Summary
Brain-derived neurotrophic factor (BDNF), a neurotrophin, enhances the survival and differentiation of several classes of neurons in vitro. To determine its essential functions, we have mutated the BDNF gene. Most homoxygote mutants die within 2 days after birth, but a fraction live for 2–4 weeks. These develop symptoms of nervous system dysfunction, including ataxia. The BDNF mutant homoxygotes have substantlaliy reduced numbers of cranlal and spinal sensory neurons. Although their central nervous systems show no gross structural abnormalities, expression of neuropeptlde Y and calcium-binding proteins is altered in many neurons, suggesting they do not function normally. In contrast with mice lacking the BDNF receptor TrkB, motor neurons appear normal in the BDNF mutant.
Introduction
Neurotrophins are important regulators of the formation and function of vertebrate nervous systems (reviewed by Korsching, 1993). Innervation of target organs is necessary for the survival of many types of neurons, and neurotrophins synthesized and secreted by target organs are crucial mediators of these interactions. In vitro and in vivo, individual neurotrophins promote the survival and differentiation of specific types of neurons. At present, four neurotrophins have been described: nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT-3), and NT-4/NT-5. The second to be discovered, BDNF was purified and cloned by Barde and coworkers (see Leibrock et al., 1989). Neuronal populations sensitive to BDNF include neural crest– and placode-derived sensory neurons, dopaminergic neurons in the substantia nigra, basal forebrain cholinergic neurons, hippocampal neurons, cerebellar granule cells, and retinal ganglion cells (reviewed by Korsching, 1993). Degeneration of motor neurons, nigral dopaminergic neurons, and basal forebrain cholinergic neurons is seen, respectively, in amyotrophic lateral sclerosis, Parkinson’s disease, and Alzheimer’s disease. This makes identification of the in vivo functions of BDNF of special interest.
The neurotrophins interact with two types of cell surface receptors (reviewed by Chao, 1992). All of the neurotrophins bind with similar affinity to the low affinity NGF receptor, p75LNGFR. A family of three receptor tyrosine kinases, designated Trk, constitutes the second class of neurotrophin receptors. NGF specifically activates TrkA; BDNF and NT-4/NT-5 activate TrkB; NT-3 activates TrkC and in addition activates TrkB and possibly TrkA in some, but not all, cellular contexts (see Ip et al., 1993b). Recently, mice with a disrupted trkB gene have been characterized (Klein et al., 1993). Homozygotes die shortly after birth. Anatomical deficiencies include substantial losses of primary sensory neurons in the trigeminal and dorsal root ganglia and of motor neurons in the facial nucleus and lumbar spinal cord.
BDNF and its receptor, TrkB, are widely expressed in the embryonic, postnatal, and adult central and peripheral nervous systems (e.g., Hofer et al., 1990; Schecterson and Bothwell, 1992; Merlio et al., 1992). Given these observations and the many activities of BDNF described in vitro and in lesioned animals, BDNF is likely to have multiple functions. To determine the requirements for BDNF in vivo, we have created a BDNF null mutation. As described below, mice homozygous for this mutation have profound deficits in multiple populations of peripheral sensory neurons. In contrast with homozygous trkB mutant mice, they do not have obvious deficits in motor neuron populations. Although the gross anatomy of the central nervous system of BDNF mutants appears normal, the expression of several neuronal markers is reduced, suggesting that BDNF is essential for normal differentiation of the central nervous system.
Results
Generation of Mice Having a Disrupted BDNF Gene
We used the DNA construct shown in Figure 1 to delete most of the sequence encoding the mature BDNF hormone (Leibrock et al., 1989) as well as sequences required for appropriate prohormone folding and processing (Suter et al., 1991). The deletion encompasses 4 of 6 cysteine residues believed to be critical in determining the structure of neurotrophins (see Figure 1A; McDonald et al., 1991) and residues absolutely required for prohormone processing and for interactions of mature neurotrophins with Trk receptors and with p75LNGFR (Suter et al., 1991; Ibañez et al., 1992, 1993). Thus, the mutation shown in Figure 1 is almost certainly a null mutation.
Figure 1. Targeted Disruption of the BDNF Locus.
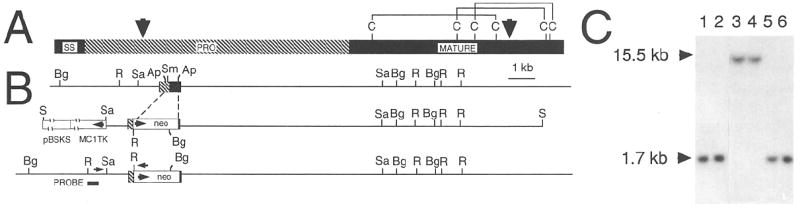
(A) Schematic showing the portion of the BDNF prohormone that is deleted and replaced by the phosphoglycerate kinase (PGK)–neo selectable marker in the BDNPOneo mutant. The signal sequence (SS), pro, and mature regions are indicated by closed, hatched, and closed bars, respectively. The two arrowheads indicate the limits of the deletion. The Cs and lines indicate the positions of cysteine residues and disulfide bonds in the mature portion of BDNF, as predicted from the structure of NGF (McDonald et al., 1991).
(B) Schematic showing the strategy used to disrupt the BDNF gene. The thin horizontal lines represent murine genomic DNA. The exon encoding BDNF is indicated by a box. The portion of the exon replaced by the PGK–neo cassette (arrow indicates direction of transcription) is indicated by broken lines. The targeting construct is shown as linearized for transfection with the thymidine kinase gene used for negative selection (open box labeled MCITK; arrow indicates direction of transcription) and pBluescript KS vector (open box labeled pBSKS). The probe used for Southern blot analysis is indicated by the closed bar. Arrows above the schematic of the mutated BDNF gene show the approximate positions of oligonucleotide primers used for polymerase chain reaction. Restriction site abbreviations: R, EcoRl; Sa, Sacl; Ap. Apal; Bg, Bglll; S, Sall.
(C) Southern blot analysis. The probe was derived from sequences outside the targeting construct (see [B]). A single restriction fragment of the size expected to result from introduction of an EcoRl restriction site into the BDNF locus by homologous recombination with the targeting construct is detected in mice homozygous for both mutant alleles (1.7 kb in mutants, 15.5 kb in wild type). EcoRl digested genomic DNA samples from mice homozygous for BDNPneo1 (lanes 1 and 2) wild type (lanes 3 and 4) and BDNFneo2 (lanes 5 and 6).
