Abstract
Objective:
Catechol-O-Methyltransferase (COMT) and Methylenetetrahydrofolate reductase (MTHFR) had been reported to relate to depression but with inconsistent results. The basal ganglia are also important in the pathophysiology of affective disorder via connections with limbic system and prefrontal cortex. The authors examined the relationship between an interaction of COMT/MTHFR polymorphisms and volumes of putamen in depressed and nondepressed elders.
Methods:
Participants included 170 depressed and 83 nondepressed subjects aged 60 years or older. Subjects completed cross-sectional assessments, including clinical evaluation, brain magnetic resonance imaging (MRI) scan, and COMT Val158Met and MTHFR C677T genotyping. Putamen volumes were measured using 1.5-Tesla whole-body MRI system. Statistical models examined the relationship between COMT/MTHFR genotype, proportional volumes of putamen and depression while controlling for age and sex.
Results:
After controlling for covariates, depressed subjects with MTHFR C/C, both the right and left putamen have smaller volumes as the number of COMT 158Val increase. The left putamen volumes of depressed subjects with COMT Met/Met are smaller as the number of MTHFR 677T increase compared to nondepressed subjects.
Conclusions:
Our findings do not support a major role for COMT or MTHFR alone. However, an epigenetic interaction of COMT Val158Met and MTHFR C677T polymorphisms may contribute to putamen volumes differences between depressed and nondepressed subjects. Further studies with a larger sample size are necessary to support a genetically based role for basal ganglia structures in the etiopathogenesis of depression.
Keywords: MTHFR, COMT, putamen, genetics, depression
INTRODUCTION
Geriatric depression is characterized by structural and functional abnormalities in the frontal lobe, basal ganglia (particularly the caudate and putamen), hippocampus, and amygdala (Beyer and Krishnan, 2002). The basal ganglia may be particularly important in the pathophysiology of affective disorder as it is connected with both the limbic system and prefrontal cortex (Steffens et al., 1998). Previous studies focusing on the basal ganglia report that depressed subjects have more severe subcortical gray-matter hyperintensities (particularly in the putamen) compared with nondepressed subjects (Tupler et al., 2002). Some studies reported age of first depressive episode was related to putamen volume (Parashos et al, 1998) and elderly depressed patients were found to have smaller caudate and putamen (Krishnan et al., 1993). The caudate and putamen are components of corticostriatal circuits involved in affect, motivation, executive and motor function (Seger, 2008), which are neurocognitive domains which commonly exhibit deficits in older depressed adults (Butters et al., 2004).
Depression is a clinically heterogeneous disorder thought to result from an interaction of multiple genes with environmental influences and developmental epigenetic components (Lesch, 2004). Although we have a growing body of research identifying genetic polymorphisms related to depression and a separate body of work examining genetic influences on brain structure and function, few studies have examined genetic influences on brain structure in context of depression. Such work is critical as we conduct association studies analyzing genetic polymorphisms with presumed functional significance in the pathophysiology of mood disorders.
Catechol-O-methyltransferase (COMT) is a methylation enzyme engaged in the degradation of norepinephrine and dopamine by catalyzing the transfer of a methyl group from S-adenosylmethionine (SAM). Biochemical studies have established different enzyme activity in patients with major depression than in nondepressed subjects (Karege et al., 1987). In a multicenter European study, an association was found between COMT Val158Met (G324A) functional polymorphism and major depressive disorder (Massat et al., 2005). The valine allele has been reported to result in a three- to fourfold higher activity as compared to the methionine allele (Lachman et al., 1996), and the Met158 presumably results in greater synaptic levels of dopamine (Lotta et al., 1995). One recent report suggests an association between the higher activity COMT 158Val/Val genotype and poorer antidepressant treatment response (Baune et al., 2008).
A second pathway of interest that has been related to depression is the B12- and folate-dependent pathway involved in the conversion of homocysteine to methionine. Elevated homocysteine levels, caused by B12 or folate deficiencies, or seen with specific genetic polymorphisms, have been associated with depression. Plausible hypotheses for this association include the observation that high homocysteine levels increase the risk of cerebral vascular disease and also adversely affect the levels of monoamine neurotransmitters (Folstein et al., 2007). The thermolabile variant of methylenetetrahydrofolate reductase (MTHFR) C677T T/T (results in a alanine to valine substitution) homozygotes have been shown to have higher homocysteine levels (Lewis et al., 2006; Sunder-Plassmann and Födinger, 2003). Two meta-analyses demonstrate the association between the MTHFR C677T variant and depression (Gilbody et al., 2007; López-León et al., 2007). Several studies reported the polymorphism of MTHFR C677T may be causally related to risk of depression (Lewis et al., 2006; Bjelland et al., 2003).
