steric hindrance with Asn108, which clashes with the p-chlorophenyl ring of pyrimethamine, the DHFR inhibitor in Fansidar™. NADPH, an essential cofactor in the conversion of H2-folate to H4-folate by DHFR, is shown bound to the DHFR active site as well.
The bifunctional malarial TS-DHFR enzyme has non-active regions which both flank and tether these two active sites together, and which have no homology to the human TS and DHFR enzymes (17). As In vitro kinetic studies have shown, the non-active site regions of parasitic TS-DHFR enzymes can play a significant role in modulating catalysis (17). In the malaria enzyme specifically, E. coli complementation studies with the P. falciparum TS and DHFR enzymes have also suggested an important role for these distant structural regions (18, 19).
The structural and kinetic properties of P. falciparum TS-DHFR therefore suggest different approaches towards novel inhibitor design. One method would involve developing new active-site inhibitors that are equally effective against WT and drug-resistant parasites. A second method would target the non-active site regions, which have no homology to the human host. To our knowledge, however, no current efforts are focused on the development of novel, non-WR99210 scaffolds, or on the extensive non-active site regions of P. falciparum TS-DHFR, to develop novel, parasite-specific antifolates.
The 89 aa-long linker (from residues 232 to 320) in P. falciparum TS-DHFR encodes significant secondary structure and at least an RNA-binding site (20), and complementation studies suggest a role for this junctional region in modulating activity and association between the TS and DHFR domains (18, 19). Therefore, the linker could serve as an interesting target for molecular docking and virtual screening studies for a potential novel, non-active-site inhibitor of P. falciparum TS-DHFR.
In this study, we performed virtual screening studies of the Maybridge Hitfinder™ library of diverse, drug-like compounds (21), that could bind with moderate (mid-micromolar) affinity to a non-active site pocket within the linker region of P. falciparum TS-DHFR. While previous molecular docking studies have been reported on a homology model of the P. falciparum DHFR (16, 22, 23) before the three-dimensional structure was available, our study involves a virtual screen of the completed crystal structure of the P. falciparum bifunctional TS-DHFR enzyme. Using in vitro enzyme screening, as well as malarial cell culture studies, we identified three lead compounds with similar activity against WT and antifolate-resistant P. falciparum parasites in culture. Significantly, these three compounds had non-WR99210 scaffolds and further computational analysis was used to evaluate their interaction with non-active site pocket as well as the DHFR active site. Using computational analysis, virtual free energy of binding calculations predicted that these lead compounds would preferentially interact with the DHFR active site, instead of the linker region screened by molecular docking. Structural studies were carried out to obtain the structure of the inhibitors bound to the bifunctional P. falciparum TS-DHFR enzyme and distinguish between the two plausible sites for inhibitor binding. Serendipity played a role since co-crystallography studies confirmed that these lead inhibitors bound to the DHFR active site for WT and drug-resistant TS-DHFR enzyme rather than the non-active pocket initially targeted by virtual screening. These structural studies also revealed the molecular mechanism for the compounds interaction with the quadruple, drug-resistant form of the TS-DHFR enzyme and offers a new avenue to exploit for future development of targeted, structure-guided antimalarial therapy.
RESULTS and DISCUSSION
We describe the identification a novel biguanide scaffold that is active in cell culture against the clinically important, antifolate-resistant mutant of P. falciparum TS-DHFR. Using enzymatic and cell culture assays to screen the ‘hits’ from our molecular docking studies, we identified three compounds, RJF 001302, RJF 00670, and RJF 00719, with around 20 μM inhibition in P. falciparum pyrimethamine-resistant DHFR enzyme in in vitro kinetic studies. Our data also showed these compounds to be active in both WT and quadruple mutant P. falciparum cell culture. As kinetic studies regarding the pattern of inhibition were inconclusive, and the compounds had a biguanide moiety somewhat akin to WR99210 (a known competitive inhibitor of P. falciparum DHFR), we pursued free energy of binding calculations using MM-GB/SA (molecular mechanics and the generalized Born model and solvent accessibility) (24). These results suggested that all three compounds preferred to bind to the DHFR active site, compared to the non-active site pocket which was originally targeted. Since we could not decisively assign the RJF compounds as non-active-site inhibitors, we proceeded with structural studies of these inhibitors complexed with P. falciparum TS-DHFR enzyme for definitive determination of the mode of binding. Interestingly, despite our virtual screening data, co-crystallization studies of inhibitor complexed with the WT and pyrimethamine-resistant, quadruple mutant enzymes confirmed that these compounds do indeed bind to DHFR active site. Importantly, however, the RJF compounds avoid steric clash with Asn108, a key residue that hinders existing antifolates like pyrimethamine from binding the drug-resistant mutant enzymes. Also, several key interactions between inhibitor and the DHFR active site, shown previously to be essential in successful antifolates like WR99210 or pyrimethamine (8), are retained by the RJF compounds, even in the quadruple mutant enzyme. Each of these points is elaborated upon below.
Virtual Screening of a Non-active Site Pocket in P. falciparum TS-DHFR
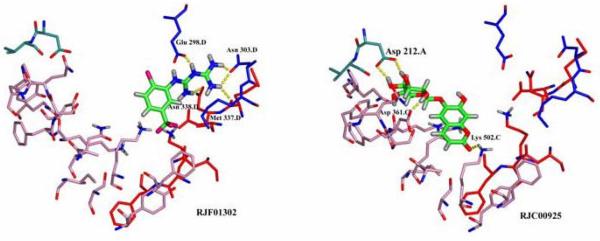
We screened 16,000 compounds from the Maybridge HitFinder™ library against the crystal structure of WT P. falciparum TS-DHFR using Glide SP (Supplementary Figure 1). The top fourteen compounds identified by this screen are shown in Supplementary Table 1, with their corresponding Glide scores and chemical structures, and a summary flowchart of the screening process is provided in Supplementary Figure 2. Overlaying the ‘top hit’ compounds in the crossover region showed that the ligands had different binding modes, implying that they occupy one of several ‘spaces’ within the non-active site pocket (Figure 2). These inhibitors were characterized with QikProp and showed favorable predicted ADME properties (25) (Supplementary Table 2).