Germline transmission of the mutation in the BDNF gene was obtained with two independent embryonic stem cell lines. DNA blot analysis indicates that in both cases a homologous recombination event replaced the BDNF coding exon with a deleted exon, as schematized in Figure 1B. As shown in Figure 1C, hybridization with a probe derived from DNA outside of the targeting construct detects a novel restriction fragment in genomic DNA from pups carrying the mutant alleles. Hybridization with other probes further indicates that both alleles, referred to as BDNPneo1 and BDNFneo2, are replacement events whose structure is shown in Figure 1.
BDNF Mutant Mice Die within the First Few Weeks of Life and Display Severe Movement Abnormalities
Mice heterozygous for either BDNPneo allele show no overt abnormalities, are fertile, and have been alive for over 1 year. Mating of heterozygotes for either allele results in the birth of homozygous mutant offspring at a frequency of approximately 25% (38 of 154), indicating that BDNF is not required for survival to term. At birth, homozygous mutants are the same size as their littermates, respond to a foot pinch, and display motor activity. Most die within 48 hr, but a few survive for up to 25 days. These lag behind littermates in growth, display periods of hyperactivity and inactivity, and have progressively more severe movement defects with age. In older animals, abnormal behaviors include difficulty in righting and spinning, ataxia, and a hunched stance, but these animals perform behaviors as complicated as grooming and chewing. The older animals have reduced and irregular breathing rates. Both mutant alleles display the same behavioral phenotype and, in all cases in which it has been examined, have identical phenotypes in the analysis that follows. Examination has revealed no apparent abnormalities outside of the nervous system.
Motor Neurons and Muscle Innervation Appear Normal in the BDNFneo Mutant
Given the movement abnormalities of the BDNPneo mutant, the motor neuron deficits in the trkB BDNF receptor mutant (Klein et al., 1993) and the ability of BDNF to support motor neuron survival, we carefully examined multiple motor neuron populations for any abnormalities (Figure 2). No significant differences were found in the appearance of the brain stem (Figures 2A–2F) or spinal cord (Figures 2G–2I) or in the number of motor neurons present in discrete populations within these structures (Table 1). Brain stem motor neurons in the mutant were not distinguishable in size from those present in wild-type animals (Table 1; Figures 2B and 2E) and express the enzyme choline-o-acetyltransferase (ChAT) (Figure 2F). Mutants have apparently normal muscle innervation, as shown in Figure 2J. As shown in Figure 2K, synapse elimination seems to occur normally in the BDNPneo mutant animals since at P15 single endplates receive single motor axons (850 endplates were analyzed). Our observations indicate that many aspects of motor neuron function are normal in the BDNPneo mutant.
Figure 2. Motor Neurons and Muscle Innervation.
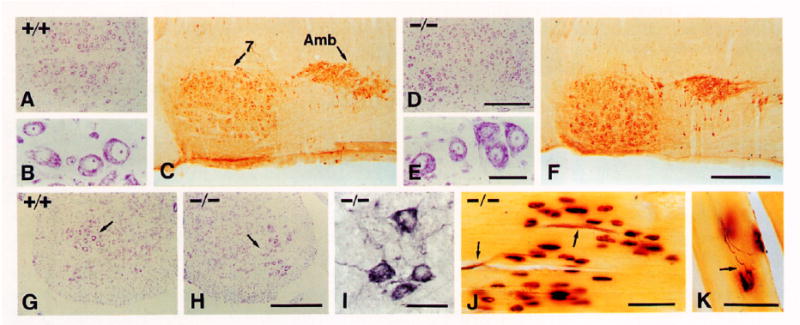
(A, B, D, E, G, and H) Cresyl violet (Nissl) stain. (C, F, and I) ChAT immunocytochemistry. (J and K) Cholinesterase and silver staining for muscle endplates and silver nitrate technique for axons in skeletal muscle.
(A–C) Facial (7) and ambiguus (Amb) nuclei of +/+ P17 mice.
(D–F) Same nuclei of −/− P17 mice.
(G and H) Ventral horns of lumbar spinal cord in +/+ (G) and −/− (H) P15 mice. Note normal appearance of Nissl-stained motor neurons (arrows) in −/− mice.
(I) ChAT-positive motor neurons in the spinal cord of a −/− P16 mouse.
(J and K) Skeletal muscle endplate regions of −/− P15 mice showing the presence of motor axons (arrows) and endplates and normal innervation by one fiber (arrow in [K]) per endplate.
Scale bars, (A and D) 300 μm; (B and E) 50 μm; (C and F) 400 μm; (G and H) 300 μm; (I) 50 μm; (J) 50 μm; (K) 50 μm.
Table 1.
Numbers of Neurons in Motor Populations
| Nucleus/Regiona | N | Wild Type | BDNFneo |
|---|---|---|---|
| Oculomotor (III) | 2 | 494 ± 1 | 464 ± 3 |
| Trochlear (IV) | 2 | 106 ± 14 | 112 ± 11 |
| Trigeminal (V) | 5 | 947 ± 84 | 981 ± 130 |
| Facial (VII) | 5 | 2129 ± 208 | 2174 ± 184 |
| Hypoglossal (XII) | 3 | 1138 ± 32 | 1185 ± 114 |
| Spinal cord (L1–L6) | 3 | 1436 ± 139 | 1364 ± 97 |
| Dorsal motor nucleus of the vagus (X)b | 3 | 1429 ± 192 | 1440 ± 56 |
No differences are found in neuron numbers between wild-type and mutant animals. Raw counts (expressed as mean ± SEM) were obtained by counting clear nucleoli in all sections and no correction factor was applied. Nucleolus size was the same in wild type (3.5 ± 1.1 μm; n = 72) and mutant (3.6 ± 0.8 μm; n = 72). Cell body size of neurons belonging to the trigeminal and facial nuclei were also similar: wild type, 31.5 ± 0.6 μm (n = 63); BDNFneo, 29.1 ± 0.5 μm (n = 74). N is the number of nuclei counted per genotype. The number of the cranial nerve with which each nucleus is associated is shown in parentheses.
Oculomotor, trochlear. and hypoglossal nuclei innervate head muscles of myotomal origin, equivalent to the body musculature innervated by spinal motor neurons. Trigeminal and facial nuclei innervate muscles derived from branchial arches.