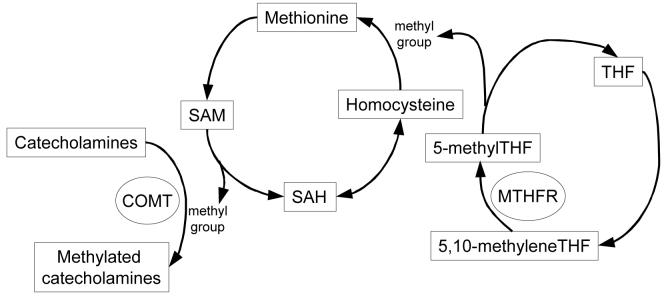
The metabolic pathways of COMT and MTHFR are interconnected (Figure 1; Mudd et al., 2001). In this study, we examined if there was an interaction between these polymorphisms and putamen volume. We hypothesized: 1) the interaction of COMT and MTHFR genotype are related to depression by influencing the putamen volume; and 2) low activity MTHFR 677T and high activity COMT 158Val would be negatively associated with the volumes of putamen in depressed subjects.
Figure 1.
The interconnected metabolic pathways of COMT and MTHFR. COMT: catechol-O-methyltransferase; MTHFR: methylenetetrahydrofolate reductase; SAH: S-adenosylhomocysteine; SAM: S-adenosylmethionine; THF: tetrahydrofolate.
METHODS
Sample
Depressed subjects were participants in the National Institute of Mental Health (NIMH) Conte Center for the Neuroscience of Depression in Late Life, located at Duke University Medical Center. Eligibility was limited to patients aged 60 years or older with a diagnosis of unipolar major depression. Exclusion criteria included: 1) another major psychiatric illness; 2) history of alcohol or drug abuse or dependence; 3) primary neurologic illness, including dementia; and 4) any contraindication to magnetic resonance image (MRI). Subjects were recruited for the study primarily through clinical referrals to the study.
Comparison subjects were community-dwelling recruited from the Aging Center Subject Registry at Duke University. Eligible comparison subjects had a nonfocal neurological examination, no self-report of neurologic or psychiatric illness, and no evidence of a depression diagnosis based on NIMH Diagnostic Interview Schedule (DIS; Robins et al., 1981).
The study protocol was approved by the Duke University Medical Center Institutional Review Board. All subjects provided written informed consent before beginning study procedures.
Clinical Evaluation
A trained interviewer administered the Duke Depression Evaluation Schedule (DDES) to each subject. The DDES, a composite diagnostic interview instrument, includes sections of the DIS assessing depression, enriched with items assessing sleep problems and the clinical features of melancholia and psychosis, dysthymia, mania, and alcohol abuse or dependence. The DDES also includes the Montgomery-Asberg Depression Rating Scale (MADRS; Montgomery and Asberg, 1979). All depressed subjects were additionally evaluated by a study geriatric psychiatrist, who reviewed entry criteria, current psychiatric symptoms, and their psychiatric history.
Subjects were excluded if they had a diagnosis of dementia or if the study geriatric psychiatrist suspected dementia at baseline. The majority of subjects had Mini-Mental State Exam (MMSE; Folstein et al., 1975) scores above 24; some severely depressed individuals had scores below 25. These subjects were followed through an acute three-month treatment phase; if the scores remained below 25, they were not included in this study.
Magnetic Resonance Image Acquisition and Analysis
MRI Acquisition
Subjects were imaged under an Institutional Review Board Approved protocol, with a 1.5 Tesla whole-body MRI system (Signa, GE Medical Systems, Milwaukee, WI) using the standard head (volumetric) radiofrequency coil. Padding was used to immobilize the head without causing discomfort. The scanner alignment light was used to adjust the head tilt and rotation so that the axial plane lights passed across the cantho-meatal line and the sagittal lights were aligned with the center of the nose. A rapid sagittal localizer scan was acquired to confirm the alignment.
A dual-echo fast spin-echo (FSE) acquisition was obtained in the axial plane for morphometry of total cerebrum and putamen. The pulse sequence parameters were TR = 4000 ms, TE = 30, 135 ms, 32 KHz (±16KHz) full imaging bandwidth, echo train length = 16, a 256 × 256 matrix, 3-mm section thickness, 1 Nex and a 20-cm FOV. The images were acquired in two separate acquisitions with a 3-mm gap between sections for each acquisition. The second acquisition was offset by 3 mm from the first so that the resulting data set consisted of contiguous sections.
Image Processing
The MR images were transferred to the Neuropsychiatric Imaging Research Laboratory (NIRL), located at Duke University Medical Center, for processing on SUN workstations, and secondary archive. Two computer programs were used to make volume measurements. Putamen volumes were determined using the GRID Program which was developed at NIRL. All other volume measurements used a NIRL-modified version of MrX Software, which was created by GE Corporate Research and Development (Schenectady, NY) and originally modified by Brigham and Women's Hospital for image segmentation (Boston, MA).
The segmentation protocol used by NIRL has been described previously (Payne et al., 2002). This was a supervised, semi-automated method that used the multiple MR contrasts available to identify different tissue classifications through a ‘seeding’ process wherein a trained analyst manually selected pixels in each tissue type that was to be identified (such as gray matter, white matter, CSF, lesions, background). Once the brain was segmented into tissue types and the non-brain tissue stripped away through a masking procedure, specific regions of interest (ROI) were assessed using tracing and connectivity functions. The cerebral hemispheres were traced, and a mask was created and applied to the segmented brain. The final step was to run a summarizing program that calculated the volume of each tissue type within the specific ROI defined by the analyst. Volumes were determined for the whole brain and cerebrum (serving as a marker of head size).