Figure 2.
Glide parameters were optimized to maximize surface binding flexibility in the non-active site pocket, as well as binding to both TS (red/pink) and DHFR (blue) domains. Here, RJF 01302 and RJC 00925 are shown binding to different regions of the non-active-site pocket.
The Maybridge HitFinder™ compound database was screened because it comprises the chemical diversity of the entire Maybridge compound library, prescreens the compounds for Lipinski rules (26), and is commercially available to over 90% purity (21). The Glide SP score is a soft scoring function used to identify ligands likely to bind to a target protein, thereby minimizing false negatives. The most heavily weighted items in determining the Glide score are van der Waals interactions and lipophilic contact, and compounds with lower scores are predicted to bind more tightly to the target protein. Scores ranged from −9.32 to −8.50, which is within the normal range of expected scores for moderate affinity (mid-micromolar) compounds from Glide SP (27).
Since this was an initial attempt at small molecule binding in the non-active site pocket, we set a large boundary condition (Supplementary Figure 1), and favored results with diversity in shape and binding mode of the inhibitor (Figure 2). QikProp was used to check important physical properties of the top ‘hits’ from this screening, as it predicts log P, log S and Caco-2 (physical properties useful in inhibitor optimization in the drug design process). Compounds with molecular weight less than 450, logP less than 5, log S greater than −5 and pCaco greater than 25 are considered to have favorable lead-compound like properties (25).
Virtual screening methods have limitations due to approximations used in the scoring function, as well as measures taken to simplify the conformational search space to reduce computation time for the thousands of screened compounds (28). Another limitation of this virtual screening study was the molecular docking of a predicted doubly charged biguanide moiety in the screened compounds, although experimental evidence suggests that these biguanides are monoprotonated at physiological pH (29, 30). Furthermore, kinetic evidence suggests that the current crystal structure of WT P. falciparum TS-DHFR may not be in its most active conformation as the TS active site is not doubly liganded (17). As this was the initial virtual screening of the non-active site pocket, these molecular docking studies were done with minimal experimental evidence about which residues in the non-active site are functionally important, or how protein conformation would adjust to inhibitor binding at the non-active site.
In Vitro Enzymatic Assays of Inhibitor Assay
Using a spectrophotometric assay, which follows the consumption of NADPH at 340 nm, we screened the ‘hits’ for inhibition of the bifunctional TS-DHFR and DHFR activities. Several compounds exhibited intrinsic absorbance or display signal attenuation at 340 nm, and the activity of these compounds was further examined under steady-state conditions using radiolabeled assay for bifunctional TS-DHFR and DHFR activity. A number of ‘hits’ also precipitated in buffer conditions, even in high/low salt conditions and in very acidic or very basic pH's, and those compounds insoluble in reaction buffer were excluded from the analysis.
The compounds were initially screened for inhibition of the bifunctional TS-DHFR reaction using both a spectrophotometric and a radiolabeled assay. As the radiolabeled assay showed a buildup of H2-folate in the reaction products at early time points for RJF 01302, we also screened these compounds for DHFR activity in the bifunctional enzyme. Of the top fourteen compounds tested, we found three active compounds to inhibit the DHFR reaction in the bifunctional enzyme – RJF 00670, RJF 01302 and RJF 00719. They did not inhibit the human DHFR enzyme at 500 μmol/L concentrations. Kinetic studies to determine a pattern of inhibition (competitive or non-competitive) in the bifunctional enzyme using both a radiolabeled and spectrophotometric assay were somewhat inconclusive.
This combination of these analyses identified three compounds of interest, all of which inhibited DHFR (but not TS) activity in the WT bifunctional TS-DHFR enzyme. Due to difficulties with expressing sufficient quantities of the full-length, bifunctional quadruple mutant (N51I/C59R/S108N/I164L) P. falciparum TS-DHFR enzyme, we used monofunctional constructs of P. falciparum WT and quadruple mutant DHFR for further in vitro enzyme assays to determine an accurate, comparative IC50 for the DHFR reaction in WT and quadruple mutant enzymes. These results are summarized in Table 1, and the corresponding curves shown in Supplementary Figure 3.
Table 1.
IC50 of RJF compounds in in-vitro enzyme and cell culture assays of WT and quadruple mutant P. falciparum TS-DHFR
| IC50 | in WT DHFR in vitro assay |
in quadruple mutant DHFR in vitro assay |
in WT parasite cell culture |
in quadruple mutant cell culture |
|---|---|---|---|---|
| RJF 00719 | 21 ± 2 μM | 40 ± 2 μM | 20 ± 2 μM | 19.9 ± 0.5 μM |
| RJF 01302 | 29.9 ± 0.5 μM | 39.1 ± 0.5 μM | 22 ± 3 μM | 20 ± 2 μM |
| RJF 00670 | 17 ± 2 μM | 20 ± 2 μM | > 50 μM | > 50 μM |
Interestingly, all three of these compounds - RJF 01302, RJF 00670, and RJF 00719 (see Footnote 2) – contain a biguanide scaffold. The IC50 of these compounds was ∼ 500 μM for the bifunctional reaction predominately reflecting the TS reaction, but ∼20 μM for the DHFR reaction, as determined by spectrophotometric assays. It is of note that other compounds containing a biguanide scaffold (see Supplementary Table 1) were not inhibitory: in some of these cases, solubility was limiting (RJF 00587, RJF 00729), but in other cases, the lack of inhibitory effect is puzzling (RJF 00600). RJF 01059 had weak activity against the DHFR (but not bifunctional TS-DHFR reaction), and was therefore not pursued for further analysis. We also tested biguanides (RDR 01651, RJF 00541 and RDR 003335) which were not identified by the Glide screen, but that were Maybridge HitFinder™ compounds related to RJF 01302. None of these latter compounds were active in in vitro enzymatic assays (data not shown). Excitingly, when RJF 00719, RJF 00670 and RJF 01302 were assayed for their activity in the monofunctional construct of P. falciparum DHFR, they showed similar inhibition in both WT enzyme, and the strongly pyrimethamine-resistant quadruple mutants of the enzyme.