The dorsal motor nucleus of the vagus contains the parasympathetic preganglionic neurons of the vagal system.
BDNPneo Mutants Exhibit Daficlts in Many Sensory Neuron Populations
In virtually every sensory ganglion examined in BDNPneo mutants, there are deficits in size or cell number (Tables 2-4). Among the cranial ganglia, the reductions in the vestibular (Figures 3A and 3B) and petrosal–nodose (Figures 3C and 3D) ganglia are especially interesting in the context of the mutant phenotype. Vestibular ganglion function is required for normal balance and locomotion. Sensory neurons located in the petrosal and nodose ganglia convey sensory information from visceral organs to nuclei located in the brain stem and are important in regulation of the parasympathetic vagal system and in limbic function. Two populations of sensory neurons, derived entirely from the neural crest (see Scott, 1992) were also reduced: the trigeminal mesencephalic nucleus and dorsal root ganglia (Table 3). Moreover, counts of myelinated axons in dorsal roots showed that there is a reduction in myelinated axon number in the BDNPneo mutant (Table 4). Myelinated axons mostly innervate muscles spindles and Golgi–tendon organs, proprioceptors important in position sense, and mechanoreceptors (see Scott, 1992). Muscle spindle afferents appear to be unaffected in the BDNPneo mutant since there was no reduction in the number of muscle spindles, which are dependent on these neurons for their differentiation (thigh semitendinosus muscle for +/+, 15; for −/−, 14; n = 2). This is consistent with the normal presence of large lightly stained neurons in the dorsal root ganglia (Figures 3E and 3F). The other major population of sensory neurons having myelinated axons, the mechanoreceptors, are likely to comprise at least in part the sensory population missing in the BDNPneo mutant.
Table 2.
Effects on Volumes of Sensory and Sympathetic Ganglia
| Cranial Ganglia | Wild Type | BDNFneo | Reduction (%) |
|---|---|---|---|
| Trigeminal (V) | 129.0 ± 8.8 (3) | 109.0 ± 10.6 (3) | 16a |
| Geniculate (VII) | 2.0 ± 0.4 (3) | 1.2 ±0.1 (3) | 40a |
| Vestibular (VIII) | 9.3 ± 0.8 (3) | 1.2 ±0.1 (2) | 87b |
| Superior–jugular (IX–X) | 4.9 ± 0.7 (3) | 4.6 ± 2.2 (3) | No significant reduction |
| Petrosal–nodose (IX–X) | 11.2 ± 0.4 (3) | 6.3 ± 0.4 (3) | 44b |
| Superior cervical (sympathetic) | 24.6 ± 2.8 (3) | 25.3 ± 3.3 (3) | No significant reduction |
The volume in 106 μm3 (mean ± SEM) of different sensory cranial ganglia and of the superior cervical ganglion (sympathetic) were calculated in P0 heads. The geniculate, petrosal–nodose, and vestibular ganglia, all of placodal origin, are substantially reduced in volume. The trigeminal ganglion, which is of mixed neural crest and placode origin, is moderately reduced in volume. No significant reduction was found in the volume of the superior–jugular sensory ganglia that are completely neural crest derived or in the superior cervical ganglion (sympathetic). The sampling number is shown in parentheses. Student’s one-tailed t test determined the significance levels.
p < 0.05.
p < 0.001.
Table 4.
Effects on Numbers of Myelinated Sensory Axons
| Dorsal Root Level | Wild Type | BDNFneo | Reduction (%) |
|---|---|---|---|
| T1 | 1926 ± 150 (3) | 1475 ± 86 (2) | 23a |
| T10 | 683 ± 46 (3) | 507 ± 44 (2) | 26b |
| L5 | 2336 ± 108 (4) | 1475 ± 55 (4) | 37c |
Counts of myelinated axons in dorsal roots from segments T1, T10, and L5 of P15 animals. The sampling number is shown in parentheses. Student’s one-tailed t test determined the significance levels.
p < 0.10.
p < 0.05.
p < 0.001.
Figure 3. Sensory Populations.
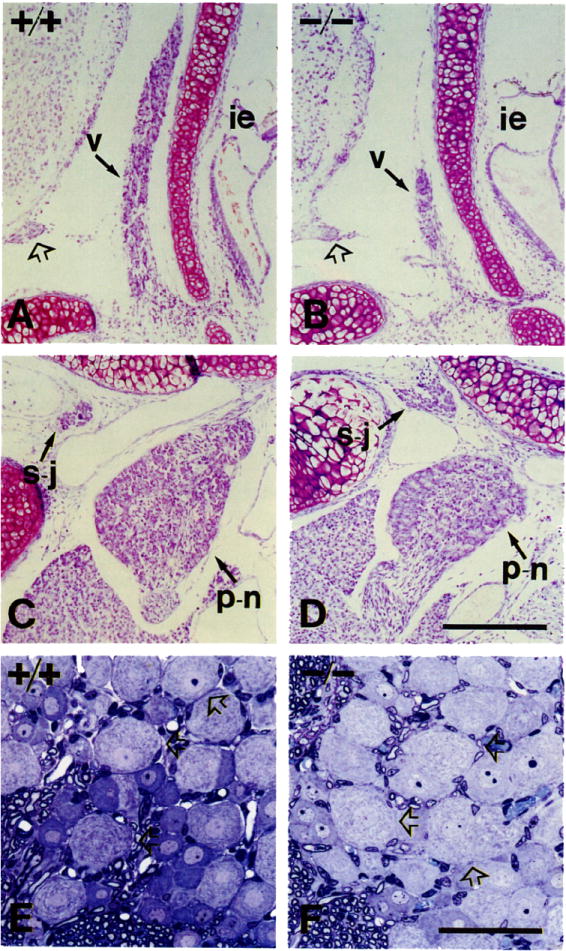
(A and B) Vestibular ganglia (v) of P0 +/+ (A) and −/− (B) mice. Symbols: ie, inner ear; open arrow, acoustic nerve.
(C and D) Superior–jugular (s–j) and petrosal–nodose (p–n) ganglia of +/+ and −/− mice. The pair petrosal–nodose (placode derived) is reduced in volume in mutant animals. The intracranial pair superior–jugular (neural crest derived) appears unaffected.
(E and F) PI5 mice L5 dorsal root ganglia sectioned in araldite and stained with toluidine blue. Note the presence of large lightly stained neurons in the mutant (open arrows), consistent with the presence of normal numbers of muscle spindles in the thigh skeletal muscles.