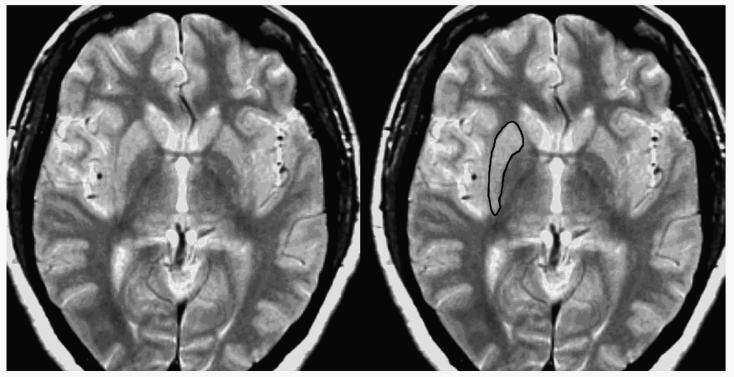
The method for measuring the putamen has also been described previously (Figure 2; Greenberg et al., 2008). Tracing began on the most inferior slice on which the putamen was separable the caudate. Hyperintensities were included if they appeared within the body of the putamen; they were excluded if they appeared along the border. The globus pallidus and the claustrum were both excluded. If the lateral border of the putamen appeared to be fused with the insular cortex, the most posterior point at which they were separable was connected to the most anterior point at which they were separable. The superior border of the putamen was defined as the most superior slice on which it was visible.
Figure 2.
This is a proton density image showing same slice with and without tracing of right putamen.
All NIRL image analysis technicians received extensive training by experienced analysts. Reliability was established by repeated measurements on multiple MR scans before raters were approved to process study data. Intraclass correlation coefficients (ICC's) attained were as follows: total brain = 0.998, left cerebral hemisphere = 0.996, right cerebral hemisphere = 0.997, total cerebrum = 0.997, left putamen = 0.8 and right putamen = 0.7 (putamen reliability was based upon two raters and 10 scans, each scan was processed twice by each rater after an interval of at least one week, scans were randomly selected for reliability so as to include depression/control subjects, as well as a range of scan qualities).
Genotyping
DNA was extracted from fresh and frozen blood and stored according to previously reported methods (Rimmler et al., 1998). An aliquot of DNA was used for COMT genotyping following methods previously published (Lachman et al., 1996) using polymerase chain reaction (PCR) amplification with a Taqman by-design assay (Applied Biosystems) that recognized the single nucleotide polymorphism (SNP) which defines the Val158Met polymorphism (rs4680). MTHFR genotyping was PCR-amplified applying a Tagman by-design assay, too. The samples were examined with an ABI7900 DNA analyzer and the genotypes determined with the SDS software package (Applied Biosystems). Greater than 95% genotyping efficiency was required before data were submitted for further analysis. We had blinded duplicates that were also used in the genotyping analyses and they were required to match 100% before we would accept the data for analysis.
Statistical Analysis
Univariate comparisons were done using T-test to compare differences means of age, MMSE and brain volumes. Chi-square tests were used to compare differences in frequencies of sex and genotypes between depressed and nondepressed subjects.
Linear regression models were fit to test for the effect that subjects' diagnostic status (depressed or nondepressed), COMT genotype and MTHFR genotype has on both the left and right putamen (proportional volumes, defined as regional volume divided by cerebrum) as outcome variables. The predictor variables were the main effects of patient status, COMT genotype and MTHFR genotype and a three-way interaction of the main effect was fit in the model with all appropriate two-way interactions. Both gene variables were fit in the model as an ordinal variable because of some small cell counts. The model was fit with age and gender as covariates.
Post-hoc analyses were done driven by inspection of the results and data from hypothesized analysis. Linear regression models were fit to test these new hypotheses. Two sets of analyses were done based on a group of each of the two genes. One set of analyses the models were fit with the COMT genotype as a dichotomized variable with Met/Met in one group and Val/Val or Val/Met in the other group. A three-way interaction was tested using the variable with MTHFR genotype as an ordinal variable and testing for interaction with patient status while including all appropriate main effects and two-way interactions and controlling for age and gender. The second set of analyses fit model with MTHFR genotype as a dichotomized variable with C/C in one group and C/T or T/T in the other group. A three-way interaction was tested using the variable with the COMT genotype as an ordinal variable and the same conditions as above.
RESULTS
The sample for this study, as given in table 1, includes 253 Caucasian adults 60 years of age or older. 170 subjects had a diagnosis of depression and 83 were nondepressed subjects. There was no difference between diagnostic groups in average age, sex representation, COMT genotype frequency, MTHFR genotype frequency, or unadjusted MRI measures. The nondepressed subjects exhibited significantly higher MMSE scores.
