Abstract
Rad60 family members contain functionally enigmatic, integral SUMO-like domains (SLDs). Intriguingly, we find that despite their divergence from SUMO, each Rad60 SLD interacts with a subset of SUMO pathway enzymes. SLD2 specifically binds the SUMO E2 conjugating enzyme (Ubc9), whereas SLD1 binds the SUMO E1 activating and E3 specificity enzymes. The molecular basis of this selectivity is revealed by our 0.97 Å crystal structure of Rad60 SLD2, which shows that apart from the conserved non-substrate SUMO:Ubc9 interface, SLD2 surface features are distinct from those of SUMO. Abrogation of the SLD2:Ubc9 FEG-motif dependent interaction results in hypersensitivity to genotoxic stress, and an increase in spontaneous aberrant replication fork-associated recombination. Our results provide a mechanistic basis for the near synonymous roles of Rad60 and SUMO in survival of genotoxic stress, and suggest unprecedented DNA damage response functions for SLDs in regulating sumoylation.
Keywords: Rad60, SUMO, Genome Stability, Ubc9
The maintenance of genome stability is critical, as genetic defects can promote tumorigenesis, aging and neurodegenerative disease 1. The genome is particularly vulnerable to spontaneous and damage-induced alterations during its replication (S phase). Replication forks can encounter various barriers to their processivity including base lesions, discontinuities in the template, transcription intermediates and tightly bound protein complexes 2,3. Thus, to ensure high-fidelity completion of replication, cells engage critical mechanisms that include cell cycle checkpoints and DNA repair 2-4.
Through their covalent conjugation to target proteins, the post-translational modifiers SUMO and ubiquitin execute key functions in suppressing toxic DNA transactions at the replication fork 5,6. SUMO and ubiquitin regulate target protein function by triggering changes in their subcellular localization, protein-protein interactions or stability 7. Conjugation of each modifier occurs via a related enzymatic cascade involving E1 (activating), E2 (conjugating) and E3 (ligase/specificity) factors 7. Besides posttranslational modification by ubiquitin, some proteins contain integral ubiquitin-like (Ubl) domains 8. These Ubl-domain proteins interface with and regulate factors involved in ubiquitin metabolism, thereby influencing many cellular processes including DNA repair 8. Notably, analogous roles for a SUMO-like domain (SLD) in the SUMO pathway remain undefined.
Identification of the fission yeast DNA repair protein Rad60 9,10 led to the recognition of a unique SLD-containing protein family, which is conserved from yeast to humans 9. Rad60 was recovered as an interactor of the replication checkpoint kinase Cds1, and in a genetic screen for factors required for the viability of cells lacking the Okazaki fragment-processing endonuclease Rad2 (Fen1 homolog; 9,10). Rad60 is essential for cell viability, and cells hypomorphic for Rad60 are sensitive to a range of genotoxic stresses, including exquisite sensitivity to the replication stalling agent hydroxyurea (HU; 9-11). Rad60 physically and functionally associates with the Smc5-6 DNA repair holocomplex, which contains a SUMO E3 ligase subunit, Nse2 9,10,12. Notably, homologous recombination (HR)-dependent toxic DNA structures accumulate in both Rad60 and Smc5-6 mutant cells following replicative stress 13. Furthermore, the viability of Rad60 and Smc5-6 mutants depends on the replication fork stabilization/processing activities of the RecQ homolog Rqh1 (human BLM) and the Holliday junction endonuclease Mus81-Eme1 9-11,14.
The N-termini of Rad60 family members are acid-rich and appear to be disordered, while their C-termini contain two SLDs (SLD1 and SLD2) 9,15. The N-terminus of Rad60 contains a phosphorylation-dependent SUMO-interacting motif (SIM), and this motif is required for cellular resistance to replicative stress 11. Demonstrating a critical cellular function for Rad60 SLDs, the temperature and genotoxin-sensitive mutants rad60-3 and rad60-1 have mutations in SLD1 that likely affect domain structure 9,10. The budding yeast Rad60 homolog, Esc2, was identified via its ability to establish silencing when targeted to particular loci 16. The role of Esc2 in silencing is likely mediated by its interaction with the NAD+-dependent histone deacetylase Sir2 17. Esc2 also plays roles similar to Rad60 in surviving replicative stress, indicating that this function is evolutionarily conserved 18. The human Rad60 homolog, Nip45, was identified as an interactor and potentiator of NF-AT-dependent transcription 19,20. The anticipated roles for Nip45 in DNA repair/replication are currently undefined.
Here, we employed structure-function analyses to define critical roles of the fission yeast Rad60 SLDs, and uncovered striking parallels with Ubls in the ubiquitin system: The two Rad60 SLDs can act as mimetics of SUMO, despite their substantial sequence divergence from SUMO, and from each other. In our analysis we determined the crystal structure of Rad60 SLD2 at 0.97 Å, revealing that Rad60 contains the β-grasp fold of Ubl proteins, despite very low sequence identity. Intriguingly, however, with the exception of the non-covalent E2 (Ubc9, also known in S. pombe as Hus5) interface, other functionally critical surface features of SUMO are not conserved in Rad60 SLD2. Structure-based mutagenesis to specifically uncouple the SLD2 interaction with the non-covalent interface of Ubc9 reveals a pivotal role for this interaction in the cellular response to genotoxic stress. Further genetic analyses implicate the SLD2-Ubc9 interaction in suppressing aberrant replication-associated HR. Rad60 SLD1 does not bind Ubc9, but it does support interactions with other SUMO pathway factors. These factors include the E1 SUMO activating enzyme component Fub2, the E3 SUMO ligase Pli1 and the recently identified SUMO-targeted Ubiquitin Ligase (STUbL). Together, these results reveal a mechanistic basis for the near synonymous roles of SUMO and Rad60 in survival of genotoxic stress, and suggest that Rad60 is a novel component/regulator of the SUMO pathway in response to DNA damage.