Although an obvious limitation of the IC50 determination was that we assayed for activity using a monofunctional DHFR enzyme, the comparative results are valid as both the WT and quadruple mutant enzyme were monofunctional in this assay, and determining inhibitor activity using a monofunctional DHFR enzyme is a well-accepted, standard assay in the P. falciparum antifolate literature (31). While standard steady-state competition assays performed using the spectrophotometric assay suggested these compounds had a non-competitive pattern of inhibition, these data were obviously limited by the inherent absorbance of the compounds at 340 nm, as well as the signal attenuation described above. Therefore, free energy of binding calculations were pursued to predict whether the RJF compounds preferred the DHFR active site to the non-active site pocket in the linker region (see below).
Cell Culture Assays of Inhibitor Activity
To determine activity in both WT and antifolate-resistant cell culture, the three RJF compounds were assayed for activity in P. falciparum parasites with either WT DHFR enzyme (TM4/8.2) or the quadruple mutant DHFR enzyme (V1/S or N51I/C59R/S108N/I164L). The P. falciparum parasite cell culture (TM4/8.2 and V1/S, respectively) in the erythrocytic stage of the life cycle was performed using a standard [3H]-hypoxanthine incorporation assay (32).
The compounds demonstrated similar activity in both WT and quadruple mutant parasites in the erythrocytic stages (Table 1). The higher IC50 of RJF 00670 in cell culture could possibly be explained by degradation or metabolic instability. More importantly, the fact that the IC50's were relatively similar for the DHFR monofunctional enzyme and cell culture assays for two of the three compounds suggested that DHFR might indeed be the major target of the RJF compounds.
Inhibition of parasite replication during erythrocytic stages is an important indicator of activity, since the majority of P. falciparum parasites in an infected human host reside within the erythrocyte. It is intra-erythrocytic replication and subsequent extravasation of erythrocytes which causes the fevers, chills, anemia, and secondary organ damage characteristic of falciparum malaria (33). However, inhibition of the liver stages (sporozoite phase) by the RJF compounds, as well as their oral bioavailability and toxicology, must be further examined in animal models of malaria (like P. bergheii). Evaluation of inhibition of the sporozoite phase of this parasite are important for determining the use of these lead compounds for malaria prophylaxis (34).
Evaluation of the Relative Free Energy of Binding to determine the Preferred Binding Site for RJF Compounds
To investigate whether that these lead compounds were binding at the non-active site pocket or the DHFR active site, relative free energies of binding were calculated based on their Glide poses. WR99210, the ligand from the crystal structure used in the docking studies (PDB id 1j3i), was used as a control. The relative free energies of binding (ΔΔ G bindactive site – nonactive site) for WR99210, RJF 01302, RJF 00670, and RJF 00719 were −33, −31, −56 and −22 kcal/mol, respectively. These computational data indicated that WR99210 and the RJF compounds all preferred binding the DHFR active site over the non-active site pocket in the linker region (Figure 3).
Figure 3.
MM/GBSA calculations of free energy of binding predict that RJF compounds bind to the active site of P. falciparum DHFR (upper panel), as confirmed by co-crystallization studies (lower panel).
Conventional docking studies could not be used to compare these binding energies, given the 45 Å distance between the sites. MM-GB/SA methods to estimate approximate relative free energies of binding are considered an efficient way to post process compounds after high-throughput, virtual screening, avoiding more time-consuming free energy simulations (24). Traditionally, energies are used relatively in one site, comparing multiple ligands. Here, multiple sites for individual ligands were examined, to determine specificity rather than to find a top lead. Known DHFR active-site inhibitor WR99210 was used as a control, as the calculations should indicate that it prefers binding the DHFR active site. In this case, trends are more important than magnitudes, due to necessary approximations made due to the distance between the two sites (around 45 Å) and the size of the bifunctional homodimer. While MM-GBSA methods tend to quantitatively overestimate binding free energies (35), the calculations provide a qualitative indication that the RJF compounds (as well as the known DHFR active-site inhibitor control, WR99210), prefer binding the active site of P. falciparum DHFR. The RJF compounds likely ranked high in the original screen of the non-active site pocket in the linker region because of their charged (protonated) states, and the multiple charged residues at both the DHFR active site and the non-active site pocket (Figure 2).
Co-crystallization of Inhibitors with P. falciparum WT and Quadruple Mutant TS-DHFR
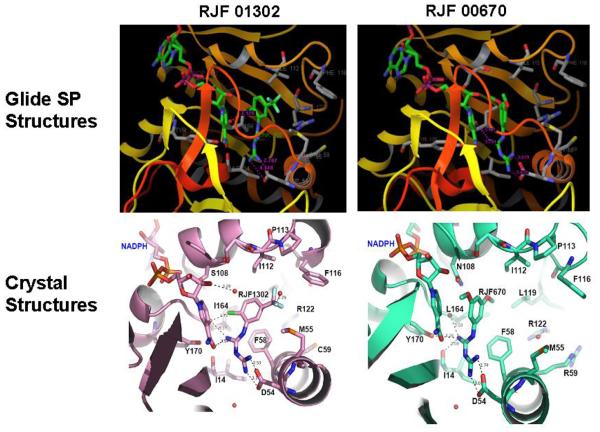
However, given that computational methods suggested that the DHFR active site was the target site, structural studies were pursued. We were able to co-crystallize the inhibitor RJF 00670 with the full-length, bifunctional, quadruple mutant of P. falciparum TS-DHFR to 2.56 Å resolution (PDB ID: 3DG8; Figure 4a), and RJF 01302 with the WT enzyme to 2.70 Å resolution (PDB ID: 3DGA, Figure 4b). We were also able to crystallize the co-complex of TS-DHFR with RJF 00719, but unfortunately could not see enough of the electron density to report the complete co-complex structure. This is likely due in part to solubility limitations with RJF00719, indicating perhaps that the binding site was not completely saturated.
