Abstract
Ranaspumin-2 (Rsn-2) is a monomeric, 11 kDa surfactant protein identified as one of the major foam nest components of the túngara frog (Engystomops pustulosus), with an amino acid sequence unlike any other protein described so far. We report here on its structure in solution as determined by high-resolution NMR analysis, together with investigations of its conformation and packing at the air-water interface using a combination of infrared and neutron reflectivity techniques. Despite the lack of any significant sequence similarity, Rsn-2 in solution adopts a compact globular fold characteristic of the cystatin family, comprising a single helix over a four-stranded sheet, in a motif not previously associated with surfactant activity. The NMR structure of Rsn-2 shows no obvious amphiphilicity that might be anticipated for a surfactant protein. This suggests that it must undergo a significant conformational change when incorporated into the air-water interface that may involve a hinge-bending, clamshell opening of the separate helix and sheet segments to expose hydrophobic faces to air while maintaining the highly polar surfaces in contact with the underlying water layer. This model is supported by direct observation of the relative orientations of secondary structure elements at the interface by infrared reflection absorption spectroscopy, and by protein packing densities determined from neutron reflectivity profiles.
Introduction
Protein foams are relatively rare in biology, but they provide an intriguing opportunity to explore an unusual form of soft matter involving interactions of biological macromolecules at air-water interfaces. Ranaspumin-2 (Rsn-2) is an 11 kDa surfactant protein that has been identified as one of the components in foam nests of the túngara frog (Engystomops pustulosus = Physalaemus pustulosus), which is commonly found in tropical regions of Central America (1). Such foam nests act as biocompatible incubators for fertilized eggs and embryos while remaining resilient against both mechanical and microbial disruption. In previous work (2) we showed that the natural foam material from this species is made up of a dilute cocktail of proteins and complex carbohydrates (∼1 mg mL−1 each) with significant surfactant properties. A subsequent sequence analysis of natural material and trials with recombinant versions revealed that some of these ranaspumins are lectins, with carbohydrate-binding properties probably related to antimicrobial protection and long-term stability of the foam (1). The functions of other proteins in the mixture are less obvious. One in particular, Rsn-2, has an amino acid sequence (GenBank AY226147) unlike any other currently known, but with an unusual amphipathic profile hinting at possible surfactant activity. This has been confirmed by experiments showing that recombinant Rsn-2 provides a significant reduction in the surface tension of aqueous solutions at concentrations as low as 10 μg mL−1.
Such surfactant activity is almost unprecedented in natural proteins, and deliberate foaming and surface interaction are usually best avoided under normal circumstances because of the denaturing effects they can have on protein structure. However, there are exceptions, including the lung surfactants that allow alveolar expansion and also act as a defense against inhaled pathogens (3), surfactin and other microbial lipopeptide surfactants (4), milk caseins (5,6), and, most notably, the hydrophobins of filamentous fungi that reduce surface tension and facilitate growth in thin water layers and at air-water interfaces (7,8). All of these systems differ in various ways from the materials we describe here. Pulmonary surfactants are made up of a mixture of proteins (∼10%) and phospholipids (∼90%) that coat the narrow airways in the lung. The surfactant activity arises mainly from the phospholipid component, with a range of surfactant and plasma proteins to facilitate spreading and other functions. The caseins are a heterogeneous class of surfactant/micelle-forming proteins that are extensively phosphorylated and have flexible, open, natively unfolded structures. Their surfactant/micelle behavior remains somewhat controversial and is complicated by significant calcium-binding activity (9). Hydrophobins are a class of small proteins (7–9 kDa) unique to filamentous fungi, with a characteristic four-disulphide bridge motif and amphipathic tertiary structure related to the self-assembly and surfactant properties of these proteins. In contrast, the relatively nonspecific foaming of denatured proteins is commonplace and widely exploited in food technology and other processes. However, this usually requires much higher protein concentrations and much more vigorous physical treatment than is the case with the specific surfactant and related proteins described here. The process is generally acknowledged to be associated with the higher hydrophobicity and/or increased viscosity of denatured protein in which physical entrapment of air bubbles is facilitated in concentrated viscous mixtures (10,11). This is usually the dominant mechanism in familiar culinary processes such as the whipping of cream or preparation of meringue from egg white, for example. Other biosurfactants, mostly of microbial origin, are predominantly lipid-based (4,12).
Rsn-2 has no sequence similarity to hydrophobins, lipopeptides, or lung surfactants, involves no associated lipid, and is active at much lower concentrations than are normally required for foaming of denatured proteins. Consequently, it represents a new class of surfactant protein. Here we describe the high-resolution NMR structure of Rsn-2 in solution, which, together with complementary surface IR and neutron reflection measurements, suggests a novel mechanism for its surface activity.
Materials and Methods
Protein preparation and characterization
Recombinant Rsn-2 was prepared as described elsewhere (1). Briefly, Rsn-2 was overexpressed as a thrombin-cleavable N-terminal His6 fusion-tagged form in Escherichia coli BL21(DE3) cells (Novagen) using the pCR T7/NT-TOPO expression vector incorporating the DNA sequence of Rsn-2 isolated from oviduct tissue of E. pustulosus after mating. Soluble protein from sonicated cells was subsequently purified by a combination of nickel affinity chromatography, thrombin cleavage, and gel-filtration chromatography, to yield homogeneous protein (as assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis) of the appropriate size as confirmed by matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Isotope-enriched (13C, 15N) protein for NMR studies was obtained using M9 minimal media (13) incorporating 15NH4Cl or 13C-D-glucose (Spectra Stable Isotopes, Andover, MA) as sole nitrogen or carbon sources during overexpression. Protein concentrations were determined from UV absorbance using a molar extinction coefficient for Rsn-2, ɛ278 = 5960 M−1 cm−1, estimated from its amino acid composition (zero Trp, four Tyr, four Phe, and two Cys) according to Gill and Von Hippel (14) and using the online ProtParam tool. Circular dichroism (CD) and differential scanning calorimetry (DSC) data for Rsn-2 in solution were obtained using Jasco (Tokyo, Japan) J-810 and MicroCal (Northampton, MA) VP-DSC instruments.