Results
Rad60 SLDs interact with different sumoylation enzymes
Rad60 mutant phenotypes appear to overlap with those of cells hypomorphic for sumoylation pathway components 10,11,13,21. Therefore, we wished to determine if Rad60 SLDs mimic certain SUMO (Pmt3) functions, despite low sequence similarity between Rad60 SLDs and SUMO. To probe for potentially conserved interfaces, we analyzed Rad60 interactions with SUMO pathway enzymes and associated factors. We first tested interaction between Rad60 and the E2 SUMO conjugating enzyme Ubc9, which has a defined non-covalent interaction interface with SUMO 22,23,24,25. GST-Rad60 fusions were expressed in a strain containing Ubc9 epitope-tagged at its endogenous locus (Ubc9-TAP). Ubc9-TAP specifically co-precipitated with GST-Rad60 SLD2 (332-406aa), as it failed to interact with either GST alone or GST-Rad60 N-terminus+SLD1 (1-325aa; Fig.1a). The interaction between Ubc9-TAP and GST-Rad60 SLD1+2 (188-406aa) appears weaker than SLD2 alone; however, this likely reflects the relatively strong expression of the GST-SLD2 construct (Fig. 1a).
Figure 1. Rad60 SLDs interact with distinct components of the sumoylation machinery.
(a) Ectopically expressed GST-Rad60 SLD2, GST-Rad60 SLD1+2, GST-Rad60 N-terminus+SLD1 (or GST alone) were induced in the Ubc9-TAP strain, and subjected to GST pull-down. Inputs and pull-downs were immunoblotted with antisera for TAP (anti-PAP) or GST (anti-GST). Ubc9 interacts specifically with GST-fusion proteins that contain Rad60 SLD2. (b) Ectopically expressed GST-Rad60 SLD2, GST-Rad60 SLD1+2, GST-Rad60 N-terminus+SLD1 (or GST alone) were induced in a Fub2-TAP background, and subjected to GST pull-down. Inputs and pull-downs were immunoblotted with antisera for PAP or GST. Unlike Ubc9, Fub2 interacts specifically with GST-fusion proteins containing Rad60 SLD1. (c) Ectopically expressed GST-Rad60 SLD2, GST-Rad60 SLD1+2, GST-Rad60 N-terminus+SLD1 (or GST alone) were induced in a Pli1-TAP strain, and subjected to GST pull-down. Inputs and pull-downs were immunoblotted with antisera for PAP or GST. As seen with Fub2, Pli1 also interacts only with GST-fusion proteins that contain Rad60 SLD1. (d) Ectopically expressed GST-Rad60 SLD2, GST-Rad60 SLD1+2, GST-Rad60 N-terminus+SLD1 (or GST alone) were induced in an Slx8-myc strain, and subjected to GST pull-down. Inputs and pull-downs were immunoblotted with antisera for myc or GST. As seen with Fub2 and Pli1, Slx8 interacts only with GST-fusion proteins that contain Rad60 SLD1. (e) Schematic depicting the interaction of Rad60 SLD1 and Rad60 SLD2 with components of the sumoylation pathway and STUbL complex.
We next examined if Rad60 SLDs interact with other components of the SUMO pathway. Fub2, a component of the Fub2/Rad31 (Uba2/Aos1) E1 heterodimer or the E3 SUMO ligase Pli1 were TAP-tagged, and the same GST-Rad60 constructs described above were expressed in these cells. Interestingly, the interactions differed from the Ubc9-TAP results; GST-Rad60 N-terminus+SLD1 and GST-Rad60 SLD1+2 both interact with Fub2-TAP (Fig. 1b) and Pli1-TAP (Fig. 1c). However, neither SLD2 nor the GST control interacts with Fub2-TAP or Pli1-TAP, indicating that these Rad60-E1/E3 interactions require Rad60 SLD1. We recently demonstrated that Rad60 interacts with the fission yeast STUbL complex (Slx8-Rfp1)21. We now find that Rad60 SLD1 is necessary and sufficient for interaction with Slx8 (Fig. 1d). Thus, together with the failure of SLD2 to interact with Slx8-myc (Fig. 1d), SLD1 contains the critical interface for the STUbL complex. Overall, the Rad60 SLDs are able to interact non-covalently with SUMO pathway enzymes and associated factors (see Fig. 1e), similar to SUMO. Intriguingly, however, distinct interaction interfaces are conserved within each SLD.