Table 1.
Demographics and Univariate Comparisons
| Depressed subjects (n = 170) |
Control subjects (n = 83) |
Total (n = 253) |
t or χ2 | p Value | |
|---|---|---|---|---|---|
| Age | 69.4 (7.5) | 69.8 (5.6) | 69.5 (6.9) | t = 0.49 | p = 0.623 |
| Sex: female | 65.9% (112) | 73.5% (61) | 68.4% (173) | χ2 = 149 | p = 0.222 |
| Baseline MADRS | 27.0 (7.9) | -------- | 27.0 (7.9) | -------- | -------- |
| MMSE scores | 28.3 (2.0) | 29.1 (1.0) | 28.6 (1.8) | t = 3.90 | p = 0.0001 |
|
| |||||
| COMT Val158Met: | χ2 = 113 | p = 0.567 | |||
| Val/Val | 25.3% (43) | 27.7% (23) | 26.1% (66) | ||
| Val/Met | 49.4% (84) | 53.0% (44) | 50.6% (128) | ||
| Met/Met | 25.3% (43) | 19.3% (16) | 23.3% (59) | ||
|
| |||||
| MTHFR C677T: | χ2 = 103 | p = 0.598 | |||
| T/T | 11.2% (19) | 10.8% (9) | 11.1% (28) | ||
| C/T | 46.5% (79) | 53.0% (44) | 48.6% (123) | ||
| C/C | 42.4% (72) | 36.1% (30) | 40.3% (102) | ||
|
| |||||
| Left Putamen | 3.61 (0.76) | 3.65 (0.63) | 3.62 (0.72) | t = −0.34 | p = 0.731 |
| Right Putamen | 3.63 (0.78) | 3.69 (0.71) | 3.65 (0.76) | t = 0.51 | p = 0.610 |
| Cerebrum | 1156.4 (130.8) | 1163.6 (123.3) | 1158.6 (128.2) | t = 0.42 | p = 0.675 |
This table shows demographic and univariate comparisons. Continuous variables presented as mean (standard deviation). Nominal variables presented as percentage (n). Age is in years. Volume measures are in milliliters. MADRS: Montgomery-Asberg Depression Rating Scale; MMSE: Mini-Mental State Exam; COMT: Catechol-O-methyltransferase; MTHFR: methylenetetrahydrofolate reductase
Proportional putamen volumes of depressed and nondepressed subjects are presented in table 2 grouped by subjects' genetic make up. A model was run to test for an interaction of subject's status, COMT genotype and MTHFR genotype in a dose response manner for the number of COMT 158Val and MTHFR 677T in each gene. Age and gender were also included in the model as covariates and were found to be significant in all models. In models of both the left and right putamen, we did not observe significant three-way interactions between diagnosis of depression, COMT genotype, and MTHFR genotype.
Table 2.
Left and Right Putamen Proportional Volumes
| Left putamen | |||
|---|---|---|---|
| Depressed subjects |
COMT = Val/Val | COMT = Val/Met | COMT = Met/Met |
| MTHFR=T/T | 0.00330 (0.00033), n=5 | 0.00314 (0.00023), n=10 | 0.00296 (0.00037), n=4 |
| MTHFR=C/T | 0.00331 (0.00015), n=24 | 0.00318 (0.00012), n=35 | 0.00307 (0.00016), n=20 |
| MTHFR=C/C | 0.00275 (0.00020), n=14 | 0.00309 (0.00012), n=39 | 0.00347 (0.00017), n=19 |
|
| |||
|
Control subjects |
COMT = Val/Val | COMT = Val/Met | COMT = Met/Met |
|
| |||
| MTHFR=T/T | 0.00292 (0.00041), n=2 | 0.00331 (0.00033), n=3 | 0.00305 (0.00029), n=4 |
| MTHFR=C/T | 0.00324 (0.00017), n=12 | 0.00311 (0.00011), n=27 | 0.00337 (0.00026), n=5 |
| MTHFR=C/C | 0.00329 (0.00019), n=9 | 0.00307 (0.00015), n=14 | 0.00305 (0.00022), n=7 |
|
| |||
| Right Putamen | |||
|
| |||
|
Depressed subjects |
COMT = Val/Val | COMT = Val/Met | COMT = Met/Met |
|
| |||
| MTHFR=T/T | 0.00305 (0.00034), n=5 | 0.00318 (0.00024), n=10 | 0.00312 (0.00039), n=4 |
| MTHFR=C/T | 0.00335 (0.00016), n=24 | 0.00323 (0.00013), n=35 | 0.00328 (0.00017), n=20 |
| MTHFR=C/C | 0.00286 (0.00021), n=14 | 0.00296 (0.00012), n=39 | 0.00342 (0.00018), n=19 |
|
| |||
|
Control subjects |
COMT = Val/Val | COMT = Val/Met | COMT = Met/Met |
|
| |||
| MTHFR=T/T | 0.00357 (0.00043), n=2 | 0.00336 (0.00035), n=3 | 0.00336 (0.00031), n=4 |
| MTHFR=C/T | 0.00309 (0.00018), n=12 | 0.00312 (0.00012), n=27 | 0.00335 (0.00027), n=5 |
| MTHFR=C/C | 0.00353 (0.00020), n=9 | 0.00316 (0.00016), n=14 | 0.00278 (0.00023), n=7 |
This table shows the proportional volumes of left and right putamen of depressed and control subjects grouped by genetic make up. Data are presented as mean (standard deviation). COMT: Catechol-O-methyltransferase; MTHFR: methylenetetrahydrofolate reductase
In order to better understand our observations of the raw data, we conducted post-hoc analyses of the data in table 2.