Solution structure determination
NMR resonance assignment of 15N,13C-labeled Rsn-2 was achieved using standard triple resonance techniques with data recorded at 308 K in 20 mM sodium phosphate, 50 mM NaCl, 0.02% w/v NaN3, pH 7.0 on a 14.1 T AVANCE spectrometer (Bruker, Karlsruhe, Germany) equipped with a Cryoprobe. Data were processed with AZARA (15) and analyzed with CCPN analysis software (16). Structures were calculated based on distance restraints derived from 15N- and 13C-edited 3D-nuclear Overhauser enhancement spectroscopy (NOESY) experiments recorded with 120 ms mixing times. Unassigned NOESY cross-peaks were used to generate ambiguously assigned restraints. Structure calculations were implemented in CNS (17) using ARIA (18) tools to filter ambiguously assigned restraints as described previously (19). The structures were refined using distance restraints representing 17 hydrogen bonds inferred from deuterium exchange experiments and the incorporation of an intramolecular disulphide bond. The final ensemble of structures was further refined in explicit solvent (20). The coordinates have been deposited in the Protein Data Bank (PDB ID: 2wgo).
Infrared reflection absorption spectroscopy
For infrared reflection absorption spectroscopy (IRRAS) measurements, deionized water with a resistivity of 18.2 MΩ cm (SG Wasseraufbereitung und Regenerierstation GmbH, Barsbüttel, Germany) was used. All experiments were performed with a Wilhelmy film balance (Riegler & Kirstein, Berlin, Germany) using a filter paper as the Wilhelmy plate. Two Teflon troughs of different sizes (300 × 60 × 3 mm and 60 × 60 × 3 mm for the reference trough) were linked by small water-filled bores to ensure equal height of the air-water interface in both troughs. The temperature of the subphase was maintained at 20 ± 0.5°C. Measurements were performed in the small trough, with the larger one serving as the reference trough. A plexiglas hood covered both troughs to minimize evaporation of water. Both troughs were filled with deionized water. IR spectra were recorded with an Equinox 55 FT-IR spectrometer (Bruker, Karlsruhe, Germany) connected to an XA 511 reflection attachment (Bruker) with an external narrow-band MCT detector using the trough system described above. The IR beam is focused by several mirrors onto the water surface and different angles of incidence can be adjusted. A computer-controlled rotating KRS-5 polarizer (>98% degree of polarization) was used to generate linearly polarized light. The trough system was positioned on a moveable platform so as to shuttle between the sample and the reference trough. This shuttle technique diminishes the spectral interferences due to the water vapor absorption in the light beam (21).
Protein adsorption experiments at the air-water interface were performed in the small reference trough by injection of a concentrated protein solution (228 μM) into the subphase. The acquisition of the IRRAS spectra started before the injection of the protein into the subphase. Initially, parallel (p)-polarized light with an angle of 40° with respect to the normal of the water surface was used. When the surface pressure reached a constant value, measurements at different angles of incidence using p-polarized light were performed.
The single-beam reflectance intensities from reference (R0) and sample (R) trough surfaces were used to calculate the reflection absorption spectrum, −log(R/R0). All spectra were recorded at a spectral resolution of 8 cm−1 using Blackman-Harris-4-term apodization and a zero-filling factor of 2. For each spectrum, 2000 or 4000 scans were co-added over a total acquisition time of ∼6–9 min. Peak positions were determined by the standard peak picking method (interpolation) supplied by the Bruker software (OPUS). Spectral calculations were performed using a Visual Basic program (22) with an implementation of the formalism published by Flach et al. (23) and Mendelsohn et al. (24).
Neutron reflection
The adsorption of recombinant Rsn-2 was investigated by specular neutron reflection, as previously described for the natural foam fluid (2). Neutron reflectivity profiles were obtained with a SURF reflectometer (Rutherford Appleton Laboratory, ISIS, Didcot, UK) according to established techniques (25–27), using a white beam source with neutron wavelengths in the range of 0.5–6.5 Å. Samples were prepared by dissolving lyophilized Rsn-2 at various concentrations (0.007–0.25 mg mL−1) in 11:1 H2O/D2O to make a null reflecting water (NRW) solution, or in D2O directly, and transferred to a Teflon trough mounted on an antivibration table in the reflectometer. As previously described (28,29), experimental neutron reflectivity profiles were analyzed by means of the optical matrix modeling formalism, in which the calculated reflectivity of an assumed layer model is compared with the measured data. The structural parameters were then varied by least-squares iteration until the best fit was found. The structural parameters used in the fitting were the number of layers, the thickness (τ), and the corresponding scattering length density (ρ) for each layer. The choice of the number of sublayers in the model depended on the extent of inhomogeneity across the interface, with two layers being sufficient to fit the data adequately here. For a mixed layer under water, the observed scattering length density (ρobs) is given by:
where ρp and ρw are the scattering length densities for protein and water, respectively, and ϕp and ϕw are their respective volume fractions in the layer (ρp is calculated from the amino acid composition; ρw is 6.35 × 10−6 Å−2 for pure D2O and zero for NRW (30)). These methods give estimates of the thickness and volume fractions of protein layers at the air-water interface with depth resolution of order 1–3 Å along an axis normal to the plane of the interface.