Rad60 SLD2 crystal structure
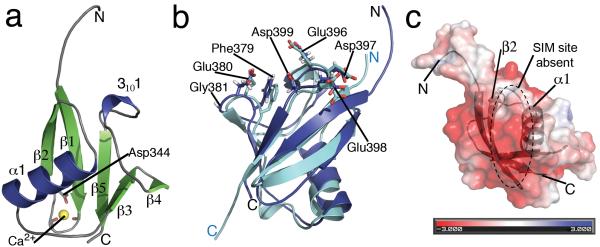
To define the observed differences in protein binding partners of Rad60 SLD2 and SUMO at the molecular level, we determined the Rad60 SLD2 protein crystal structure, at 0.97 Å ultra-high resolution (Fig. 2a). Rad60 SLD2 residues 332-406 (as used in Fig. 1) form a β-GRASP fold, as suggested from weak sequence similarity to SUMO 9. Notably, a search of the protein databank within the SCOP classified β-grasp fold indicated that the Rad60 SLD2 crystal structure is the highest-resolution structure determined for this protein fold to date. In our structure the N-terminal Hexa-His tag and all of the Rad60 SLD2 residues were clearly observable in the electron density maps (Supplementary Fig. 1 provides an example of map quality) except the surface exposed C-terminal Asp406 that is likely disordered (Fig. 2a). Electron density was also observed indicating the presence of a Ca2+ ion from the crystallization solution. The Ser341 and Ser343 backbone oxygen’s, the Asp344 side chain carboxylate, two water molecules and the Glu373 side chain carboxylate from a symmetry related molecule bind this metal ion. A search of the protein databank database using the Dali Server 26, for similarity in backbone structure, confirmed that Rad60 SLD2 belongs to the Ubl superfamily of proteins. The Rad60 SLD2 fold is most similar to the S. cerevisiae Dsk2p Ubl domain, structurally superimposing with 1.8 Å RMSD, a Z-score of 12.4 and sharing 16% sequence identity. Dsk2p Ubl domain likely scores well due to a similar high model quality at a resolution of 1.15 Å (PDB code: 2BWF). However, structure-based sequence alignment shows that SUMO homologues, rather than Dsk2p, have greater sequence identities to SLD2. This is in keeping with the function of Dsk2p, which acts in the ubiquitin but not SUMO pathway. Structural superimposition of the Rad60 backbone alpha carbon atoms with human SUMO-1, -2 & -3 and budding yeast Smt3p determines that Rad60 SLD2 fold overlays with 2.07, 2.46, 2.13 and 2.61 Å RMSD to these proteins, respectively (SUMO-1 structural superimposition is shown in Fig. 2b). Sequence identities are 20, 24, and 25% for human SUMO-1, -2 and -3 respectively, and 13% for Smt3p. The fission yeast SUMO homologue, Pmt3, has not been structurally determined, but aligns with 24% sequence identity (sequence alignments of the SUMO homologues with SLD2 are depicted in Supplementary Fig. 2). Interestingly, we observe key similarities and differences between the SUMO homologues and Rad60 SLD2 surfaces, which likely account for their selective interactions with SUMO pathway factors. In SUMO moieties, acidic motifs support complex formation in the non-covalent SUMO:Ubc9 structure, by interacting with a conserved basic stretch of amino acids on the Ubc9 surface 22,23,24. This includes a Phe-Glu-Gly (FEG) motif on the β-3/β-4 loop of SUMO-1 (well conserved in SUMO paralogues), where the acidic amino acid has water-mediated interactions with the Ubc9 backbone. A second cluster of acidic side chains is present on the 310helix/β-5 loop and strand, which have direct hydrogen bond interactions with the Ubc9 surface. Interestingly, these carboxylate side chain motifs are not conserved in the Dsk2p Ubl domain sequence, being replaced by predominantly basic side chains (Supplementary Fig. 2). The acidic side chain of the FEG motif occurs at an important interface, as mutation of this residue to Arg markedly perturbed the non-covalent SUMO interaction with Ubc9 but did not affect the covalent interaction22. In Rad60 SLD2 these acidic interface motifs are well conserved, including the β-3/β-4 loop FEG motif, and the cluster of acidic 310helix/β-5 loop residues (Fig. 2b and Supplementary Figs. 2 and 3), maintaining the negative charge of this non-covalent Ubc9 binding region. The corresponding key basic stretch of amino acids in Ubc9, which form the interface with budding yeast Smt3p 24, are completely conserved in the fission yeast Ubc9 homologue. This confirms that the interaction interface is well-maintained between these highly diverged yeast species. Moreover, multiple sequence alignments of SLD2 in Rad60 family proteins indicate that both the FEG motif and the acid cluster are well conserved throughout, and the acid side chain of the FEG motif (Glu380 in Rad60) is strictly conserved as Asp or Glu (Supplementary Fig. 3), suggesting a conserved Ubc9 interaction interface in the Rad60 family. Interestingly, neither the acid side chain cluster nor the FEG motif are maintained in SLD1 of Rad60 family members (Supplementary Fig. 3), providing a rationale for SLD2-specific Ubc9 binding. The lack of a C-terminal tail in the Rad60 SLD2 structure is one key difference with SUMO (Fig. 2b). The C-terminus is important for E1:SUMO complex formation 27, suggesting why Rad60 SLD2 does not bind the E1 Fub2/Rad31 in our assay (Fig. 1). Also distinct in our Rad60 SLD2 structure is the lack of a positively charged cleft that is formed between β-strand 2 and α-helix 1 in SUMO-1, -2 & -3, which is non-covalently bound by SIMs of partner proteins 28. In the Rad60 SLD2 structure we observe that the groove at β-strand 2/α-helix 1 is absent, with side chains Pro351, Phe354, Arg362, Glu366 occluding the cleft, and this protein surface region has a net negative charge (Fig. 2c). This indicates that Rad60 SLD2 does not function to bind SIM sequences, such as those in the Pli1 E3 ligase or STUbLs 21.
Figure 2. Rad60 SLD2 Crystal Structure.