Scale bars, (A–D) 200 μm; (E and F) 50 μm.
Table 3.
Effects on Numbers of Sensory Neurons
| Sensory Population | Wild Type | BDNFneo | Reduction (%) |
|---|---|---|---|
| Trigeminal | 36,122 ± 4,267 (3) | 26,433 ± 4,140 (3) | 27a |
| Petrosal–nodose | 3,884 ± 95 (3) | 2,222 ± 135 (3) | 43b |
| Trigeminal mesencephalic nucleus | 528 ± 24 (5) | 323 ± 17 (5) | 39a |
| L4 dorsal root ganglion | 4,120 ± 357 (2) | 2,733 ± 160 (2) | 34a |
Neuron numbers (mean ± SEM) were estimated for the trigeminal (P0), the petrosal–nodose (P0), the trigeminal mesencephalic nucleus (P15), and L4 dorsal root ganglion (P15). The trigeminal mesencephalic nucleus nucleolus size (wild type, 3.4 ± 0.9 μm [n = 65]; BDNFneo, 3.5 ± 1.1 μm [n = 48]) and cell body size (wild type, 32.5 ± 0.8 μm [n = 54]; BDNFneo [−/−], 31.2 ± 0.6 μm [n = 43]) were not affected by the genotype. The sampling number is shown in parentheses. Student’s one-tailed t test determined the significance levels.
p < 0.05.
p < 0.001.
Many Aspects of Braln Development Occur In the BDNFneo Mutant
Considering the ability of BDNF to support many different types of central nervous system neurons in vitro and its widespread expression in the brain, we have uncovered surprisingly few deficits in the brains of mutants (see Table 5). The cerebellum has a grossly normal histological appearance (Figures 4A and 4B), and Purkinje cells have well-differentiated dendritic arbors (Figures 4C–4E). An external granule layer is apparent in P17 mutants but not in P17 wild-type mice (Figures 4A and 4B) but was not apparent in a P23 BDNPneo mutant (data not shown), indicating that granule cell migration is retarded, but not prevented, in the mutant. As determined by anti-tyrosine hydroxylase (TH), there appears to be a normal complement of dopaminergic neurons in the substantia nigra (Figures 4F and 4G), and their projection to the striatum appears normal (see Table 5). Basal forebrain cholinergic neurons are present and express ChAT (Figures 4H and 4I), p75LNGFR, and TrkA (Table 5). In Nissl-stained sections of mutant neocortex, all six layers are present and intracortical organization is at least partially preserved, as shown by the presence of barrels in the somatosensory cortex (data not shown). The hippocampal formation appears generally normal in mutants, as determined by examination of Nissl-stained sections and immunocytochemistry (Table 5). Expression of the neurotransmitter γ-aminobutyric acid (GABA) appeared normal in the cerebral cortex and hippocampal formation.
Table 5.
Analysis of the Brain of BDNFneo Mutants
| Brain Structure | Staining | N | Resulta | Remarks |
|---|---|---|---|---|
| Cerebellum | Nissl | 5 | Normal | Delayed granule cell migrationb (Figure 4B) |
| Calbindin | 3 | Normal | Normal Purkinje cell morphology (see Figure 4E) | |
| Parvalbumin | 3 | Normal | ||
| PEP-19 | 4 | Normal | ||
| Substantia nigra | TH | 6 | Normal | See Figures 4F and 4G |
| Calbindin | 3 | Reduced | Innervating fibers from striatum (see striatum) | |
| Parvalbumin | 3 | Reduced | Slight reduction in cells stained | |
| Striatum | GABA | 3 | Normal | Medium-sized spiny neurons |
| ChAT | 6 | Normal | Interneurons | |
| TrkA | 5 | Normal | Interneurons | |
| TH | 6 | Normal | Innervating fibers from substantia nigra | |
| NPY | 3 | Normal | Neurons and fibers | |
| Calbindin | 3 | Reduced | Substantially reduced (see Figures 6H and 6I) | |
| PEP-19 | 3 | Normal | Most neurons stained | |
| Basal forebrain | ChAT | 6 | Normal | See Figures 4H and 4I |
| TrkA | 4 | Normal | ||
| LNGFR | 5 | Normal | ||
| Hippocampal formation | Nissl | 5 | Normal | |
| GABA | 3 | Normal | Interneurons | |
| ChAT | 6 | Normal | Innervating fibers from basal forebrain | |
| NPY | 3 | Reduced | Interneurons (Figures 5C and 5D; see cerebral cortex) | |
| Catbindin | 3 | Reduced | Interneurons (see cerebral cortex) | |
| Parvalbumin | 3 | Reduced | Interneurons (Figures 6D–6G; see cerebral cortex) | |
| PEP-19 | 3 | Normal | ||
| Cerebral cortex | Nissl | 5 | All layers | Reduction in thicknessb |
| GABA | 3 | Normal | Interneurons | |
| NPY | 3 | Reduced | 65% ± 10% (see Figures 5A and 5B)c | |
| Calbindin | 3 | Reduced | 83% ± 13%c | |
| Parvalbumin | 3 | Reduced | 50% ± 11% (see Figures 6A–6C)c | |
| PEP-19 | 3 | Normal |
Brain structures containing neuronal populations reported to be affected by BDNF or to express its receptor, TrkB. The table shows the results obtained for P15–P20 animals after conventional staining with cresyl violet (Nissl) or immunocytochemical staining using antibodies to ChAT, GABA, TH, NPY, calbindin, parvalbumin, PEP-19, TrkA, and pZSLNGFR (LNGFR). The number of animals analyzed is indicated (N). An equal number of age-matched wild-type littermates, processed at the same times, served as controls.
Normal means normal histological appearance in Nissl-stained sections or apparently normal presence and labeling intensity of immunoreactive neurons, fibers, or both. Reduced means that a reduction was readily apparent by eye examination.
Possible nutritional deficits also involved.
To document reductions in staining intensity further, immunoreactive neurons in the cerebral cortex and hippocampal formation were counted in equivalent sections from wild-type and mutant animals. The percentage of positive cells found in mutant compared with wild-type brains is shown (mean ± SEM).
Figure 4. The Cerebellum, Substantia Nigra, and Basal Forebrain.