Results
Recombinant Rsn-2 expresses well as a soluble protein in E. coli and has solution characteristics typical of a thermally stable globular protein with Tm = 80 (±1) °C (from DSC) and secondary structure (from CD) indicating a predominantly β-sheet (35%) conformation, with ∼14% helix, 20% turn, and 31% unordered structure (see the Supporting Material for details). This was confirmed by a more detailed structural analysis, described below. As reported elsewhere (1) and confirmed here, recombinant Rsn-2 shows strong surfactant activity. In addition, dilute solutions of Rsn-2 (≤1 mg mL−1) foam readily upon agitation (C. D. Mackenzie and A. Cooper, unpublished observation), yielding foamy masses that are superficially similar to the natural túngara frog foam nests. In this section we describe the solution structure of Rsn-2 determined by high-resolution NMR, together with results from IRRAS and neutron reflection from surfaces of aqueous Rsn-2 solutions that give information about protein packing and structural distribution at the air-water interface relevant to surfactant properties.
Rsn-2 structure in solution
The 3D structure of Rsn-2 in aqueous solution was determined using NOE data from 2D {1H,1H}- and 3D 15N- and 13C-edited NOESY experiments in combination with hydrogen-bond restraints derived from hydrogen/deuterium exchange data. Representative NMR data and structural statistics are given in Fig. S3 and Table S2, respectively, in the Supporting Material. The structure is well defined over the regular secondary structure elements, as illustrated in Fig. 1. In solution, Rsn-2 adopts a mixed α/β structure comprising a four-stranded antiparallel β-sheet with a kinked α-helix resting across one of its faces perpendicular to the strands (Fig. 1). A comparison with the SCOP database (31) (http://scop.mrc-lmb.cam.ac.uk/scop/) reveals that Rsn-2 fits firmly within the cystatin/monellin superfamily and has a cystatin-like fold (see Fig. S4). The N- and C-terminal regions (residues <17 and >86) are poorly defined in the calculated structures, reflecting their dynamic nature as confirmed by analysis of 15N T1, T2 and {1H,15N}heteronuclear NOE data (data not shown). Although the highly charged C-terminal region of the protein 88-K(D)6GY-96 is dynamic, as would be expected for such a sequence, NOESY cross-peaks are observed that indicate that the C-terminal tyrosine appears to bind back to the face of the β-sheet opposite to the helix.
Figure 1.
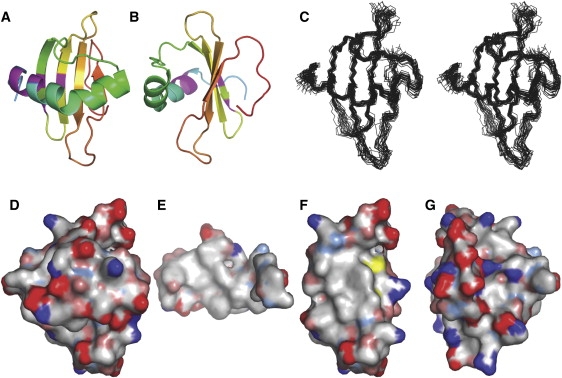
Solution structure of Rsn-2. (A) Ribbon diagram illustrating the Rsn-2 fold and (B) rotated 90° about the vertical axis. Residues showing conformational heterogeneity in the NMR spectra are indicated in magenta. (C) Stereo-pair superposition of the backbone structures of the 25 lowest-energy NMR structures. (D–G) Hydrophobic surface maps of the entire Rsn-2 molecule, front face (D) and back face (G), with the inner face of the helical segment (E) and the inner face of the sheet segment (F). Color codes: white, hydrophobic; red, negative; blue, positive; and yellow, sulfur. Figures prepared using PyMOL (DeLano Scientific, San Francisco, CA).
The majority of cross-peaks were well resolved and the samples were stable throughout data acquisition, but an additional set of low-intensity peaks were observed in the NMR spectra that could be sequence-specifically assigned as representative of alternative conformations of a cluster of residues (Leu20, Phe21, Gly22, Lys23, Thr49, Glu51, and Val58) that are close in the 3D structure (Fig. 1 and Fig. S3). Although because of sample degradation we cannot entirely rule out chemical heterogeneity, these additional peaks did not change with time and are present in different sample preparations. The structural heterogeneity in this region could arise from the presence of a small population of other conformational isomers. This may be significant in relation to the proposed surfactant mechanism (see Discussion).