Figure 4.
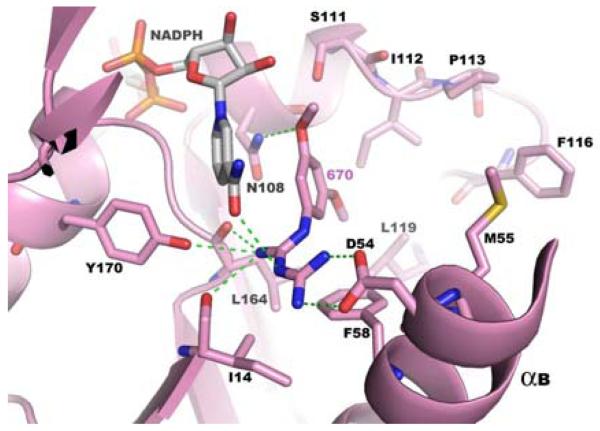
Figure 4a Co-crystallization studies show RJF 00670 to be bound at the DHFR active site of the quadruple mutant of P. falciparum TS-DHFR enzyme. The DHFR active site of this co-crystal was in its fully liganded conformation as NADPH was also bound. Hydrogen bonds between the inhibitor and the enzyme active site are shown with green dashed lines.
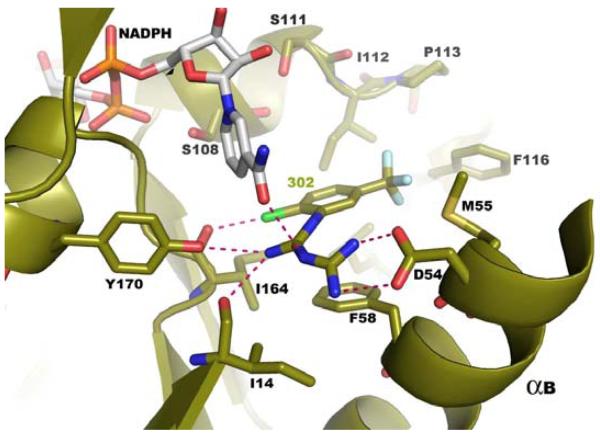
Figure 4b Co-crystallization studies show RJF 01302 to be bound at the DHFR active site of the WT P. falciparum TS-DHFR enzyme. The DHFR active site of this co-crystal was in its fully liganded conformation as NADPH was also bound. Hydrogen bonds between the inhibitor and the enzyme active site are shown with green dashed lines.
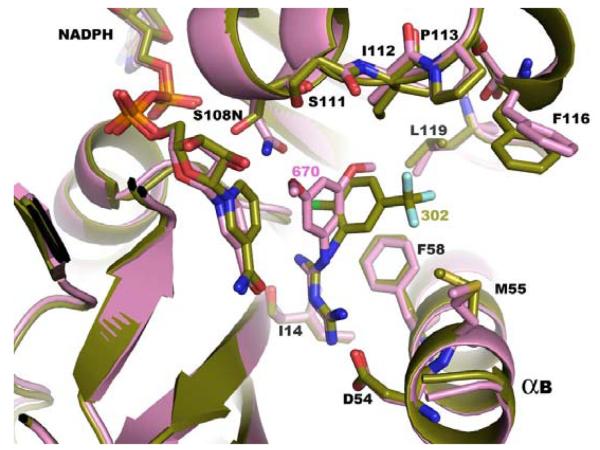
Figure 4c Superimposition of the X-ray crystal structures of RJF 00670 (shown in purple) and of RJF 01302 (shown in green) bound to the DHFR active site of P. falciparum TS-DHFR.
Both RJF01302 and RJF00670 co-crystallized at the active site of the DHFR enzyme, indicating that virtual free energy of binding studies, which predicted binding of RJF compounds at the DHFR active site, corresponded well with the crystallography data (Figure 3). The complete electron density of both ligands was observed, despite the relatively high B factors. The protein-ligand interactions formed between the active site residues of DHFR and the inhibitors were calculated using LigPlot and are shown in Supplementary Table 3. We also performed an analysis of the common interactions formed between WR99210 and the quadruple mutant DHFR, as well as between pyrimethamine and the WT DHFR, and compared these interactions to those formed by the RJF compounds. These interactions are summarized in Supplementary Table 3 and their implications are discussed below. Data collection and refinement statistics are shown in Table 2, and Fo-Fc maps are shown in Supplementary Figures 4 and 5.
Table 2.
Data Collection and Refinement Statistics for Co-crystal Structures of RJF 00670 bound to the quadruple mutant P. falciparum TS-DHFR, and RJF 01302 bound to the WT P. falciparum TS-DHFR. Numbers in parentheses are for the highest resolution bin. Asterisk indicates number of residues in favored, allowed and disallowed regions by Ramachandran plot.