(a) The 0.97 Å crystal structure of Rad60 SLD2 forms a β-GRASP fold that is composed of 5 β-strands, an α-helix and a 310helix. A Ca2+ ion, yellow sphere, is bound by two backbone oxygen’s and the Asp344 side chain carboxylate (in addition to two water molecules and the Glu373 side chain carboxylate from a symmetry related molecule). (b) Structural superimposition of Rad60 SLD2, depicted in dark blue, and SUMO-1 depicted in cyan (SUMO crystal structure taken from PDB code 2UYZ). A conserved non-covalent Ubc9 binding face is observed in Rad60 SLD2, with well-conserved residues depicted as sticks, and labeled for Rad60. These residues include the β3/β4-loop FEG motif, residues 379-381, where the equivalent acid side chains make water mediated interaction with Ubc9 in the human and budding yeast structures. Also well-conserved are carboxylate residues in the 310-helix β-strand 5 region, including residues Glu396 and Asp399, whose equivalents make direct hydrogen bond contacts with Ubc9 surface residues in the human and budding yeast SUMO:Ubc9 complex structures. (c) Electrostatic surface diagram for Rad60 SLD2 that is depicted partially transparent and beneath which is the SLD2 secondary structure, shown in dark grey. The α-helix 1 β-strand 2 region shows a net negative charge and the SIM-binding pocket highlighted on this structure, which occurs in this region of SUMO, is absent (red, -3.0 KTe-1, blue +3.0 KTe-1).
Abrogating the Rad60 SLD2:Ubc9 interface
Our structural analyses indicate that Rad60 SLD2 shares key surface residues of SUMO that constitute a non-covalent SUMO:Ubc9 interface 22,23,24, but does not contain the required region for covalent interaction. To test this observation, we mutated Rad60 SLD2 surface residue Glu380 to Arg (SLD2E380R) and determined the effect on Rad60 SLD2-Ubc9 interaction. The equivalent SUMO-1 mutation, Glu67Arg, abolished Ubc9 binding without impacting the SUMO-1 structure, or the formation of a covalent Ubc9:SUMO-1 complex at the active site of Ubc9 22. Strikingly, a GST-Rad60 SLD2E380R construct did not detectably interact with Ubc9-TAP (Fig. 3a), unlike the wild-type SLD2 control. To confirm that the observed interaction between SLD2 and Ubc9 is direct, we tested it in vitro using bacterially expressed proteins. GST-Ubc9 specifically bound wild-type 6His-Rad60 SLD2, but not 6His-Rad60 SLD2E380R (Fig. 3b). Thus, a direct interaction exists between Ubc9 and SLD2, which as observed in vivo (Fig. 3a), is abrogated by the SLD2E380R mutation.
Figure 3. Analyzing the Rad60 SLD2:Ubc9 interface.
(a) Ectopically expressed GST-Rad60 SLD2, GST-Rad60 SLD2E380R or GST alone were induced in the Ubc9-TAP strain, and subjected to GST pull-down. Inputs and pull-downs were immunoblotted with antisera for PAP or GST. Despite similar levels of protein induction, GST-Rad60 SLD2E380R did not pull down Ubc9. (b) Bacterially expressed GST-Ubc9, 6His-Rad60 SLD2 or 6His-Rad60 SLD2E380R were combined as indicated and subjected to GST pull-down, followed by immunoblotting with antisera for His or GST. Wild-type 6His-Rad60 SLD2 interacted robustly with GST-Ubc9 in vitro, whereas 6His-Rad60 SLD2E380R did not, even when loaded in excess of wild-type SLD2. (c) Immunoblot of the indicated strains with anti-GFP. (d) GST, GST-Rad60 SLD2 or GST-Rad60 SLD2E380R were expressed in an slx8-1 mutant and immunoblotted with antisera for GFP or GST. Induction of wild-type GST-Rad60 SLD2 inhibited chain formation, whilst overexpression of GST-Rad60 SLD2E380R had no effect on the slx8-1 mutant phenotype. (e) The indicated strains were serially diluted onto selective media, which either repressed (+B1), or induced (-B1) ectopic expression of GST alone, GST-Rad60 SLD2 or GST-Rad60 SLD2E380R. All plates were incubated at 32 °C. (f) Total lysates from the indicated strains were immunoblotted with antisera for GST or Cdc2 (loading control). Despite similar levels of protein induction to wild-type GST-Rad60 SLD2, GST-Rad60 SLD2E380R overexpression was not toxic in an rqh1Δ mutant.
The non-covalent SUMO:Ubc9 interface is critical for the formation of poly-SUMO chains 22,23,24. Thus, if SLD2 and SUMO bind the same site on Ubc9, ectopically expressed Rad60 SLD2 should compete for the SUMO:Ubc9 interface and block poly-SUMO chain formation. We utilized an in vivo assay based on the accumulation of SUMO chains in slx8-1 STUbL mutant cells 21. High molecular weight SUMO chains accumulate in slx8-1 cells expressing GFP-SUMO (Pmt3), but not in GFP-SUMOallR that lacks internal lysine residues essential for SUMO chain formation (Fig. 3c; 21). Chain formation in this assay is dependent on the major E3 SUMO ligase Pli1 (Fig. 3c). Notably, expression of wild-type GST-Rad60 SLD2 strongly attenuated SUMO chain formation, whereas GST-Rad60 SLD2E380R expression had little effect on SUMO chain abundance (Fig. 3d).
We previously reported that expression of the SLD-containing Rad60 C-terminus kills cells that lack Rqh1 11. We repeated this assay and found that wild-type GST-Rad60 SLD2, but not GST-Rad60 SLD2E380R, is toxic to rqh1Δ cells (Fig. 3e), despite similar levels of protein expression (Fig. 3f). This suggests that binding of Ubc9 by Rad60 SLD2 mediates its toxicity in rqh1Δ cells.