For the first set of analyses, groups were created to compare subjects with MTHFR C/C to those with MTHFR C/T or T/T. For both the left and right putamen, models were fit to test for interaction between diagnostic group, the variable of constructed MTHFR groups, and COMT as an ordinal variable, while age and sex were included as covariates. The models were significant for both the right putamen (beta = −0.00071, p = 0.0078) and the left putamen (beta = −0.00056, p = 0.0244) proportional volume (Table 3). So in depressed subjects with MTHFR C/C, both the right and left putamen exhibit significantly smaller volumes as the number of COMT 158Val increase.
Table 3. Post-hoc analyses (1).
MTHFR C/C vs. MTHFR C/T or T/T (dichotomized variable) COMT genotype as an ordinal variable
| Left putamen | ||||
|---|---|---|---|---|
| Predictor | Beta | F | df | p-value |
| Depressed/ Control | −0.00013 | 0.71 | 1 | 0.4016 |
| COMT | −0.00004 | 0.04 | 1 | 0.8435 |
| MTHFR | −0.00021 | 0.10 | 1 | 0.7581 |
| Depressed*COMT | 0.00016 | 0.97 | 1 | 0.3249 |
| Depressed*MTHFR | 0.00052 | 2.69 | 1 | 0.1024 |
| COMT*MTHFR | 0.00017 | 0.84 | 1 | 0.3608 |
| Depressed*COMT*MTHFR | −0.00056 | 5.13 | 1 | 0.0244 |
|
| ||||
| Right putamen | ||||
|
| ||||
| Predictor | Beta | F | df | p-value |
|
| ||||
| Depressed/ Control | −0.00005 | 1.68 | 1 | 0.1965 |
| COMT | −0.00011 | 0.12 | 1 | 0.7282 |
| MTHFR | −0.00051 | 2.26 | 1 | 0.1338 |
| Depressed*COMT | 0.00013 | 2.89 | 1 | 0.0904 |
| Depressed*MTHFR | 0.00053 | 2.49 | 1 | 0.1161 |
| COMT*MTHFR | 0.00049 | 0.95 | 1 | 0.3305 |
| Depressed*COMT*MTHFR | −0.00071 | 7.19 | 1 | 0.0078 |
Age and sex were included as covariates
For the second set of analyses, groups were created to compare COMT Met allele homozygous subjects with COMT Val allele carriers. For both the left and right putamen, models were fit to test for interaction between diagnostic group, the variable of constructed COMT groups, and MTHFR as an ordinal variable, while age and sex were included as covariates. The models for the left putamen (beta = −0.00063, p = 0.0396) but not the right putamen (p = 0.1462) proportional volume was found to be significant (Table 4). The left putamen volumes of depressed subjects homozygous for the COMT Met allele are smaller as the number of MTHFR 677T alleles increase.
Table 4. Post-hoc analyses (2).
COMT Met/Met vs. COMT Val/Met or Val/Val (dichotomized variable) MTHFR genotype as an ordinal variable
| Left putamen | ||||
|---|---|---|---|---|
| Predictor | Beta | F | df | p-value |
| Depressed/ Control | −0.00043 | 0.97 | 1 | 0.3268 |
| COMT | −0.00022 | 0.21 | 1 | 0.6441 |
| MTHFR | −0.00015 | 0.59 | 1 | 0.4424 |
| Depressed*COMT | 0.00058 | 3.95 | 1 | 0.0479 |
| Depressed*MTHFR | 0.00049 | 1.44 | 1 | 0.2320 |
| COMT*MTHFR | 0.00023 | 0.33 | 1 | 0.5686 |
| Depressed*COMT*MTHFR | −0.00063 | 4.28 | 1 | 0.0396 |
|
| ||||
| Right putamen | ||||
|
| ||||
| Predictor | Beta | F | df | p-value |
|
| ||||
| Depressed/ Control | −0.00038 | 0.64 | 1 | 0.4237 |
| COMT | −0.00038 | 0.60 | 1 | 0.4388 |
| MTHFR | −0.00018 | 0.59 | 1 | 0.4423 |
| Depressed*COMT | 0.00051 | 2.63 | 1 | 0.1062 |
| Depressed*MTHFR | 0.00038 | 0.77 | 1 | 0.3799 |
| COMT*MTHFR | 0.00035 | 0.49 | 1 | 0.4856 |
| Depressed*COMT*MTHFR | −0.00048 | 2.12 | 1 | 0.1462 |
Age and sex were included as covariates
To investigate whether our findings were specific to the putamen or reflective of a genetic influence on the broader striatum, we performed similar analyses on a measure of caudate volume obtained from previously published methods (Beyer et al., 2004). We found no statistically significant relationship between caudate volume and the COMT polymorphism, the MTHFR polymorphism, or a gene-gene interaction (data not shown).