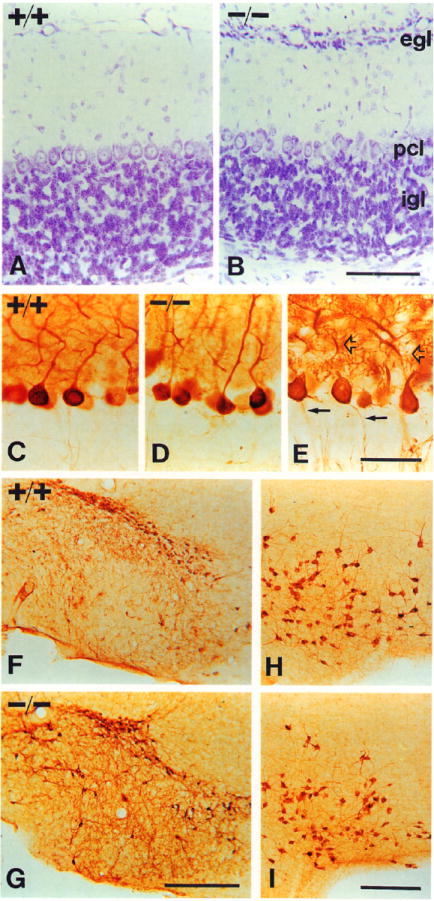
All panels are photomicrographs of PI7 littermates.
(A and B) Cresyl violet stain.
(A) Cerebellum of a +/+ mouse.
(B) Cerebellum of a −/− mouse. Note the persistence of an external granule cell layer, indicating a delay or deficit in granule cell migration. Abbreviations: egl, external granule cell layer; pcl, Purkinje cell layer; igl, inner granule cell layer.
(C and D) Purkinje cell morphologies revealed by PEP-19 immunocytochemistry in +/+ (C) and −/− (D) mice.
(E) Purkinje cells of a −/− mouse stained for calbindin. Note normal dendritic (open arrows) and axonal (closed arrows) morphology.
(F and G) Substantia nigra of +/+ (F) and −/− (G) mice, immunostained for TH. Note similar numbers and appearance of TH-expressing neurons.
(H and I) Basal forebrain of +/+ (H) and −/− (I) mice, stained with ChAT antibodies. Note normal numbers of neurons and expression levels of ChAT in the mutant animal.
Scale bars, (A and B) 100 μm; (C–E) 50 μm; (F and G) 400 μm; (H and I) 200 μm.
In cultured cortical neurons, expression of neuropeptide Y (NPY) is enhanced by BDNF (Nawa et al., 1993). In mutant animals, expression of NPY is reduced at 15–20 days of age in all layers of the cerebral cortex, particularly in layer VI, and in the hippocampus (Figure 5). Since NPY is expressed by GABAergic interneurons in these structures and GABAergic neurons are present in mutants (Table 5) BDNF very likely affects peptide expression, but not survival, of these cells. Notably, in another forebrain structure, the striatum, expression of NPY appears normal in mutants (Table 5).
Figure 5. NPY Expression.
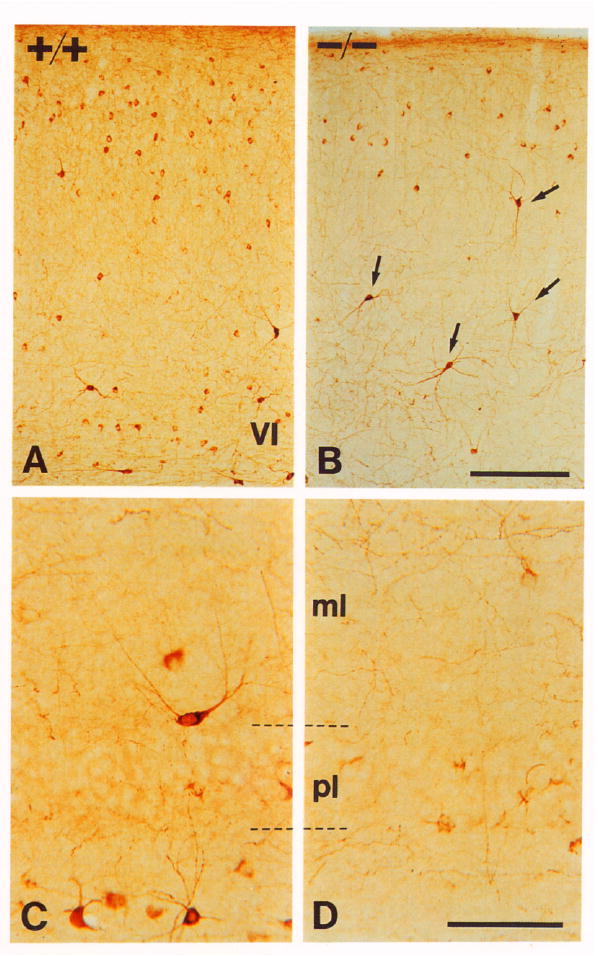
(A and B) Cerebral cortex stained with NPY antibodies of +/+ (A) and −/− (B) littermates. Note that there is some reduction in numbers of intermediate to small neurons expressing NPY in the mutant animal. However, the large neurons expressing high levels of NPY (arrows) persist.
(C and D) CA1 region of the hippocampus of P17 wild-type (C) and mutant (D) animals. The number of immunoreactive interneurons is reduced in all areas of the hippocampal formation Note that cells in mutant animals express very low levels of peptide. Abbreviations: pl, pyramidal cell layer; ml, molecular layer.
Scale bars, (A and B) 200 μm; (C and D) 50 μm.
Expresslon of Calcium-Binding Proteins Is Reduced in Discrete Brain Locstlons In the BDNFneo Mutant
The calcium-binding proteins calbindin-D28K and parvalbumin, thought to have roles in buffering intracellular calcium, are expressed in subpopulations of neurons within the brain (reviewed by Baimbridge et al., 1992). The expression of calbindin and parvalbumin are reduced in specific brain regions in the BDNPneo mutant (see Table 5). Parvalbumin expression is reduced in multiple forebrain regions in the BDNPneo mutant. In the cerebral cortex, many fewer parvalbumin-expressing cells are visible (Figures 6A and 6B); these are almost absent in frontal and visual cortices (Figure 6C). Parvalbumin staining is also substantially reduced in cells of the hippocampus of the mutant (Figures 6D–6G). Parvalbumin is expressed by a subset of cortical and hippocampal GABAergic neurons (Baimbridge et al., 1992). These cells were apparently normal in abundance in the cerebral cortex and hippocampus (Table 5). Thus, BDNF most likely is important in regulating differentiation, but not survival, of these neurons.