Rsn-2 adsorption at the air-water interface
IRRAS was used to detect the presence of the protein and its secondary structure in the adsorbed state at the air-water interface in a Langmuir trough. A comparison of the amide I and II bands spectra taken with p-polarized light with different angles of incidence gives information on the orientation of the peptide, as p-polarized light probes the transition dipole moment components parallel and perpendicular to the surface. The measured signal can lead to positive and/or negative bands depending on the angle of incidence and orientation of the transition dipole moment with respect to the interface. At a given angle of incidence below the Brewster angle (53° for air/water), negative intensities suggest a transition dipole moment oriented preferentially in the plane of the surface, and positive bands reflect a perpendicular orientation. The adsorption kinetics of Rsn-2 at the air-water interface were monitored by IRRAS after injection of 165 μL of a 228 μM (2.5 mg mL−1) stock solution into the small trough, giving a 2.6 μM (0.029 mg mL−1) subphase concentration (Fig. 2). Rsn-2 shows relatively fast adsorption kinetics in comparison with nonsurfactant proteins (26), reaching a final surface pressure plateau value of 18 mN m−1 within 3 h, even without stirring of the subphase. During the whole adsorption process, the amide I and II bands of Rsn-2, together with the OH stretching vibrational band, increase in intensity (Fig. 2 B) until constant absorbance values are observed at the surface pressure plateau. The amide I band is a superposition of amide I bands at 1655 cm−1 and 1640 cm−1, which represent the α-helical and β-sheet secondary structures in the protein. The same holds for the amide II band, which consists of amide II bands at 1545 cm−1 and 1524 cm−1. The constant wavenumbers of the amide I and II band maxima indicate no change in the proportion of the secondary structure elements of Rsn-2 during the whole adsorption process. An additional weak band at ∼1680 cm−1, indicative of antiparallel β-sheets, is observed as a shoulder, especially for spectra 1–3 shown in the inset of Fig. 2 B. This observation agrees with the NMR structure of Rsn-2, which shows four antiparallel strands (Fig. 1). The intensity of the OH stretching band in IRRA spectra depends on the film thickness and its optical constants (32–34). Although there is not (yet) sufficient information about optical constants to allow quantitative estimates, the observations reported here are qualitatively consistent with an increase in average layer thickness and/or buildup of (oriented) Rsn-2 concentration at the air-water interface during the adsorption process up to the surface pressure plateau.
Figure 2.
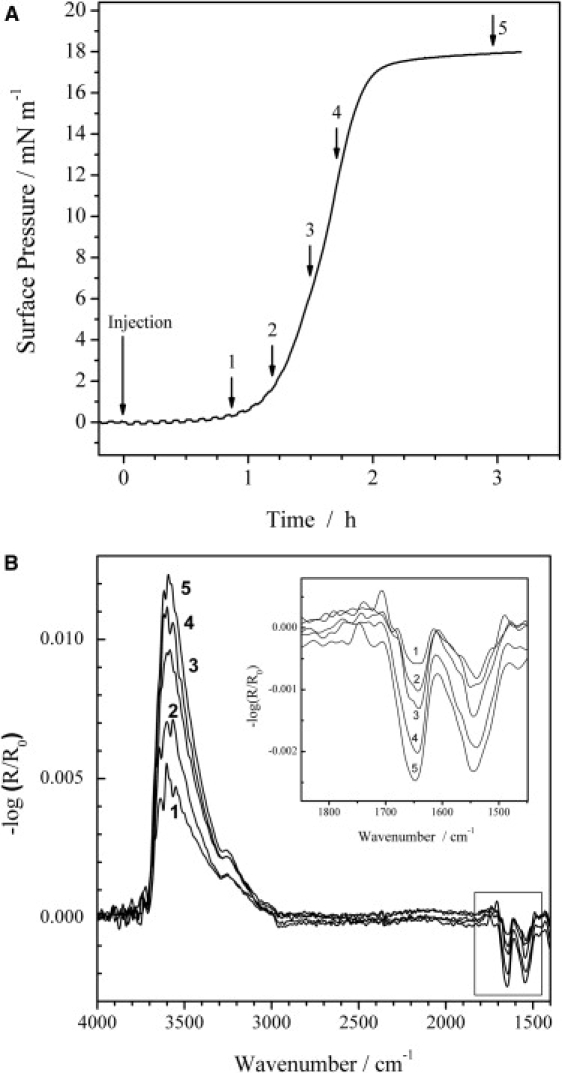
(A) Surface pressure versus time course for Rsn-2 adsorption to the air-water interface (2.6 μM Rsn-2 subphase concentration) and (B) IRRA spectra of the Rsn-2 film at the respective positions of the surface pressure versus time curve given in A. All spectra were recorded with p-polarized light and an angle of incidence of 40°. The inset shows an enlarged representation of the amide I and II region.
Determination of orientation from amide I and II band simulations
To elucidate the orientation of the protein from IRRAS measurements, the NMR structure of the protein was used as a basis for the amount of secondary structure elements (18 amino acids in α-helical and 29 amino acids in β-sheet conformation). The orientation of both secondary structure elements relative to the air-water interface was investigated using p-polarized light at different angles of incidence (32, 36, 40, 60, 64, and 68°) at surface pressure values in the pressure plateau region (Fig. 3 A). Since p-polarized light probes the dipole moment components parallel and perpendicular to the surface, the sign of the band reverses around the Brewster angle (Fig. 3 A) and the resulting changes in band shape, intensity, and position allow the orientation of secondary structure elements to be determined, as previously demonstrated for peptides and lipids (22,35–37). We used the same approach here to simulate amide I and II band contours for p-polarized light at different angles of the incident light (32, 36, 40, 60, 64, and 68°) taking into account the relative orientations of the secondary structure elements as proposed for Rsn-2. Anticipating the hinge-bending, clamshell-opening mechanism for surfactant activity in Rsn-2 (see Discussion), we modeled various configurations for comparison. For the two secondary structure elements, a Lorentzian band shape with a reduced halfwidth of 20 cm−1 was used to improve the clarity of the amide I and II bands located at 1655 cm−1 and 1545 cm−1 for the α-helix and at 1640 cm−1 and 1524 cm−1 for the β-sheet, respectively. The different contributions of the α-helical and β-sheet structure elements for the assumed parallel orientation relative to the air-water interface were weighted according to the number of amino acids in the elements, and then averaged amide I and II bands were simulated. In the simulation, a uniaxial distribution of the secondary structures relative to the surface normal to the air-D2O interface was assumed. The amide I band of an α-helix has two transition moments: the major one, which is almost parallel to the long axis of the helix with an angle of ∼36° relative to the helix axis, and the degenerate one, which is perpendicular to it. Since the second transition has the same frequency but much lower intensity, we used only the major transition for our calculations. In contrast, the amide II band of an α-helix has a transition moment perpendicular to the long axis of the helix. Considering the uniaxial symmetry of the helix and a uniaxial distribution of the vibrational dipole moments around the helix axis, the twist angle was set to 45° for the α-helical element. For the β-sheet structure, the transition dipole moment of the amide I mode is oriented along the interchain hydrogen bonds perpendicular to the strands, whereas the amide II mode has a transition moment perpendicular to the interchain hydrogen bonds parallel to the strands.