| RJF 00670 co-crystallized with quadruple mutant TS- DHFR |
RJF 01302 co-crystallized with WT TS-DHFR |
|
|---|---|---|
| Space group | P212121 | P212121 |
| Unit cell parameter (Å) | 56.516 155.404 165.183 90 90 90 |
57.377 156.015 164.567 90 90 90 |
| Resolution (Å) | 50-2.56 (2.65-2.56) | 30-2.70 (2.80-2.70) |
| No. observed / unique reflections |
189,861 (45,686) | 205,888 (41,531) |
| Completeness | 98.0% (95.0%) | 99.8% (99.9%) |
| I/sigma I | 19.86 (6.36) | 17.18 (4.41) |
| Rmerge | 6.6% (26.5%) | 8.0% (29.9%) |
| No. of molecules or asymmetric units |
2 | 2 |
| Refinement resolution (Å) |
30-2.56 | 30-2.70 |
| Rfac / Rfree | 0.198 / 0.239 | 0.205 / 0.256 |
| Reflection used in refinement |
45,613 (95.4%) | 41385 (99.7%) |
| Working set | 43,291 (90.5 % ) | 39,304 (94.7 % ) |
| Test set | 2,322 (4.9 % ) | 2,081 (5.0 % ) |
| Number of glycines | 48 | 48 |
| Most favoured regions* | 843 | 836 |
| Additional allowed regions* |
147 |
150 |
| Generously allowed regions* |
10 |
14 |
| Disallowed regions* | 0 | 0 |
| Average B factors of Enzyme (Ǻ2) |
45.96 |
44.32 |
| Average B factors of RJF, chains A and B (Ǻ2) |
50.1 and 89.2 |
79.0 and 89.2 |
| Average B factors of NADPH, chains A and B (Ǻ2) |
48.8 and 89.6 |
34.3 and 75.9 |
While these results were unanticipated by our initial virtual screening studies of the linker region, a low-micromolar, novel scaffold which binds to the DHFR active site of, and have activity in, both WT and quadruple mutant P. falciparum TS-DHFR remains an important finding. When the co-crystal structures of RJF 00670 and RJF 01302 are superimposed (Figure 4c), and the hydrophobic, van der Waals and H-bond interactions formed between inhibitor and DHFR active site are examined closely, several interesting observations are made.
First, and most importantly, both RJF 00670 and RJF 01302 are not sterically hindered by Asn 108, the point mutation essential to pyrimethamine resistance. (RJF 01302 was co-crystallized with the WT enzyme, and this was determined by aligning this structure to the active site of the quadruple mutant.) That the RJF compounds also avoid this steric clash - just as WR99210 but unlike pyrimethamine -, suggests a common mechanism for overcoming antifolate resistance in malarial DHFR, and is encouraging with respect to their potential as promising antifolate scaffolds for further lead optimization (Figure 5).
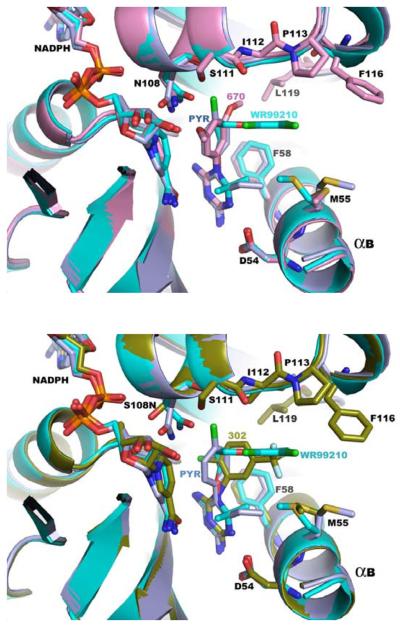
Figure 5.
Overlays of the active sites of P. falciparum DHFR demonstrating that RJF compounds overcome drug resistance by avoiding steric clash with Asn 108.
RJF 01302 (green, bottom panel), RJF 00670 (purple, top panel), pyrimethamine (navy blue) and WR99210 (cyan) are shown bound to the DHFR active site. The structures overlaid are from pdb codes 1J3K (quadruple mutant with WR99210), 1J3J (double mutant with pyrimethamine) and the co-crystal structures described in this paper.
Second, in the antifolate inhibitor literature on P. falciparum TS-DHFR, there has been much analysis to discern the interactions that are essential between enzyme and inhibitor. In addition to Ser 108, another such important residue (which lies at the opposite ends of the hydrophobic substrate space) that interacts with WR99210 and pyrimethamine is Asp 54 (on α helix B) (8). Chemically, the RJF compounds vary from WR99210 by lacking a cyclic guanidinium ring and the dimethyl group. Despite that, both RJF 00670 and RJF 01302 form hydrogen bonds with the carboxyl side chain of Asp 54 and the backbone carbonyl of Ile 164 (another common mutation in the quadruple mutant of pyrimethamine-resistant DHFR). Furthermore, as shown in Supplementary Table 3, the RJF compounds also make polar interactions with Ile14, Tyr170, and Cys15, and van der Waals contact with residues Phe58, Ile112, Leu119, Met 55, Cys15 and Ala16 at the DHFR active site. Despite being about 100 g/mol smaller in molecular weight than WR99210, the RJF compounds thus form many of the same interactions considered to make WR99210 and the pterin ring of the natural substrate H2F bind so tightly and specifically within the substrate space of the quadruple mutant DHFR.
A third critical observation is that the aromatic ring of the RJF compounds forms very few interactions with the residues of the active site (Figure 4a and b). This finding reaffirms the usefulness of the RJF series as potential lead compounds since functional groups can be added easily to this ring, especially in a position meta- or para- to the bisimine.
Do these inhibitors specifically target P. falciparum DHFR?
Several lines of evidence suggest that P. falciparum DHFR is at least one of the primary targets of the RJF compounds. First, the IC50 in cell culture and in enzyme assays are approximately equivalent, suggesting that DHFR may be the primary target of these inhibitors. Also, these compounds co-crystallize at the active site of both WT and quadruple-mutant P. falciparum DHFR. Though the RJF series are small lead compounds, it should be noted that the human DHFR enzyme is not inhibited by these compounds at 500 μmol/L concentrations. “Folate effect” studies are often used for to demonstrate folate pathway inhibition by sulfa drugs. However, the utility of similar folate antagonism studies remains unclear for target validation of DHFR inhibitors in P. falciparum, given the complexities of folate salvage and catabolism pathways in the Plasmodium parasite, and the tremendous folate reserve of Plasmodium parasites even after several reinvasions (36). For example, folate can be cleaved to pterin-6-aldehyde and p-amino-benzoylglutamate, byproducts utilized in folate biosynthesis, while exogenous intact folates are also alternatively be salvaged and utilized by the parasite (37, 38). P. falciparum may have putative folate transporters, whose candidate genes, MAL8P1.13 and PF11_0172, we have previously identified (39). For example, folic acid and folinic acid could not be used to reverse the effect of even pyrimethamine in P. knowlesi (40) and of WR99210 in P. falciparum, though the refractory effect depended on the strain and experimental approach used (41-43). Consequently, experiments using the “folate effect” to validate targets of inhibition such as DHFR are often difficult to interpret for the purpose of drawing definitive conclusions.