DISCUSSION
To our knowledge, this is the first study to examine the interaction of COMT genotype and MTHFR genotype on basal ganglia volumes in geriatric depression. We found a significant interaction between putamen volume and depression diagnosis, COMT genotype, and MTHFR genotype. Our findings do not support a major role for COMT and MTHFR genotype alone, but provide initial evidence for genetic epistasis, where the combination of both polymorphisms and putamen volume might increase susceptibility of depression.
There were no significant associations between putamen volumes and either genotype of MTHFR or COMT in our study. However, post-hoc analyses found that when depressed subjects with MTHFR C/C, both the right and left putamen have smaller volumes as the number of COMT 158Val increase. Genetic association studies are not consistent regarding the involvement of COMT 158Val or 158Met allele in the etiology of depressive disorder (Massat et al., 2005; Ohara et al., 1998). The results of our study may partially explain why COMT analyses have been inconsistent in previously studies in relationship with depressive disorder (Funke et al., 2005; Kunugi et al., 1997), which contained a mixture of MTHFR C/C, C/T and T/T.
The MTHFR polymorphism could regulate COMT function via methylation of the COMT promoter region (Roffman et al., 2008; Sasaki et al., 2003; Friso et al., 2002). The activity of MTHFR C/C is much higher than C/T or T/T. MTHFR C/C would presumably result in hypermethylation of the COMT promoter region and reduced COMT expression. Under this low expressive condition, it is possible that the influence of COMT Val158Met polymorphism may become more obvious.
The relationship between MTHFR and COMT polymorphism may be complex. One link between these pathways and depression may lie in alterations in striatal dopamine structure or function. Striatal dopamine levels are associated with working memory and motor speed in healthy older adults (Landau et al., 2008). This observation supports a study associating psychomotor speed in depression with caudate volume and MTHFR C677T genotype (Naismith et al., 2002) and a study observing an interactive effect of these COMT and MTHFR polymorphisms on executive function in schizophrenia (Roffman et al., 2008). The low-activity MTHFR 677T allele could increase homocysteine level and may have an indirect and negative effect on COMT activity through increase the SAH (a strong inhibitor of COMT) levels (Yi et al., 2000). Moreover, the low-activity MTHFR would result in SAM depletion and negatively influence COMT function due to insufficiency of methyl groups necessary for the successive methylation processes (Finkelstein, 1990).
Some previous studies showed MTHFR C677T polymorphism was associated with risk of depression (Gilbody et al., 2007; López-León et al., 2007; Bjelland et al., 2003). But other researches reported no association between MTHFR and depressive disorder (Kunugi et al., 1998; Gaysina et al., 2007). In this study, we found that depressed subjects with COMT Met/Met, the left putamen volumes were smaller as the number of MTHFR 677T increase. COMT 158Met allele produces a form of COMT that has reduced activity (Lachman et al., 1996). The low-activity COMT Met/Met polymorphism would result in greater levels of synaptic neurotransmitter (Lotta et al., 1995) and display lower homocysteine levels (Tunbridge et al., 2008). It is possible that low-activity COMT Met/Met is more sensitive to the levels of SAM or methyl groups. The role of MTHFR C677T polymorphism may become more prominent due to it is the key regulatory enzyme to determine the SAM levels for COMT methylation (Muntjewerff et al., 2008). However, this is inconsistent with the finding that the effect of MTHFR C677T polymorphism is more important under higher level of homocysteine or COMT 158Val allele in other studies (Roffman et al., 2008; Tunbridge et al., 2008; Friso et al., 2002). The relationship between MTHFR genotype and depressive disorder may become more complicated when combining the effects of polymorphisms of other potential interactive genes.
The primary limitation of this study is a small sample size, especially for a gene interaction study. Other limitations include its cross-sectional nature that limits our ability to make conclusions regarding etiology and the mechanism behind this relationship. As in any study of older individuals, there is also the potential for confounding by early, comorbid dementia; however our method of assessment lowers the risk.
Our results, although preliminary, support that the interactions of MTHFR and COMT polymorphisms are related to putamen volume differences between depressed and nondepressed subjects. This requires further study and replication with larger sample size to support a role for basal ganglia structures in the etiopathogenesis of depression. Additional work may verify whether MTHFR genotype dependent methylation of COMT promoter region, with consequent alteration in neurotransmitters, underlies this effect. Combining MRI studies with genetics has the potential to localize abnormalities in metabolism and neurotransmitter and provide a better integrated model of depression.