Figure 3.
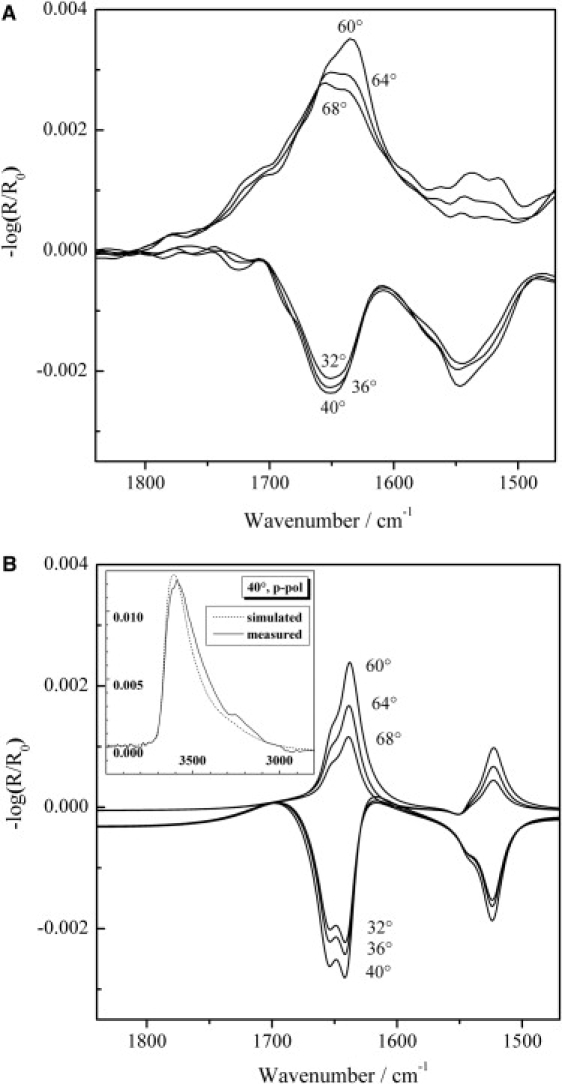
(A) IRRA spectra of Rsn-2 (2.6 μM) adsorbed at the air-water interface acquired with p-polarized light at various angles of incidence (32, 36, 40, 60, 64, and 68°). The surface pressure was 18 mN/m. (B) Simulations of IRRA spectra of α-helical and β-sheet elements of the Rsn-2 protein with a parallel orientation relative to the air-water interface. The calculation was performed for p-polarized light and different angles of incidence for the amide I and II band regions and for the OH stretching vibrational band. The inset shows a comparison of measured (solid line) and simulated (dotted line) OH stretching vibrational bands for an incident angle of 40° and p-polarized light.
Fig. 3 B shows simulated IRRA spectra for an open clamshell orientation of the protein at different angles of incidence and p-polarized light. In this simulation a parallel orientation of both secondary structures relative to the air-water interface was assumed, with the β-sheet lying flat on the water surface. (Note that bands in the simulated spectra appear sharper than the experimental spectra due to the narrower linewidths assumed in the simulation. This facilitates the analysis because the intensity changes of the two bands due to the α-helix and the β-sheet structure can be seen more easily.) A comparison of the experimental and simulated spectra (Fig. 3) reveals a good agreement of the band intensities for the amide I and II bands, which confirms the proposed model of parallel oriented secondary structure elements relative to the air-water interface.
Several positions of the α-helix and the β-sheet relative to each other are possible. The secondary structures can lie either on top of each other or side by side as in the proposed model of the open clamshell. Information about this arrangement at the air-water interface can be obtained from the intensity of the OH-stretching vibrational band, which is related to the film thickness. The inset of Fig. 3 B shows a comparison of measured and simulated OH stretching vibrational bands for an incident angle of 40° and p-polarized light. The measured OH band intensity corresponds to a value typical for β-sheet structures of peptides lying flat at the air-water interface (35) (A. Erbe, A. Kerth, M. Dathe, and A. Blume, unpublished results). The simulated OH stretching vibrational band was calculated for the open clamshell model, in which the α-helix and the β-sheet are lying side by side at the air-water interface. In this case, the film thickness is significantly lower than for the closed clamshell model with both secondary structure elements lying on top of each other. The simulations show that the open clamshell model seems to be the more likely arrangement.