Figure 6. Parvalbumin and Calbindin Expression.
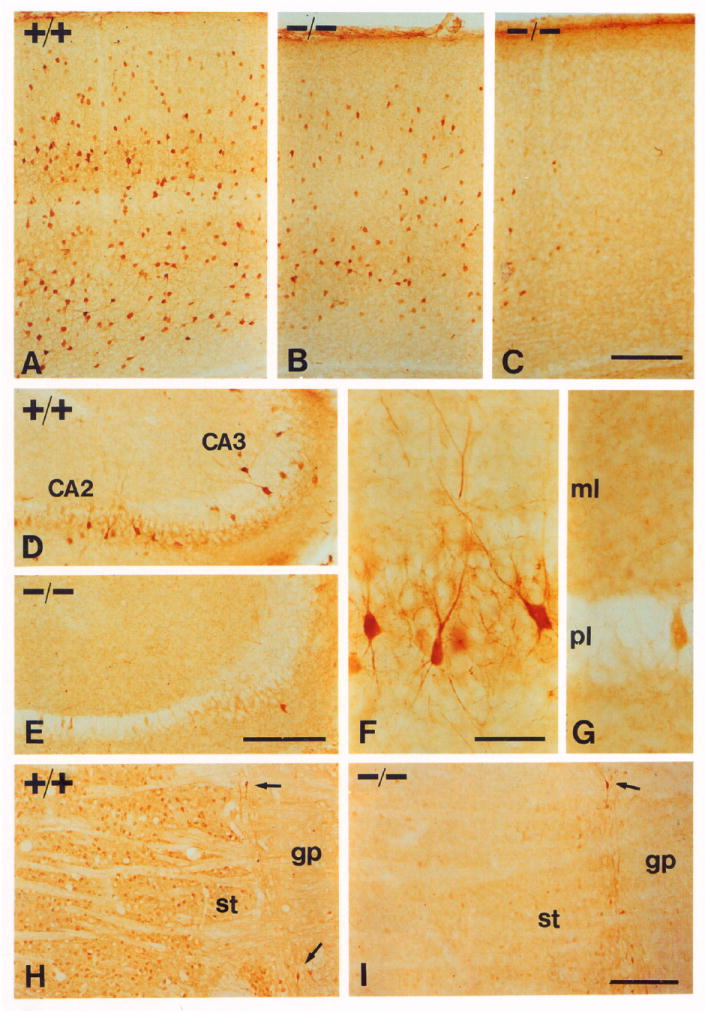
Sections from P17 mice stained with parvalbumin (A–G) or calbindin-D28K (H and I) antibodies.
(A–C) Parvalbumin staining in the cerebral cortex.
(A) Somatosensory cortex from a wild-type mouse.
(B) The same area from a −/− mouse.
(C) Occipital visual cortex from a −/− mouse.
(D–G) Hippocampus sections from a +/+ mouse (D and F) and from a −/− mouse (E and G), documenting absence of neurons expressing parvalbumin in a −/− mouse. Abbreviations: ml, molecular layer; pl, pyramidal cell layer.
(H and I) Calbindin staining in the striatum (st) of +/+ (H) and −/− (I) mice, showing that the reduction is almost complete. Positive neurons between the globus pallidus (gp) and the striatum (arrows) appear the same.
Scale bars, (A–E) 200 μm; (F and G) 50 μm; (H and I) 200 μm.
BDNF stimulates expression of calbindin in cultured hippocampal neurons (see Ip et al., 1993a). The expression of calbindin appears generally normal in many brain regions in the BDNFneo mutant (Table 5). However, in the cerebral cortex and hippocampus, calbindin expression is reduced in the mutant. In addition, its expression is substantially diminished in the striatum and striatal projection to the substantia nigra (Figures 6H and 6I; data not shown). The vast majority of calbindin-positive cells in the rat striatum are medium-sized spiny GABAergic neurons (Celio, 1990). In BDNFneo mutant animals, the striatum appears histologically normal and GABAergic neurons appear to be present at normal density (Table 5). Thus, it is likely that BDNF is necessary for differentiation, but not survival, of these neurons. In contrast with the reductions of calbindin and parvalbumin expression in specific brain structures, the expression of a putative calcium-binding protein, PEP-19 (Ziai et al., 1988), appeared normal in these and all other structures (Table 5).
Discussion
Several conclusions can be drawn from the present work. First, BDNF is essential for normal development of multiple cell types in the nervous system, including neurons in multiple sensory ganglia, the cerebral cortex, the hippocampus, and the striatum. Second, BDNF is not essential for all functions suggested by previous observations. For example, analysis of the BDNFneo homozygotes suggests that BDNF is not essential for survival of motor neurons, cerebellar granule cells, dopaminergic neurons in the substantia nigra, cholinergic neurons in the basal forebrain, or retinal ganglion cells (latter not shown). Finally, our data regarding reductions in neuropeptide and calcium-binding protein expression suggest that BDNF regulates postnatal brain differentiation, as suggested previously by BDNF and trkB mRNA expression studies.
As shown above, disruption of the BDNF gene results in a profound loss of sensory neurons. Sensory neuron populations derived from ectodermal placodes are particularly affected, consistent with previous observations on the activity of BDNF on a large proportion of these neurons in primary culture (reviewed by Korsching, 1993). Cell losses in the vestibular ganglion were particularly striking and are consistent with previous observations concerning BDNF expression in the sensory epithelia innervated by vestibular neurons and the responsiveness of these cells to BDNF in vitro (see, e.g., Pirvola et al., 1992). Vestibular deficits are likely to explain in part the abnormal behavior of BDNFneo mutant mice, including the observed difficulty in remaining upright, in ataxia, and in spinning. The cell losses in the petrosal and nodose ganglia could explain the lethality of this mutation. Sensory neurons in these ganglia relay information from the heart, lungs, great vessels, and gut to the central nervous system. This sensory information is used in regulation of heart rate, blood pressure, respiratory rate, bronchodilation, gut motility, and many other aspects of visceral function. The observed deficits in cranial and dorsal root ganglia are also likely to affect the viability of the mutant. Sensory neurons in these ganglia are extraordinarily diverse, differing in sensory modality and many other properties (see Scott, 1992). Our studies suggest that muscle spindle afferents are unaffected in the BDNPneo mutant but that mechanoreceptors are likely to be affected.