Summary and Implications for Potential Novel Antifolates based on the RJF Scaffold
Biguanides are routinely used clinically and are recognized as safe, well-tolerated and highly effective anti-diabetic therapy agents (44). Additionally, guanidines have been implicated before in the development of E. coli DHFR inhibitors (45, 46), and at least one cyclic biguanide has been shown to have anti-Plasmodial activity at 10 μmol/L when tested in the wildtype 3d7 strain (47). However, this is the first report of guanidine scaffolds co-crystallized with a drug-resistant mutant of the malarial TS-DHFR enzyme. Our data both demonstrate that RJF compounds have similar activity in cell culture in both WT and antifolate-resistant parasites, and potentially suggest a common mechanism for how antifolates overcome drug resistance in malarial DHFR. Furthermore, these co-crystal structures can enable structure-guided development of novel inhibitors that maximize key active site interactions by the addition of functional groups to the RJF scaffold. Future directions will focus on testing these inhibitors in human cell culture, as well as developing subtractive in silico filters to select non-active inhibitors that do not bind the DHFR active site.
In the face of growing mortality, new antimalarials are urgently needed. RJF compounds therefore hold the exciting potential of serving as novel scaffolds for development of targeted, structure-guided antimalarial therapy.
METHODS
Additional details for each section are available in Supplementary Methods.
Molecular Docking Studies
With close examination of the linker region in the structure of P. falciparum TS-DHFR (PDB structural ID: 1j3i), we identified a pocket within the linker region defined by contributing residues from the linker, DHFR and TS domains, with the rationale that binding of a small molecule to this linker might disrupt DHFR and/or TS function (Figure 2). The Maybridge Hitfinder Library™ (Maybridge, United Kingdom), containing a subset of 16,000 molecules representing the chemical diversity of the entire Maybridge database (21), was virtually screened against this pocket using Glide Standard Precision v 3.5 (Schrödinger, New York), a third generation molecular docking program (27). Additional details have been previously reported (48).
In vitro Enzymatic Assays of Inhibitor Activity
The ‘hits’ from the Glide molecular docking studies were screened using a spectrophotometric assay for the P. falciparum TS-DHFR enzyme, as well as an assay using tritiated substrate and rapid chemical quench methodology for TS-DHFR activity, both performed under steady-state conditions and previously described (17). The IC50 was determined by plotting rate versus inhibitor concentration and determining inhibitor concentration at half maximal velocity (vmax). Detailed methods have been previously published (17).
Approximate relative free energy of binding to determine the preferred binding site
Due to similarity of the active compounds identified by in vitro screening, to known active-site inhibitors, computational techniques were used to determine relative binding affinities. Based on experimental results (29, 30), we determined that all three compounds should be in a monoprotonated ionization state, in contrast to the bisprotonated ionization state generated by LigPrep. The correct forms of the three RJF compounds and known competitive inhibitor WR99210 were redocked in both the active site, and in the non-active site pocket of linker region. Relative free energies of binding were then calculated based on complexes from this second docking. Additional details are outlined in Supplementary Methods.
Cell Culture Assays of Inhibitor Activity
The RJF compounds were tested in erythrocytic stages of both P. falciparum parasite with WT and N51I/C59R/S108N/I164L quadruple mutant of DHFR. Detailed methods were as previously described (49, 50).
Co-crystallization Studies and Data Analysis
The RJF compounds were co-crystallized in the absence of the H2-folate, the natural substrate of DHFR. RJF 01302 was co-crystallized with the WT P. falciparum TS-DHFR enzyme, and RJF 00670 was co-crystallized with the quadruple mutant. We also attempted to co-crystallize the RJF compounds in the presence of cycloguanil, an active-site inhibitor of DHFR. Blocking the active site of DHFR was attempted to see if the RJF compound would bind at an alternative binding site such as the non-active site pocket in the linker region. Otherwise, methods for co-crystallization of compounds with P. falciparum TS-DHFR, x-ray diffraction data collection, and analysis to determine the actual binding site of the compound were as previously published (51). Atomic coordinates have been deposited in the Protein Data Bank.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH Grant AI 44630 (to KSA), an NIH Medical Student Training Program grant to the Yale M.D.-Ph.D. Program (to TD), a Doctoral Research Award from the Canadian Institutes of Health Research (to TD), an international research scholarship from the Howard Hughes Medical Institute (to SK), NIH Grants AI 44616 and GM32136 (to WLJ), and a grant from Medicines for Malaria Ventures MMV99/0099 (to YY). We gratefully acknowledge the use of the National Synchrotron Radiation Research Center Beamline BL13B1 (Taiwan). Also, we would like to thank Jeff Saunders (Schrödinger, Portland, Oregon), Raymond Chung (Patent and Trade Office, Washington D.C.) and Walter (Eddie) Martucci for helpful discussions in the preparation of this manuscript.
The following abbreviations were used
- TS-DHFR
thymidylate synthase-dihydrofolate reductase (this is a functional designation as dihydrofolate is produced at TS and utilized at DHFR; this enzyme is also commonly referred to as DHFR-TS in the literature because the DHFR domain is N-terminal to TS)
- dUMP
deoxyuridine monophosphate
- CH2H4-folate
methylene tetrahydrofolate
- H2-folate
dihydrofolate
- H4-folate
tetrahydrofolate
- NADPH
nicotinamide adenine dinucleotide phosphate
- HPLC
high-performance liquid chromatography
- WT
wildtype
Footnotes
The RJF compounds were developed in the chemistry laboratory of Ray J. Froude at Maybridge (now Thermofisher Scientific, Tintagel) in the early 1980's. They were originally produced as intermediates en route to synthesizing aminotriazenes, and later developed into a “series” of drug-like molecules (1).