Neutron reflection
Accumulation of Rsn-2 at the air-water interface was confirmed by neutron reflection from the surfaces of protein solutions (0.007–0.25 mg mL−1; 0.6–2.3 μM) in both D2O and NRW. Reflectivity profiles are illustrated in Fig. 4 together with theoretical fits to a two-layer interface model using parameters given in Table 1. In all cases the reflectivity data consistently fit best to a two-layer model, with a thin but dense top layer (8–10 Å thick) above the water surface regardless of concentration, and a looser but thicker (20–40 Å) concentration-dependent layer under the air-water interface. For the thin upper layer, the observed scattering length density (ρ) is close to that anticipated for pure protein, regardless of solvent (D2O or NRW) or concentration in the subphase. This implies that this layer is made up of a hydrophobic region of closely packed protein molecules with a high protein volume fraction (>90%; see Table 1), projecting into the air, with solvent water excluded. This follows because, even in pure D2O, which has a higher scattering length density than protein, the measured ρobs is less than the calculated ρp, indicating that protein in this layer is interspersed with a low scattering medium (ρair = 0) rather than solvent water. The second sublayer is much less dense, with ρ values closer to solvent, and protein volume fractions varying from 10% to 30% depending on the concentration. This may be visualized (at low concentrations) as more hydrophilic regions of the protein molecules projecting from the dense upper layer into the bulk solvent. At higher concentrations, this layer may become deeper and more populated by other molecules competing for space at the interface (see Discussion for more on this).
Figure 4.
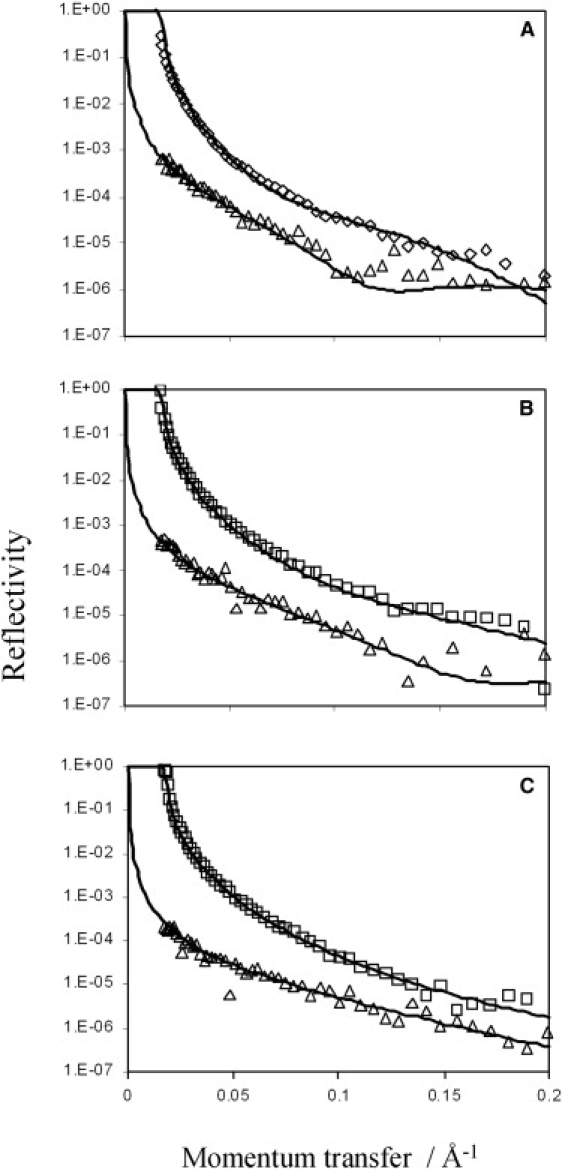
Neutron reflectivity data for Rsn-2 at the air-water interface in D2O (□, ⋄) or NRW (Δ) at pH 7 and protein concentrations of (A) 0.25 mg mL−1, (B) 0.05 mg mL−1, and (C) 0.007 mg mL−1. The solid lines are theoretical fits to a two-layer model with parameters given in Table 1.
Table 1.
Best-fit neutron reflection parameters for two-layer surface adsorption of Rsn-2 in D2O or NRW
| Solvent | Subphase concentration/mg mL−1 | Layer thickness τ/Å | ρobs∗/10−6 | ρp†/10−6 | Volume fraction‡ϕp | Surface excess/mg m−2 | Area per molecule/Å2 | |
|---|---|---|---|---|---|---|---|---|
| Layer 1 | D2O | 0.007 | 8 | 3.0 | 3.23 | 0.93 | 1.02 | 1782 |
| NRW | 0.007 | 8 | 1.8 | 1.92 | 0.94 | 1.03 | 1765 | |
| D2O | 0.05 | 8 | 3.0 | 3.23 | 0.93 | 1.02 | 1781 | |
| NRW | 0.05 | 8 | 1.8 | 1.92 | 0.94 | 1.03 | 1765 | |
| D2O | 0.25 | 10 | 3.0 | 3.23 | 0.93 | 1.28 | 1425 | |
| NRW | 0.25 | 10 | 1.8 | 1.92 | 0.94 | 1.29 | 1412 | |
| Layer 2 | D2O | 0.007 | 20 | 6.0 | 3.23 | 0.11 | 0.31 | 5900 |
| NRW | 0.007 | 16 | 0.3 | 1.92 | 0.16 | 0.34 | 5295 | |
| D2O | 0.05 | 30 | 6.0 | 3.23 | 0.11 | 0.46 | 3933 | |
| NRW | 0.05 | 25 | 0.4 | 1.92 | 0.21 | 0.72 | 2541 | |
| D2O | 0.25 | 40 | 5.6 | 3.23 | 0.24 | 1.32 | 1377 | |
| NRW | 0.25 | 35 | 0.6 | 1.92 | 0.31 | 1.50 | 1210 | |
Neutron scattering length density from experimental fits.