Effects of BDNF addition on motor neurons in vitro and in vivo have suggested strongly that BDNF is a physiological regulator of their survival and development (see Korsching, 1993). Many fewer motor neurons are present in mice lacking the BDNF receptor TrkB (Klein et al., 1993). Despite these suggestive precedents, in homozygous BDNPneo mice, all examined populations of motor neurons appear to be present in normal numbers and to have differentiated normally by several criteria. Similarly, the BDNPneo mutant does not have the substantial morphological abnormalities in brain structures that might have been predicted from earlier data. Among many possibilities, NT-3 or NT-4/NT-5, both of which have been shown to activate the TrkB BDNF receptor (see Ip et al., 1993b), may compensate for the loss of BDNF.
The observed reductions in neuropeptide and calcium-binding protein expression in specific regions of the BDNPneo mutant brain suggest that BDNF may directly regulate the differentiation of many brain neurons. Expression of NPY was normal in one forebrain structure, the striatum, but reduced in cerebral cortex and hippocampus of 15-, 17-, and 20-day-old BDNPneo mutants and did not appear to rise significantly in the older animals, suggesting that maturation of neuropeptide expression is not simply delayed, but is permanently affected.
Calcium-binding proteins may be required in normal quantities for normal neuronal functioning since intracellular calcium regulates many aspects of neuronal metabolism (Baimbridge et al., 1992). The deficiency in calbindin expression in the BDNPneo mutant striatum may result in abnormal function in the striatonigral pathway, which could account for some of the observed abnormal motor behaviors. Parvalbumin expression was specifically reduced in the cerebral cortex (with especially severe region-specific reductions) and in the hippocampus of BDNFneo mutants. Parvalbumin is localized to a subset of GABAergic neurons in the cerebral cortex and hippocampus. In hippocampus, expression levels correlate with high metabolic and firing rates (see Baimbridge et al., 1992). If, as seems likely, parvalbumin modulates GABAergic neuron function by buffering calcium, the observed deficits may prevent these neurons from functioning normally.
In conclusion, the phenotype of the BDNFneo mutant mouse demonstrates that this factor is absolutely required for development of some peripheral sensory neurons. In contrast, requirements for BDNF may be more modest in the brain. Anterior brain structures have been added progressively in vertebrate evolution. The phenotype of the BDNFneo mutant suggests that the events underlying differentiation of the brain involve more subtlety than those regulating more ancient parts of the vertebrate nervous system.
Experimental Procedures
DNA Constructs
The human BDNF gene (Jones and Reichardt, 1990) was used to make a probe for screening a tAE5/129 mouse genomic library in EMBL3, provided by M. Burmeister. Genomic DNA fragments were used to create constructs in which the BDNF coding exon is interrupted by the PGK–neo selectable marker. PMC1–tk was placed at one end of the genomic fragment for use in the positive–negative selection scheme. The Smal restriction site in the BDNF coding exon (Hofer et al., 1990) is used as 0.0 in description of the construct. In a series of cloning steps, PGK–neo was inserted between genomic DNA fragments spanning from the Sacl site at −1.1 kb to the Apal site at −0.1 kb and from the Apal site at +0.4 kb to a Sall site at +14 kb (from the EMBL3 vector polylinker) by blunting the Apal mouse genomic restriction sites and the EcoRl and Hindlll restriction sites flanking PGK–neo. PMBD/KO5 was created by transferring the resulting genomic fragments flanking the PGK–neo gene to pBS/MC1–tk. pBS/MC1–tk was created by inserting a Klenow-blunted Hindlll–Sall fragment from pMC1–tk into the blunted Kpnl site of pBluescript KS(−).
Cell Culture, Embryo Microinjection, and Animal Husbandry
The D3 embryonic stem cell line was cultured in embryonic stem medium on mitotically inactivating SNL76/7 cells (Joyner, 1993). Plasmid pMBDIKO5 was linearized with Sall and electroporated with D3 cells. After 24 hr of growth in nonselective medium and 10 days growth in selective medium (180 μg/ml geneticin and 2 μM gangcyclovir, respectively) colonies were picked and transferred to feeder layers on 96-well plates; 384 clones were picked from plates resulting from transfection with pMBDIKO5. Clones with homologous recombinants were identified by PCR analysis, verified by Southern analysis, and used to inject C57BL/6 blastocysts as described (Joyner, 1993). Germline transmission was obtained by mating male mosaics to C57BL/6 females.
Histological Methods
Animals were anesthetized and fixed by perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer. P0 heads and P15 brains and spinal cords were dehydrated in ethanol, embedded in paraffin, serially sectioned at 7 μm, and stained with 0.1% cresyl violet. The volume of each ganglion was measured using a morphometric analysis program (3DHVEM, HVEM Laboratory, Boulder, Colorado). The number of neurons was estimated by calculating cell density in 1 out of each 10 sections. In P15 animals, all motor and trigeminal mesencephalic nucleus neurons having a clear nucleolus were counted in every section and no correction factor was applied. Dorsal roots were postfixed with 2% osmium tetroxide, dehydrated in ethanol, embedded in araldite, sectioned at 1 μm, and stained with 1% toluidine blue.
For immunocytochemistry in P15–P20 animals, sagittal sections at 60 μm were obtained with a vibratome and collected in TBS (10 mM Tris buffer [pH 7.5] and 150 mM NaCI). Sections were incubated in 3% hydrogen peroxide and 10% methanol in TBS and blocked in a TBS solution containing 0.4% Triton X-100, 1% glycine, 3% bovine serum albumin, and 10% normal goat serum and left overnight at 4°C in the same blocking solution containing the primary antibody: polyclonal antibodies against ChAT at a dilution of 1:7,500 to 1:10,000; against GABA (Incstar), 1:3,000; against NPY (Incstar), 1:3,000; against PEP-19 (Ziai et al., 1988), 1:30,000; against TrkA, 1:1,000 (Clary et al., submitted); and against p75LNGFR, 1:1,000 (Weskamp and Reichardt, 1991); and monoclonal antibodies against TH (Incstar), 1:2,000; and against calbindin and parvalbumin (Sigma Chemicals), 1:5,000. The ABC method (ABC elite kit, Vector Labs) was used for immunoreactivity detection. Peroxidase was reacted with 0.05% diaminobenzidine tetrahydrochloride and 0.003% hydrogen peroxide. For staining of spinal cords, they were cryoprotected with 30% sucrose, frozen with dry ice, sectioned transversally at 15 μm, and reacted as above. Muscle innervation was analyzed using a combination of silver and cholinesterase staining for axons and endplates (Namba et al., 1967).