REFERENCES
- 1.Thermofisher Scientific Inc. 2008. [Google Scholar]
- 2.Greenwood BM, Bojang K, Whitty CJ, Targett GA. Malaria. Lancet. 2005;365:1487–98. doi: 10.1016/S0140-6736(05)66420-3. [DOI] [PubMed] [Google Scholar]
- 3.Rieckmann KH. The chequered history of malaria control: are new and better tools the ultimate answer? Ann Trop Med Parasitol. 2006;100:647–62. doi: 10.1179/136485906X112185. [DOI] [PubMed] [Google Scholar]
- 4.Greenwood D. Conflicts of interest: the genesis of synthetic antimalarial agents in peace and war. J Antimicrob Chemother. 1995;36:857–72. doi: 10.1093/jac/36.5.857. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization . WHO Press; Geneva: 2006. [Google Scholar]
- 6.Centers for Disease Control and Prevention 2004 [Google Scholar]
- 7.Centers for Disease Control and Prevention 2004 [Google Scholar]
- 8.Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, Taylor P, Walkinshaw MD, Yuthavong Y. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10:357–65. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization . WHO Regional Office for Africa; Brazzaville: 2004. [Google Scholar]
- 10.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardarian A, Douglas KT, Read M, Sims PF, Hyde JE, Chitnumsub P, Sirawaraporn R, Sirawaraporn W. Pyrimethamine analogs as strong inhibitors of double and quadruple mutants of dihydrofolate reductase in human malaria parasites. Org Biomol Chem. 2003;1:960–4. doi: 10.1039/b211636g. [DOI] [PubMed] [Google Scholar]
- 12.Warhurst DC. Resistance to antifolates in Plasmodium falciparum, the causative agent of tropical malaria. Sci Prog. 2002;85:89–111. doi: 10.3184/003685002783238906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt SY, Detering C, Varani G, Jacobus DP, Schiehser GA, Shieh HM, Nevchas I, Terpinski J, Sibley CH. Identification of the optimal third generation antifolate against P. falciparum and P. vivax. Mol Biochem Parasitol. 2005;144:198–205. doi: 10.1016/j.molbiopara.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Fogel GB, Cheung M, Pittman E, Hecht D. Modeling the inhibition of quadruple mutant Plasmodium falciparum dihydrofolate reductase by pyrimethamine derivatives. J Comput Aided Mol Des. 2008;22:29–38. doi: 10.1007/s10822-007-9152-9. [DOI] [PubMed] [Google Scholar]
- 15.Yuthavong Y. Basis for antifolate action and resistance in malaria. Microbes Infect. 2002;4:175–82. doi: 10.1016/s1286-4579(01)01525-8. [DOI] [PubMed] [Google Scholar]
- 16.Rastelli G, Pacchioni S, Sirawaraporn W, Sirawaraporn R, Parenti MD, Ferrari AM. Docking and database screening reveal new classes of Plasmodium falciparum dihydrofolate reductase inhibitors. J Med Chem. 2003;46:2834–45. doi: 10.1021/jm030781p. [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta T, Anderson KS. Probing the Role of Parasite-Specific, Distant Structural Regions on Communication and Catalysis in the Bifunctional Thymidylate Synthase-Dihydrofolate Reductase from Plasmodium falciparum. Biochemistry. 2008;47:1336–45. doi: 10.1021/bi701624u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shallom S, Zhang K, Jiang L, Rathod PK. Essential protein-protein interactions between Plasmodium falciparum thymidylate synthase and dihydrofolate reductase domains. J Biol Chem. 1999;274:37781–6. doi: 10.1074/jbc.274.53.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wattanarangsan J, Chusacultanachai S, Yuvaniyama J, Kamchonwongpaisan S, Yuthavong Y. Effect of N-terminal truncation of Plasmodium falciparum dihydrofolate reductase on dihydrofolate reductase and thymidylate synthase activity. Mol Biochem Parasitol. 2003;126:97–102. doi: 10.1016/s0166-6851(02)00240-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K, Rathod PK. Divergent regulation of dihydrofolate reductase between malaria parasite and human host. Science. 2002;296:545–7. doi: 10.1126/science.1068274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thermofisher Scientific Inc. [Google Scholar]
- 22.Toyoda T, Brobey RK, Sano G, Horii T, Tomioka N, Itai A. Lead discovery of inhibitors of the dihydrofolate reductase domain of Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Biochem Biophys Res Commun. 1997;235:515–9. doi: 10.1006/bbrc.1997.6814. [DOI] [PubMed] [Google Scholar]
- 23.McKie JH, Douglas KT, Chan C, Roser SA, Yates R, Read M, Hyde JE, Dascombe MJ, Yuthavong Y, Sirawaraporn W. Rational drug design approach for overcoming drug resistance: application to pyrimethamine resistance in malaria. J Med Chem. 1998;41:1367–70. doi: 10.1021/jm970845u. [DOI] [PubMed] [Google Scholar]
- 24.Guimaraes CR, Cardozo M. MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization. J Chem Inf Model. 2008;48:958–70. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Adv Drug Deliv Rev. 2002;54:355–66. doi: 10.1016/s0169-409x(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 26.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 27.Glide v 3.5. Schrodinger, LLC; New York: [Google Scholar]
- 28.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–5. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amigó J, Martínez-Calatayud JM, Cantarero A, Debaerdemaeker T. Molecular structure of 1-(2-sulfoethyl)biguanide. Acta Cryst. 1988;C44:1452–1454. [Google Scholar]
- 30.Hariharan M, Rajan SS, Srinivasan R. Structure of metformin hydrochloride. Acta Cryst. 1989:911–913. [Google Scholar]