Rad60:Ubc9 is critical for survival of genotoxic stress
In order to test the in vivo significance of the Rad60:Ubc9 interaction, we generated a fission yeast strain in which the endogenous rad60 locus was replaced with the rad60E380R allele. The rad60E380R strain was viable, but exhibited a slow growth phenotype and heterogeneity in cell length; reflecting elevated levels of spontaneous DNA damage and “constitutive” activation of the DNA damage checkpoint in these cells (Fig. 4a and data not shown). Importantly, rad60E380R cells are not temperature sensitive for growth and Rad60E380R is expressed at the same levels as wild-type Rad60 (Fig. 4b and Supplementary Fig. 4). We assayed rad60E380R cellular phenotypes and found them to be exquisitely sensitive to replication fork stalling by HU, and other agents that can impede normal replication fork passage, including methylmethane sulfonate, camptothecin, or ultra-violet irradiation (Fig. 4a). These phenotypes of rad60E380R are recessive and fully complemented by expression of wild-type rad60 (Fig. 4a). Thus, the Rad60:Ubc9 interface is critical for cellular survival to genotoxic agents.
Figure 4. The rad60E380R mutant is sensitive to genotoxic stress.
(a) Serial dilutions of the indicated strains were either untreated (No drug), or treated with the indicated concentrations of hydroxyurea (HU), methylmethane sulfonate (MMS), camptothecin (CPT), or ultra-violet (UV) irradiated. All plates were incubated at 32 °C. The rad60E380R mutant is hypersensitive to all of the genotoxic agents shown. (b) Serial dilutions of the indicated strains were either untreated, or treated with the indicated concentrations of hydroxyurea (HU), or ultra-violet (UV) irradiated, and incubated at 32 °C. An additional untreated plate was incubated at 36 °C. Combining the rad60E380R and rad60-3 mutations suppresses the phenotypes observed in either single mutant.
Given that Rad60 SLD1 and SLD2 bind distinct SUMO pathway proteins, we investigated whether their tandem arrangement in a single Rad60 protein was functionally critical. Initially, we analyzed strains expressing hypomorphic Rad60 that was mutated in either SLD1 or SLD2. As expected rad60-3 (SLD1 mutant) and rad60E380R (SLD2 mutant) cells are hypersensitive to genotoxic stress (Fig. 4b). Markedly, simultaneous expression of both mutants at endogenous levels in a single strain suppresses the phenotypes associated with either single mutant, e.g. the temperature sensitivity of rad60-3 and the UV sensitivity of rad60E380R (Fig. 4b). This suggests that each wild-type Rad60 SLD is able to compensate for their mutant counterpart in a different Rad60 protein i.e. trans-complementation.
Key roles of the Rad60:Ubc9 complex in genome stability
The growth phenotypes and DNA damage checkpoint activation observed in rad60E380R cells indicates that spontaneous DNA damage occurs as a result of abrogating the Rad60:Ubc9 interface. To better define the nature and source of these DNA lesions, we determined genetic interactions between rad60E380R and various DNA repair mutants. We found that rad60E380R requires Rqh1 (human BLM) and Mus81-Eme1 for viability (Fig. 5a & b & Supplementary Fig. 5a). Rqh1 both suppresses “illicit” HR and dissolves Holliday junctions (HJs), which are the end points of certain HR pathways 29. The Mus81-Eme1 endonuclease functions in a pathway parallel to Rqh1 for resolution of HJs, which are byproducts of restarting collapsed replication forks during mitotic growth 30-33. The synthetic lethality of rad60E380R combined with rqh1Δ or mus81Δ, therefore indicates that in rad60E380R cells there is an increase in spontaneous HR-dependent processes, including replication fork restart. Consistently, rad60E380R cells also require Rhp51 (Rad51) for viability (Fig. 5c & Supplementary Fig. 5b). We previously showed that in cells lacking the Smc5-6 subunit Nse6, toxic Rad51-dependent recombination structures formed in response to UV irradiation, which can stall replication forks and induce HR 14. Interestingly, the rad60E380R nse6Δ double mutant is inviable (Fig. 5d), which again indicates elevated levels of spontaneous recombination in rad60E380R cells.
Figure 5. Genetic interactions between rad60E380R and mutants in the HRR, or sumoylation pathways.
Representative tetrad dissections are shown when rad60E380R was mated against: (a) rqh1Δ, (b) mus81Δ, (c) rhp51Δ, (d) nse6Δ, (e) pli1Δ, (f) nse2-SA, (g) slx8-1 and (h) ulp2Δ. For each cross a key depicts the genotypes present within each tetrad, which are denoted by various shaped white lines placed around each colony. Combining rad60E380R with these mutants results in extreme synthetic sickness, or is a lethal event. (i) Schematic of the lethal (solid black lines) and synthetic-sick (dashed grey lines) interactions involving rad60E380R and the indicated mutants.
The non-substrate interaction between Ubc9 and Rad60 SLD2, and the SLD1 interactions with SUMO E1 and E3 suggests that Rad60 is a novel component/regulator of the SUMO pathway. To further explore this possibility, we tested for genetic interactions between rad60E380R and mutants defective in SUMO metabolism. Interestingly, rad60E380R is lethal when combined with mutations in either of the two SUMO E3 ligases, pli1Δ or nse2-SA (Fig. 5e & f). This could indicate that Rad60 functions as a third and novel type of SUMO E3 ligase, or perhaps facilitates the functions of both Pli1 and Nse2 34. We do not favor the latter interpretation because global sumoylation levels in rad60E380R cells appear normal, whereas pli1Δ cells lack > 90% of total sumoylation (35; data not shown). However, a role for Rad60 as a critical new “hub” of SUMO metabolic processes is apparent. This Rad60 function is further supported by genetic interactions observed between rad60E380R and mutations in the STUbL subunit Slx8 (slx8-1) and the SUMO protease Ulp2 (ulp2Δ). Both STUbL and Ulp2 mutant cells accumulate SUMO conjugates due to defects in the degradation or desumoylation of sumoylated proteins, respectively 36-38. Notably, rad60E380R is synthetic sick/lethal in combination with either ulp2Δ or slx8-1 (Fig. 5g & h & Supplementary Fig. 5c). Overall, these results highlight the complex interplay between Rad60 and the SUMO pathway, which may be anticipated based on the physical interactions of Rad60 with multiple regulators of SUMO metabolism.