In summary, an epigenetic interaction of MTHFR C677T and COMT Val158Met polymorphisms may contribute to putamen volumes differences in depressive disorder. It is not clear if interactions between the MTHFR C677T and COMT Val158Met polymorphisms are related to other critical issues of late-life depression such as cognitive deficits, treatment outcomes, or mortality. Future studies examining these issues should test for potential interactions between these polymorphisms and basal ganglion associated with depression as well as examine for a relationship with dietary and metabolic factors related to these pathways, such as folate, vitamin B12 status, and homocysteine levels. The results of this study will provide important knowledge about the possible pathophysiology of depression and pave the way for further advanced research.
Acknowledgments
This study was supported by National Institute of Mental Health grants P50 MH60451, R01 MH54846, K24 MH70027, and K23 MH65939. The authors would like to thank Cynthia R. Key, Daniel L. Greenberg, and Denise F. Messer for image analysis, and Brian D. Boyd for computer programming and support.
Footnotes
- There were no significant independent relationships of the COMT Val158Met or MTHFR C677T polymorphisms on putamen volume in older depressed or nondepressed subjects
- There was a significant effect of the epigenetic interaction between the COMT Val158Met and MTHFR C677T polymorphisms on putamen volume, which was also related to a diagnosis of depression.
All authors declare no conflict of interest.
REFERENCES
- Baune BT, Hohoff C, Berger K, et al. Association of the COMT val158met Variant with Antidepressant Treatment Response in Major Depression. Neuropsychopharmacology. 2008;33:924–932. doi: 10.1038/sj.npp.1301462. [DOI] [PubMed] [Google Scholar]
- Beyer JL, Krishnan KR. Volumetric brain imaging findings in mood disorders. Bipolar Disord. 2002;4:89–104. doi: 10.1034/j.1399-5618.2002.01157.x. [DOI] [PubMed] [Google Scholar]
- Beyer JL, Kuchibhatla M, Payne M, et al. Caudate volume measurement in older adults with bipolar disorder. Int J Geriatr Psychiatry. 2004;19:109–114. doi: 10.1002/gps.1030. [DOI] [PubMed] [Google Scholar]
- Bjelland I, Tell GS, Vollset SE, et al. Folate, vitamin B12, homocysteine, and the MTHFR 677C>T polymorphism in anxiety and depression: the Hordaland Homocysteine Study. Arch Gen Psychiatry. 2003;60:618–626. doi: 10.1001/archpsyc.60.6.618. [DOI] [PubMed] [Google Scholar]
- Butters MA, Whyte EM, Nebes RD, et al. The nature and determinants of neuropsychological functioning in late-life depression. Arch Gen Psychiatry. 2004;61:587–595. doi: 10.1001/archpsyc.61.6.587. [DOI] [PubMed] [Google Scholar]
- Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- Folstein M, Liu T, Peter I, et al. The homocysteine hypothesis of depression. Am J Psychiatry. 2007;164:861–867. doi: 10.1176/ajp.2007.164.6.861. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state” a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Malhotra AK, Finn CT, et al. COMT genetic variation confers risk for psychotic and affective disorders: a case control study. Behav Brain Funct. 2005;1:19. doi: 10.1186/1744-9081-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaysina D, Cohen S, Craddock N, et al. No association with the 5,10-methylenetetrahydrofolate reductase gene and major depressive disorder: Results of the depression case control (DeCC) study and a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2007 doi: 10.1002/ajmg.b.30665. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Gilbody S, Lewis S, Lightfoot T. Methylenetetrahydrofolate reductase (MTHFR) genetic polymorphisms and psychiatric disorders: a HuGE review. Am J Epidemiol. 2007;165:1–13. doi: 10.1093/aje/kwj347. [DOI] [PubMed] [Google Scholar]
- Greenberg DL, Messer DF, Payne ME, et al. Aging, gender, and the elderly adult brain: an examination of analytical strategies. Neurobiol Aging. 2008;29:290–302. doi: 10.1016/j.neurobiolaging.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karege F, Bovier P, Gaillard JM, et al. The decrease of erythrocyte catechol-O-methyltransferase activity in depressed patients and its diagnostic significance. Acta Psychiatr Scand. 1987;76:303–308. doi: 10.1111/j.1600-0447.1987.tb02899.x. [DOI] [PubMed] [Google Scholar]
- Krishnan KR, McDonald WM, Doraiswamy PM, et al. Neuroanatomical substrates of depression in the elderly. Eur Arch Psychiatry Clin Neurosci. 1993;243:41–46. doi: 10.1007/BF02191522. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Vallada HP, Hoda F, et al. No evidence for an association of affective disorder with high- or low-activity allele of catechol-o-methyltransferase gene. Biol Psychiatry. 1997;42:282–285. doi: 10.1016/S0006-3223(96)00366-6. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Fukuda R, Hattori M, et al. C677T polymorphism in methylenetetrahydrofolate reductase gene and psychoses. Mol Psychiatry. 1998;3:435–437. doi: 10.1038/sj.mp.4000390. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Morrow B, Shprintzen R, et al. Association of codon 108/158 catechol-O-methyltransferase gene polymorphism with the psychiatric manifestations of velo-cardio-facial syndrome. Am J Med Genet. 1996;67:468–472. doi: 10.1002/(SICI)1096-8628(19960920)67:5<468::AID-AJMG5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, et al. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- Landau SM, Lal R, O'Neil JP, et al. Striatal Dopamine and Working Memory. Cereb Cortex. 2008 doi: 10.1093/cercor/bhn095. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP. Gene-environment interaction and the genetics of depression. J Psychiatry Neurosci. 2004;29:174–184. [PMC free article] [PubMed] [Google Scholar]