Calculated from amino acid sequence data, assuming appropriate H/D exchange.
Calculated using ρobs = ϕpρp for layer 1 (in air) and ρobs = ϕpρp + ϕwρw for layer 2 (in water).
Discussion
The NMR structure of Rsn-2 in solution is surprising and, at first sight, gives no obvious clue to the origins of its surfactant activity. Despite the lack of any amino acid sequence similarity, the Rsn-2 fold is similar to that found in the cystatin (protease inhibition) family, in a motif not previously associated with surfactant behavior. However, although the natural foam mix does exhibit potent protease inhibition (1), recombinant Rsn-2 does not exhibit any such activity (1), and cystatins are not known to be surfactants. The outer surface of Rsn-2 appears quite uniformly polar (Fig. 1), with no indication of any hydrophobic patch that might be anticipated to confer amphiphilicity, and the protein is soluble and shows no evidence of aggregation in solution. Both NMR and DSC data, supported by gel-filtration chromatography (not shown), are consistent with Rsn-2 acting as a monomer in solution. In this regard it differs significantly from the hydrophobins, which have an entirely different fold and show prominent hydrophobic surface patches comprising up to 20% of the accessible surface area, with a natural tendency to polymerize and form self-assembling aggregates in solution (7,8).
The NMR structure that we have determined here is, of course, for the protein in solution, not at the interface, and this led us to speculate that Rsn-2 might undergo some unfolding or other conformational change upon incorporation into the surface layer. This is an attractive hypothesis since it would potentially rationalize a number of apparently conflicting observations. It is consistent with the neutron reflection data showing, albeit at much lower resolution than the solution structure, that Rsn-2 at the air-water interface forms a densely packed hydrophobic layer with a thickness (8–10 Å) incompatible with the globular structure seen in bulk solution. Similar packing is seen at interfaces with small β-hairpin peptides (25), where neutron reflection studies of a 14-mer β-hairpin adsorbed on the surface of water revealed an 8–10 Å well-packed monolayer floating on the water surface, consistent with the single β-sheet formation revealed by other methods. But there was no indication in this case of any loose hydrophilic sublayer, presumably because the hydrophobic β-hairpin studied was very short, with only a small hydrophilic connecting loop. Surfactant activity with other proteins is often attributed to surface denaturation; however, in the case of Rsn-2, complete unfolding at the interface seems unlikely in view of its high thermal stability in solution. Moreover, surface IR (IRAAS) measurements show that the proportions of protein secondary structure elements do not change during the surface absorption process (Fig. 2), and therefore any conformational change at the interface must preserve the major secondary structural features.
Examination of the Rsn-2 solution structure suggests that we can rationalize these observations by supposing a minimal model involving a simple hinge-bending, clamshell-like opening of Rsn-2 at the interface, with the helix and sheet regions swinging apart to expose the hydrophobic faces previously hidden from solvent (Fig. 1). Broad precedence for this kind of interaction with hydrophobic surfaces can be found in the insect lipophorins, lipid transport proteins that are thought to require a conformational switch opening of the helix-bundle structure for binding to lipid surfaces (38,39). In the case of Rsn-2, this mechanism would allow the protein to present a hydrophobic face to the air while maintaining the hydrophilic surfaces of the protein in contact with the underlying water. Simple molecular modeling shows that this is feasible and consistent with the neutron and IR reflection data, with one possible view illustrated in Fig. 5. Determination of interfacial structures is inevitably speculative at this stage, and we should be wary of overinterpretation, but the proposed model is compatible with all the observations made so far and provides a rational basis for further experimentation. The nonpolar patches exposed by opening of the clamshell face to the air, leaving polar groups still in contact with underlying water, allow the protein molecules to form a thin monolayer consistent with neutron reflection data. The secondary structure elements lie in the plane of the interface, with no change in helix or sheet content, as required by IRRAS observations. Furthermore, the increase in sublayer depth at higher protein concentrations seen in neutron reflection experiments may be attributed to the accumulation of (partially open?) Rsn-2 molecules in dynamic equilibrium between the bulk and the surface layer. We would therefore anticipate the presence of a population of conformational substates or intermediates in the zone adjacent to the interface as part of the mechanism of surfactant layer formation, and there are clues to this in the NMR data. However, in relation to the structure in bulk solution, as described above, there is evidence from NMR spectra for a small population of conformational isomers in solution that may represent precursors to a hinge-bending, clamshell opening. The residues most affected lie at the lips of the Rsn-2 structure (Fig. 1), where the largest initial interresidue movements would occur (assuming the “hinge” is in the loop connecting the helix and sheet substructures). The thermodynamic driving force for such conformational change may be associated with the amphipathic nature of the Rsn-2 sequence. The N- and C-terminal regions of the protein are relatively unstructured and dynamic in solution, but one might envisage that, as a molecule approaches the air-water interface, the predominantly nonpolar N-terminus would exhibit a strong affinity for the air, whereas the highly polar C-terminus would have a much greater affinity for water. This would serve to orient the Rsn-2 molecule as it approached the surface, and the opposing affinities of the N- and C-termini would provide a stretching force to pull apart the jaws of the structure as it approached the surface. Such hypotheses are, of course, difficult to probe directly since we currently lack sufficiently high-resolution methods to study interfacial processes of this kind, but they can be tested by appropriate protein mutagenesis.