Discussion
Members of the Rad60 protein family contain two SLDs, and we have discovered that these domains support distinct functional interactions. Our structural analysis revealed that despite substantial sequence divergence from SUMO, SLD2 has maintained key surface residues for interfacing with Ubc9. Critical to the SLD2:Ubc9 interaction is an acidic residue within a “FEG” motif that is well conserved in all Rad60 family members (Supplementary Fig. 1b). SLD2 has lost the ability to bind non-covalent SIMs due to changes in the region that forms the SIM-binding pocket in SUMO (see Supplementary Fig. 1b; 28). In SLD1, a reciprocal situation is found, where SLD1 lacks the Ubc9 interface but has retained the SIM-binding site of SUMO (Supplementary Fig. 1b). Notably, this interface configuration is conserved throughout the Rad60 family, underscoring the importance of these interactions. There are some intriguing parallels to be drawn between Rad60 and Isg15, a protein that contains tandem Ubl domains. The C-terminal Ubl domain binds an E2 conjugating enzyme, whereas the N-terminal Ubl domain binds E3 ligases 39. Isg15, unlike Rad60, is covalently conjugated to target proteins; however, the selective interactions of each Ubl domain echo those of Rad60 SLDs.
Our analyses indicate that Rad60 SLD2 interacts with Ubc9 at the same non-covalent interface used by SUMO to promote poly-SUMO chain formation 22-25. This interface is not proximal to the Ubc9 active site that recognizes target proteins containing a consensus sumoylation site 7. Consistent with this, we do not detect sumoylation of Rad60 (data not shown), indicating that Rad60 is not a SUMO target. Instead, we envisage Rad60 executes at least one of the following three functions: (i) attenuate SUMO chain formation as observed for SLD2 (Fig. 3d), (ii) recruit SUMO-charged Ubc9 to mediate sumoylation of specific target proteins, or (iii) sequester Ubc9 in an inactive complex, to globally downregulate sumoylation. We do not favor the latter possibility, as global sumoylation appears normal in rad60E380R cells (data not shown). However, attenuation of SUMO chain formation is consistent with rad60E380R cells requiring the SUMO chain-attenuating activities of Ulp2 and STUbLs for viability (Fig. 5g & h).
Global sumoylation appears intact in the nse2-SA SUMO E3 ligase mutant, and similar to rad60E380R, these cells exhibit sensitivity to genotoxic stress 12. Thus, similar to Nse2, Rad60 acting as a novel E3 ligase could mediate sumoylation of a subset of key DNA repair factors. This putative function would utilize a novel mechanism, wherein Rad60 SLD1 binds SIMs 21 and SLD2 recruits a Ubc9-SUMO thiolester complex to sumoylate targets. Recent data on the SIM-containing BLM and Usp25 proteins provides support for this proposed sumoylation mechanism. The SIMs of BLM and Usp25 recruit SUMO thiolester-charged Ubc9 complexes, thereby promoting cis-sumoylation of SIM-adjacent lysine residues in each protein 40,41. Interestingly, BLM and Usp25 SIM motifs also provide SUMO paralog selectivity, resulting in preferential modification of each protein by SUMO-2/3 over SUMO-1 40,41. This type of selectivity could occur with SIM-Rad60 SLD1 interactions.
The spectrum of genotoxin sensitivity and genetic interactions identified for rad60E380R indicates that the SLD2:Ubc9 complex suppresses replication fork-associated DNA damage. In particular, the extreme synthetic sickness detected between rad60E380R and rhp51Δ or mus81Δ implicates the SLD2:Ubc9 interaction in preventing replication fork collapse. This is because Rhp51 and Mus81 are critical for restarting collapsed replication forks 33. Therefore, Rad60-interacting proteins, which we hypothesize are sumoylation targets, likely include factors responsible for coordinating replication fork passage with DNA template repair. Rqh1 is one such factor, whose budding yeast homolog Sgs1 is sumoylated and plays a key role in preventing aberrant recombination at perturbed replication forks 5,32. Intriguingly, Sgs1 sumoylation is independent of known SUMO E3 ligases and may therefore be a target of the budding yeast Rad60 homolog 5. However, we observed that rad60E380R is lethal in combination with deletion of Rqh1, indicating that Rad60 also interacts with or regulates other proteins that act on parallel pathways for mitigating replication-associated DNA damage.
Notably, the E3 ubiquitin ligase Parkin contains a Ubl domain that binds the ubiquitin-interacting motif (UIM) of Eps15 and thereby stimulates Eps15 ubiquitination 42. Furthermore, via a mechanism analogous to the SIM-mediated sumoylation of BLM and Usp25 40,41, UIM-dependent monoubiquitination of proteins occurs 43. Our analysis of Rad60 suggests that it may execute analogous functions in the SUMO pathway, acting to stimulate sumoylation of certain target proteins. Overall, we have identified novel and physiologically important interactions between the SUMO-like protein Rad60 and the SUMO conjugation machinery. These interactions reveal a mechanistic basis for the extensive overlapping phenotypes of cells defective for Rad60 or the SUMO pathway. Rad60 acts as a SUMO mimetic, which is unprecedented in the SUMO pathway, but bears striking functional similarity to Ubl domains in ubiquitin metabolism.