- 31.Yuthavong Y, Yuvaniyama J, Chitnumsub P, Vanichtanankul J, Chusacultanachai S, Tarnchompoo B, Vilaivan T, Kamchonwongpaisan S. Malarial (Plasmodium falciparum) dihydrofolate reductase-thymidylate synthase: structural basis for antifolate resistance and development of effective inhibitors. Parasitology. 2005;130:249–59. doi: 10.1017/s003118200400664x. [DOI] [PubMed] [Google Scholar]
- 32.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–8. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster FL. Cultivation of plasmodium spp. Clin Microbiol Rev. 2002;15:355–64. doi: 10.1128/CMR.15.3.355-364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Andrade-Neto VF, da Silva T, Lopes LM, do Rosario VE, de Pilla Varotti F, Krettli AU. Antiplasmodial activity of aryltetralone lignans from Holostylis reniformis. Antimicrob Agents Chemother. 2007;51:2346–50. doi: 10.1128/AAC.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kormos BL, Benitex Y, Baranger AM, Beveridge DL. Affinity and specificity of protein U1A-RNA complex formation based on an additive component free energy model. J Mol Biol. 2007;371:1405–19. doi: 10.1016/j.jmb.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, Wang Q, Aspinall TV, Sims PF, Hyde JE. Transfection studies to explore essential folate metabolism and antifolate drug synergy in the human malaria parasite Plasmodium falciparum. Mol Microbiol. 2004;51:1425–38. doi: 10.1111/j.1365-2958.2003.03915.x. [DOI] [PubMed] [Google Scholar]
- 37.Krungkrai J, Webster HK, Yuthavong Y. De novo and salvage biosynthesis of pteroylpentaglutamates in the human malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol. 1989;32:25–37. doi: 10.1016/0166-6851(89)90126-6. [DOI] [PubMed] [Google Scholar]
- 38.Wang P, Brobey RK, Horii T, Sims PF, Hyde JE. Utilization of exogenous folate in the human malaria parasite Plasmodium falciparum and its critical role in antifolate drug synergy. Mol Microbiol. 1999;32:1254–62. doi: 10.1046/j.1365-2958.1999.01437.x. [DOI] [PubMed] [Google Scholar]
- 39.Massimine KM, Doan LT, Atreya CA, Stedman TT, Anderson KS, Joiner KA, Coppens I. Toxoplasma gondii is capable of exogenous folate transport. A likely expansion of the BT1 family of transmembrane proteins. Mol Biochem Parasitol. 2005;144:44–54. doi: 10.1016/j.molbiopara.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 40.McCormick GJ, Canfield CJ, Willet GP. Plasmodium knowlesi: in vitro evaluation of antimalarial activity of folic acid inhibitors. Exp Parasitol. 1971;30:88–93. doi: 10.1016/0014-4894(71)90074-9. [DOI] [PubMed] [Google Scholar]
- 41.Kinyanjui SM, Mberu EK, Winstanley PA, Jacobus DP, Watkins WM. The antimalarial triazine WR99210 and the prodrug PS-15: folate reversal of in vitro activity against Plasmodium falciparum and a non-antifolate mode of action of the prodrug. Am J Trop Med Hyg. 1999;60:943–7. doi: 10.4269/ajtmh.1999.60.943. [DOI] [PubMed] [Google Scholar]
- 42.Yeo AE, Seymour KK, Rieckmann KH, Christopherson RI. Effects of folic and folinic acids in the activities of cycloguanil and WR99210 against Plasmodium falciparum in erythrocytic culture. Ann Trop Med Parasitol. 1997;91:17–23. doi: 10.1080/00034983.1997.11813107. [DOI] [PubMed] [Google Scholar]
- 43.Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A. 1997;94:10931–6. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stumvoll M, Haring HU, Matthaei S. Metformin. Endocr Res. 2007;32:39–57. doi: 10.1080/07435800701743828. [DOI] [PubMed] [Google Scholar]
- 45.Mayer S, Daigle DM, Brown ED, Khatri J, Organ MG. An expedient and facile one-step synthesis of a biguanide library by microwave irradiation coupled with simple product filtration. Inhibitors of dihydrofolate reductase. J Comb Chem. 2004;6:776–82. doi: 10.1021/cc049953+. [DOI] [PubMed] [Google Scholar]
- 46.Summerfield RL, Daigle DM, Mayer S, Mallik D, Hughes DW, Jackson SG, Sulek M, Organ MG, Brown ED, Junop MS. A 2.13 A structure of E. coli dihydrofolate reductase bound to a novel competitive inhibitor reveals a new binding surface involving the M20 loop region. J Med Chem. 2006;49:6977–86. doi: 10.1021/jm060570v. [DOI] [PubMed] [Google Scholar]
- 47.Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ., Jr. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–6. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 48.Lyons TM. Department of Chemistry. Yale University; New Haven: 2006. p. 121. [Google Scholar]
- 49.Kamchonwongpaisan S, Quarrell R, Charoensetakul N, Ponsinet R, Vilaivan T, Vanichtanankul J, Tarnchompoo B, Sirawaraporn W, Lowe G, Yuthavong Y. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J Med Chem. 2004;47:673–80. doi: 10.1021/jm030165t. [DOI] [PubMed] [Google Scholar]
- 50.Ponmee N, Chuchue T, Wilairat P, Yuthavong Y, Kamchonwongpaisan S. Artemisinin effectiveness in erythrocytes is reduced by heme and heme-containing proteins. Biochem Pharmacol. 2007;74:153–60. doi: 10.1016/j.bcp.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chitnumsub P, Yuvaniyama J, Vanichtanankul J, Kamchonwongpaisan S, Walkinshaw MD, Yuthavong Y. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductasethymidylate synthase from Plasmodium falciparum. Acta Crystallogr D Biol Crystallogr. 2004;60:780–3. doi: 10.1107/S0907444904001544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.