- Lewis SJ, Lawlor DA, Davey Smith G, et al. The thermolabile variant of MTHFR is associated with depression in the British Women's Heart and Health Study and a meta-analysis. Mol Psychiatry. 2006;11:352–360. doi: 10.1038/sj.mp.4001790. [DOI] [PubMed] [Google Scholar]
- López-León S, Janssens AC, González-Zuloeta Ladd AM, et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002088. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- Massat I, Souery D, Del-Favero J, et al. Association between COMT (Val158Met) functional polymorphism and early onset in patients with major depressive disorder in a European multicenter genetic association study. Mol Psychiatry. 2005;10:598–605. doi: 10.1038/sj.mp.4001615. [DOI] [PubMed] [Google Scholar]
- Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–389. doi: 10.1192/bjp.134.4.382. [DOI] [PubMed] [Google Scholar]
- Mudd SH, Levy HL, Kraus JP. Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- Muntjewerff JW, Gellekink H, den Heijer M, et al. Polymorphisms in catechol-O-methyltransferase and methylenetetrahydrofolate reductase in relation to the risk of schizophrenia. Eur Neuropsychopharmacol. 2008;18:99–106. doi: 10.1016/j.euroneuro.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Naismith S, Hickie I, Ward PB, et al. Caudate nucleus volumes and genetic determinants of homocysteine metabolism in the prediction of psychomotor speed in older persons with depression. Am J Psychiatry. 2002;159:2096–2098. doi: 10.1176/appi.ajp.159.12.2096. [DOI] [PubMed] [Google Scholar]
- Ohara K, Nagai M, Suzuki Y, et al. Low activity allele of catechol-o-methyltransferase gene and Japanese unipolar depression. Neuroreport. 1998;9:1305–1308. doi: 10.1097/00001756-199805110-00009. [DOI] [PubMed] [Google Scholar]
- Parashos IA, Tupler LA, Blitchington T, et al. Magnetic-resonance morphometry in patient with major depression. Psychiatry Res. 1998;84:7–15. doi: 10.1016/s0925-4927(98)00042-0. [DOI] [PubMed] [Google Scholar]
- Payne ME, Fetzer DL, MacFall JR, et al. Development of a semi-automated method for quantification of MRI gray and white matter lesion in geriatric subjects. Psychiatry Res: Neuroimaging. 2002;115:63–77. doi: 10.1016/s0925-4927(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Rimmler J, McDowell JG, Slotterback BD, et al. Development of a data coordinating center (DCC): data quality control for complex disease studies. Am J Hum Genet. 1998;(suppl 63):A240. [Google Scholar]
- Robins LN, Helzer JE, Croughan J, et al. National Institute of Mental Health Diagnostic Interview Schedule. Its history, characteristics, and validity. Arch Gen Psychiatry. 1981;38:381–389. doi: 10.1001/archpsyc.1981.01780290015001. [DOI] [PubMed] [Google Scholar]
- Roffman JL, Weiss AP, Deckersbach T, et al. Interactive effects of COMT Val108/158Met and MTHFR C677T on executive function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30684. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Kaneuchi M, Sakuragi N, et al. Multiple promoters of catechol-O-methyltransferase gene are selectively inactivated by CpG hypermethylation in endometrial cancer. Cancer Res. 2003;63:3101–3106. [PubMed] [Google Scholar]
- Seger CA. How do the basal ganglia contribute to categorization? Their roles in generalization, response selection, and learning via feedback. Neurosci Biobehav Rev. 2008;32:265–278. doi: 10.1016/j.neubiorev.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens DC, Tupler LA, Krishnan KR. Magnetic resonance imaging signal hypointensity and iron content of putamen nuclei in elderly depressed patients. Psychiatry Res. 1998;83:95–103. doi: 10.1016/s0925-4927(98)00032-8. [DOI] [PubMed] [Google Scholar]
- Sunder-Plassmann G, Födinger M. Genetic determinants of the homocysteine level. Kidney Int. 2003;(Suppl 84):S141–144. doi: 10.1046/j.1523-1755.63.s84.52.x. [DOI] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Warden DR, et al. Polymorphisms in the Catechol-O-methyltransferase (COMT) gene influence plasma total homocysteine levels. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30700. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Tupler LA, Krishnan KR, McDonald WM, et al. Anatomic location and laterality of MRI signal hyperintensities in late-life depression. J Psychosom Res. 2002;53:665–676. doi: 10.1016/s0022-3999(02)00425-7. [DOI] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, et al. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]