Acknowledgments
We thank N. Killeen, D. Hanahan, G. Martin, and D. Littman for help with embryonic stem cell and mouse methodologies; D. Morrissey, A. Barton, P. Pahlavan, and I. Sauerwald for assistance; P. O’Hara, L. Prentice, M.-L. Wong, and D. Agard for equipment use and advice; A. Basbaum, D. Lowenstein, W. Mobley, J. Lavail, W. Holtzman, and D. Sretavan for helpful suggestions; and F. Gage and J. Morgan for antibodies. This work was supported by United States Public Health grant MH48200 and by the Howard Hughes Medical Institute. K. R. J. was a fellow of the American Cancer Society; I. F. is a Fulbright scholar (Ministerio de Educación y Ciencia, Spain).
References
- Baimbridge KG, Celio MR, Rogers JH. Calcium-binding proteins in the nervous system. Trends Neurosci. 1992;15:303–308. doi: 10.1016/0166-2236(92)90081-i. [DOI] [PubMed] [Google Scholar]
- Celio MR. Calbindin D-28kand parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophin receptors: a window into neuronal differentiation. Neuron. 1992;9:583–593. doi: 10.1016/0896-6273(92)90023-7. [DOI] [PubMed] [Google Scholar]
- Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9:2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez CF, Ebendal T, Barbany G, Murray RJ, Blundell TL, Persson H. Disruption of the low affinity receptor-binding site in NGF allows neuronal survival and differentiation by binding to the trk gene product. Cell. 1992;69:329–341. doi: 10.1016/0092-8674(92)90413-7. [DOI] [PubMed] [Google Scholar]
- Ibañez CF, Ilag LL, Murray-Rust J, Persson H. An extended surface of binding to Trk tyrosine kinase receptors in NGF and BDNF allows the engineering of a multifunctional pan-neurotrophin. EMBO J. 1993;12:2281–2293. doi: 10.1002/j.1460-2075.1993.tb05882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip NY, Li Y, Yancopoulos GD, Lindsay RM. Cultured hippocampal neurons show responses to BDNF, NT-3, and NT-4, but not NGF. J Neurosci. 1993a;13:3394–3405. doi: 10.1523/JNEUROSCI.13-08-03394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip NY, Stitt TN, Tapley P, Klein R, Glass DJ, Fandl J, Greene LA, Barbacid M, Yancopoulos GD. Similarities and differences in the way neurotrophins interact with the Trk receptors in neuronal and nonneuronal cells. Neuron. 1993b;10:137–149. doi: 10.1016/0896-6273(93)90306-c. [DOI] [PubMed] [Google Scholar]
- Jones KR, Reichardt LF. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci USA. 1990;87:8060–8064. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL. Gene Targeting: A Practical Approach. Oxford: IRL Press; 1993. [Google Scholar]
- Klein R, Smeyne RJ, Wurst W, Long LK, Auerbach BA, Joyner AL, Barbacid M. Targeted disruption of the trkB neurotrophin receptor gene results in nervous system lesions and neonatal death. Cell. 1993;75:113–122. [PubMed] [Google Scholar]
- Korsching S. The neurotrophic factor concept: a reexamination. J Neurosci. 1993;13:2739–2748. doi: 10.1523/JNEUROSCI.13-07-02739.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibrock J, Lottspeich F, Hohn A, Hofer M, Hengerer B, Masiakowski P, Thoenen H, Barde YA. Molecular cloning and expression of brain-derived neurotrophic factor. Nature. 1989;341:149–152. doi: 10.1038/341149a0. [DOI] [PubMed] [Google Scholar]
- McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL. New protein fold revealed by a 2.3-Å resolution crystal structure of nerve growth factor. Nature. 1991;354:411–414. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- Merlio JP, Ernfors P, Jaber M, Persson H. Molecular cloning of rat trkC and distribution of cells expressing messenger RNAs for members of the trk family in the rat central nervous system. Neuroscience. 1992;51:513–532. doi: 10.1016/0306-4522(92)90292-a. [DOI] [PubMed] [Google Scholar]
- Namba T, Nakamura T, Grob D. Staining for nerve fiber and cholinesterase activity in fresh frozen sections. Am J Clin Pathol. 1967;47:74–75. doi: 10.1093/ajcp/47.1.74. [DOI] [PubMed] [Google Scholar]
- Nawa H, Bessho Y, Carnahan J, Nakanishi S, Mizuno K. Regulation of neuropeptide expression in cultured cerebral cortical neurons by brain-derived neurotrophic factor. J Neurochem. 1993;60:772–775. doi: 10.1111/j.1471-4159.1993.tb03216.x. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Palgi J, Lehtonen E, Arumae U, Saarma M. Brain-derived neurotrophic factor and neurotrophin 3 mRNAs in the peripheral target fields of developing inner ear ganglia. Proc Natl Acad Sci USA. 1992;89:9915–9919. doi: 10.1073/pnas.89.20.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schecterson LC, Bothwell M. Novel roles for neurotrophins are suggested by BDNF and NT-3 expression in developing neurons. Neuron. 1992;9:449–463. doi: 10.1016/0896-6273(92)90183-e. [DOI] [PubMed] [Google Scholar]
- Scott SA. Sensory Neurons: Diversity, Development, and Plasticity. New York: Oxford University Press; 1992. [Google Scholar]
- Suter U, Heymach JJ, Shooter EM. Two conserved domains in the NGF propeptide are necessary and sufficient for the biosynthesis of correctly processed and biologically active NGF. EMBO J. 1991;10:2395–2400. doi: 10.1002/j.1460-2075.1991.tb07778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weskamp G, Reichardt LF. Evidence that biological activity of NGF is mediated through a novel subclass of high afflnity receptors. Neuron. 1991;6:649–663. doi: 10.1016/0896-6273(91)90067-a. [DOI] [PubMed] [Google Scholar]
- Ziai MR, Sangameswaran L, Hempstead JL, Danho W, Morgan JI. An immunochemical analysis of the distribution of a brain-specific polypeptide, PEP-19. J Histochem. 1988;51:1771–1776. doi: 10.1111/j.1471-4159.1988.tb01158.x. [DOI] [PubMed] [Google Scholar]