Methods
S. pombe strain construction
We used standard fission yeast culture methods as described earlier 44. Supplementary Table 1 describes all of the S. pombe strains utilized in this study. The Ubc9, Fub2 and Pli1-epitope-tagged strains were constructed using PCR-based gene replacements, utilizing the kanMX6 resistance cassette containing an in frame C-terminal TAP tag 45. The rad60E380R mutant was created using Quikchange (Stratagene).
S. pombe extract preparation and precipitations
GST-pull-down experiments were as described previously 11. Co-precipitated proteins were resolved on either 4-20% (w/v) gradient Tris-Glycine, or 3-8% (w/v) Tris-Acetate SDS-PAGE gels depending on the size of the epitope-tagged protein (Invitrogen, Carlsbad, CA) and immunoblotted for the myc, TAP and GST epitopes using anti-myc (9E10, Covance), Peroxidase-anti-peroxidase (PAP, Sigma-Aldrich) and anti-GST (rabbit polyclonal) antibodies. Where indicated, anti-cdc2 antiserum was used as a loading control (kindly provided by Steve Reed, The Scripps Research Institute, San Diego, CA).
Analyzing total levels of sumoylated S. pombe proteins in vivo
The indicated strains were cultured in the appropriate media and then incubated at 36 °C for 7 hours to inactivate slx8-1. Cells were then harvested and total lysates prepared using denaturing buffer (8M Urea, 0.5% (v/v) Nonidet P-40, 50mM Sodium Phosphate and 50mM Tris-HCL pH 8.0) with 1x Complete protease inhibitor mix EDTA-free (Roche, Indianapolis, IN), equal quantities of protein were then resolved on 3-8% (w/v) gradient Tris-Acetate gels and immunoblotted using antisera raised against GFP (Roche, Indianapolis, IN) and GST (rabbit polyclonal).
In vitro pull-down experiments
GST-Ubc9, 6His-Rad60 SLD2 and 6His-Rad60 SLD2E380R were individually purified from BL21 (DE3) pLysS cell lysates. GST-Ubc9 and 6His-Rad60 SLD2, or GST-Ubc9 and 6His-Rad60 SLD2E380R were then combined in vitro. Inputs (5%) were taken, and the remainder of the samples subjected to GST pull-down. Inputs and pull-downs were resolved using 4-20% (w/v) gradient Tris-Glycine gels, and immunoblotted using antisera raised against His (Clontech, CA) and GST.
Crystallization, data collection and analysis
Initial crystallization conditions for the purified recombinant Rad60 SLD2, residues 332-406, were discovered from a PEG/ion screen using Fluidigm Corporation Topaz® 1.96 microfluidic crystallization chips (Fluidigm Corporation, South San Francisco, CA, USA), and the Hauptman-Woodward high throughput robotic crystallization screen 46. Multiple initial hits were repeated on a larger scale by hand, using vapor-diffusion crystallization experiments. The closest initial condition to our optimized conditions was 0.2 M calcium acetate, 100 mM HEPES pH 7.5 and 27% (w/v) polyethylene glycol 4K, in the Fluidigm chips. Optimized larger crystals of Rad60332-406 protein, buffered in 50 mM Tris, pH 8.0, 50 mM NaCl and 0.5 mM EDTA, were produced in 100 mM HEPES, pH 7.75, 24% (w/v) polyethylene glycol 4K and 0.2 M calcium acetate precipitant solution at RT. Flash-frozen single crystals, supplemented with 20% (w/v) ethylene glycol cryoprotectant in the mother liquor, were used for x-ray diffraction data collection at 100K. A data set was collected from a single crystal that diffracted to 0.97 Å, using a wavelength of 0.81 Å, on a MarMosaic-325 CCD detector at beamline 11.1 SSRL, Stanford, Paolo Alto, CA, USA. The Rad60 SLD2 data set was indexed, merged and scaled using HKL2000 47. Crystals belonged to the cubic space group I213, having cell dimensions 81.5, 81.5, 81.5 Å, and 90, 90, 90°. Successful crystallographic phasing occurred through the use of an extensive molecular replacement (MR) search, on a computer cluster containing 288 dual processor servers with a total 576 3.4 GHz Intel XEON-EMT CPUs. Multiple SUMO and SUMO homolog structures, and homology models were used for the MR search, using Phaser 48 of the CCP4 package. Over 150,000 independent searches were conducted and each one was coupled to an initial refinement step. One MR search, using a crystal structure of SUMO-1 (PDB code 2UYZ) as a starting model with the best initial R-factor scores, provided a suitable phasing model. One molecule is present in the asymmetric unit of the crystal, and an initial Rad60 SLD2 model was built into σA-weighted 2Fo-Fc and Fo-Fc maps using XFIT 49 and was followed by cycles of refinement using SHELX 50 (Electron density map stereodiagram shown in Supplementary Fig. 1a). Due to the ultra-high resolution data, refinement of the final model included anisotropic thermal displacement parameters (B-factors) and the inclusion of hydrogen atoms. In these ultra-high quality electron density maps Ile25, His47 and Glu69 side chains have clear alternate conformations. Both Molprobity 51 and Procheck 52 were used to analyze the quality of the final model. The final 0.97 Å Rad60 SLD2 structure has a Molprobity score within the 99th percentile and a Ramachandran plot determined that 96% of residues are in the allowed region and 4% of residues are in the additionally allowed region. For Structural Statistics see Table 1. The structural superimpositions of Rad60 SLD2 were conducted with the SUMO homologues, using SEQUOIA 53. The closest RMSD deviation to Rad60 SLD2 were observed from chain B of 2UYZ of human SUMO-1, 1WM3 chain A of human SUMO-2, human SUMO-3 from chain B of 2DO7 and chain B in 1EUV of budding yeast Smt3p.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Crystal 1 name | |
|---|---|
| Data collection | |
| Space group | I21 3 |
| Cell dimensions | |
| a, b, c (Å) | 81.5, 81.5, 81.5 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50 - 0.97 |
| Rsym or Rmerge | 3.5 (41.4) |
| I / σI | 30.0 (2.8) |
| Completeness (%) | 95.0 (100.0) |
| Redundancy | 3.5 (3.5) |
| Refinement | |
| Resolution (Å) | 50 - 0.97 |
| No. reflections | 47977(4983) |
| Rwork / Rfree | 14.5 / 16.9 |
| No. atoms | |
| Protein | 1440 |
| Ligand/ion | 1 |
| Water | 119 |
| B-factors | |
| Protein | 18.7 |
| Ligand/ion | 18.9 |
| Water | 27.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.3 |
Data was collect on a single crystal, and in the refinement 5% of unique reflections were removed as a test set for Rfree calculation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Benoit Arcangioli (Institut Pasteur, France), Paul Russell (TSRI, U.S.A), Stephanie Pebernard (TSRI, U.S.A) and Grazia Raffa (Università di Roma La Sapienza, Italy) for generously providing fission yeast strains, additional reagents and The Scripps Cell Cycle Group for support and encouragement. This study was funded in part by NIH grants GM068608 & GM081840 awarded to M.N.B. and CA104660 to J.A.T.
Accession codes. Protein Data Bank: Coordinates for Rad60 SLD2 have been deposited with accession code XXXX.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. American Society for Microbiology Press; Washington, DC: 2006. 2006. [Google Scholar]
- 2.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6:900–13. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 5.Branzei D, et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–22. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 6.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–32. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann-Petersen R, Gordon C. Integral UBL domain proteins: a family of proteasome interacting proteins. Semin Cell Dev Biol. 2004;15:247–59. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Boddy MN, et al. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol Cell Biol. 2003;23:5939–46. doi: 10.1128/MCB.23.16.5939-5946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morishita T, Tsutsui Y, Iwasaki H, Shinagawa H. The Schizosaccharomyces pombe rad60 Gene Is Essential for Repairing Double-Strand DNA Breaks Spontaneously Occurring during Replication and Induced by DNA-Damaging Agents. Mol Cell Biol. 2002;22:3537–48. doi: 10.1128/MCB.22.10.3537-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffa GD, Wohlschlegel J, Yates JR, 3rd, Boddy MN. SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J Biol Chem. 2006;281:27973–81. doi: 10.1074/jbc.M601943200. [DOI] [PubMed] [Google Scholar]
- 12.Andrews EA, et al. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–96. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyabe I, Morishita T, Hishida T, Yonei S, Shinagawa H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol Cell Biol. 2006;26:343–53. doi: 10.1128/MCB.26.1.343-353.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, 3rd, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–30. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–80. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 17.Cuperus G, Shore D. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics. 2002;162:633–45. doi: 10.1093/genetics/162.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohya T, Arai H, Kubota Y, Shinagawa H, Hishida T. A SUMO-like domain protein, Esc2, is required for genome integrity and sister chromatid cohesion in Saccharomyces cerevisiae. Genetics. 2008;180:41–50. doi: 10.1534/genetics.107.086249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge MR, et al. NF-AT-Driven interleukin-4 transcription potentiated by NIP45. Science. 1996;274:1903–5. doi: 10.1126/science.274.5294.1903. [DOI] [PubMed] [Google Scholar]
- 20.Mowen KA, Schurter BT, Fathman JW, David M, Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol Cell. 2004;15:559–71. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 21.Prudden J, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–18. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duda DM, et al. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol. 2007;369:619–30. doi: 10.1016/j.jmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatham MH, et al. Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry. 2003;42:9959–69. doi: 10.1021/bi0345283. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–9. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–51. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S, Doherty KM, Brosh RM., Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–37. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–48. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 31.Boddy MN, et al. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–66. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–9. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 33.Roseaulin L, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–87. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xhemalce B, et al. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc Natl Acad Sci U S A. 2007;104:893–8. doi: 10.1073/pnas.0605442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 2004;23:3844–53. doi: 10.1038/sj.emboj.7600394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–8. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–20. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 38.Kosoy A, Calonge TM, Outwin EA, O’Connell MJ. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem. 2007;282:20388–94. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- 39.Chang YG, et al. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J Biol Chem. 2008;283:13370–7. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- 40.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–9. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, et al. Small Ubiquitin-related Modifier (SUMO) Binding Determines Substrate Recognition and Paralog-selective SUMO Modification. J Biol Chem. 2008;283:29405–15. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fallon L, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–42. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 43.Hoeller D, et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–8. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 45.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targetting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 46.Luft JR, et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–9. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Sweet RM, Carter CW Jr., editors. Macromolecular Crystallography. Vol. 276. Quid Ltd.; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 50.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 53.Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J Mol Biol. 1999;288:427–39. doi: 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.