Figure 5.
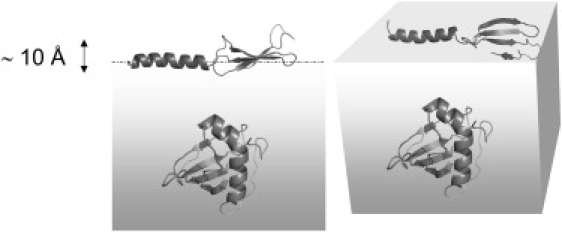
Cartoon indicating how Rsn-2 molecules might lie in a more extended conformation at the air-water interface compared with the compact closed form in solution. This would expose nonpolar faces of the helix and sheet segments to the air while maintaining more polar contacts with the underlying water.
In the biological context, surfactant proteins must satisfy several, sometimes-conflicting criteria. They must be active at low concentrations, for reasons of biochemical economy. They should be soluble, without significant aggregation, during synthesis, transport, and storage stages, yet capable of incorporating into surfaces when required. They must achieve this without detriment to biological membranes and other sensitive tissues with which they might make contact. Rsn-2 seems to fulfill these requirements in a novel fashion. In contrast to surfactin and other lipopeptide surfactants and antibiotics (4), Rsn-2 appears to have no disruptive effect on biological membranes (as evidenced by its presence in the natural foam incubation medium, and the ease of its recombinant overproduction here in bacterial cells). Soluble globular proteins do not normally interact specifically with membranes, and, as we have shown here, the structure of Rsn-2 in solution lacks any obvious hydrophobic feature that might penetrate lipid bilayers. Similarly, in the proposed unfolded state, which is predominant at the air-water interface, the structure is too large and not obviously predisposed to membrane insertion. This has obvious biological advantages, but cannot be the whole story. As previously mentioned, dilute aqueous solutions of recombinant Rsn-2 can be whipped up to yield foams that are superficially similar to túngara frog nest foam in the wild. However, unlike the natural material, which is stable for many days under tropical conditions (2), the Rsn-2 foam collapses after a few minutes under laboratory conditions. This is typical also of foams made from conventional detergents, and reflects the intrinsic thermodynamic instability of foam structures in the absence of additional stabilizing factors (11), suggesting that additional components need to be included to fully explain the properties of the natural frog foam nests. We have shown elsewhere (1) that the natural foam fluid of this species is a rich cocktail that contains not only Rsn-2 but also a range of other proteins with properties that might allow their incorporation into the air-water interface, but with additional properties related to carbohydrate binding, antimicrobial activity, and other biological defense mechanisms. The carbohydrate-binding properties of the other ranaspumins is particularly relevant here because, as we previously proposed (1,2), this could give rise to a mixed interfacial layer containing not only Rsn-2 but also other proteins linked to an underlying carbohydrate matrix, providing longer-term mechanical stability and resistance to dehydration in the natural foam. This is consistent with our earlier neutron reflection studies of the natural foam mixture (2) that revealed a more complex, thicker interfacial layer structure than that seen here with Rsn-2 alone. Consequently, we are seeing the emergence of a fascinating synergy of interactions in which the novel structural surfactant properties of Rsn-2, although fascinating in their own right, are merely part of the first stages in the construction of a stable, biocompatible foam.
Supporting Material
Four figures, two tables, and references are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00792-9.
Supporting Material
Acknowledgments
We thank Dr. R. I. Fleming for the Rsn-2 clones, Dr. S. Kelly and Mr. T. Jess for CD analysis, and Mrs. M. A. Nutley for DSC and additional technical assistance.
This work was supported by grants from the Wellcome Trust, the UK Biotechnology and Biological Sciences Research Council (to A.C., M.W.K., B.O.S., and J.R.L.), and Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie (to A.M. and A.B.).
References
- 1.Fleming R.I., Mackenzie C.D., Cooper A., Kennedy M.W. Foam nest components of the túngara frog: a cocktail of proteins conferring physical and biological resilience. Proc. R. Soc. B. 2009;276:1787–1795. doi: 10.1098/rspb.2008.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper A., Kennedy M.W., Fleming R.I., Wilson E.H., Videler H. Adsorption of frog foam nest proteins at the air-water interface. Biophys. J. 2005;88:2114–2125. doi: 10.1529/biophysj.104.046268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Head J.F., Mealy T.R., McCormack F.X., Seaton B.A. Crystal structure of trimeric carbohydrate recognition and neck domains of surfactant protein A. J. Biol. Chem. 2003;278:43254–43260. doi: 10.1074/jbc.M305628200. [DOI] [PubMed] [Google Scholar]
- 4.Lu J.R., Zhao X.B., Yaseen M. Biomimetic amphiphiles: biosurfactants. Curr. Opin. Colloid Interface Sci. 2007;12:60–67. [Google Scholar]
- 5.Holt C. Structure and stability of bovine casein micelles. Adv. Protein Chem. 1992;43:63–151. doi: 10.1016/s0065-3233(08)60554-9. [DOI] [PubMed] [Google Scholar]
- 6.Horne D.S. Casein structure, self-assembly and gelation. Curr. Opin. Colloid Interface Sci. 2002;7:456–461. [Google Scholar]
- 7.Hakanpaa J., Paananen A., Askolin S., Nakari-Setala T., Parkkinen T. Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile. J. Biol. Chem. 2004;279:534–539. doi: 10.1074/jbc.M309650200. [DOI] [PubMed] [Google Scholar]
- 8.Sunde M., Kwan A.H.Y., Templeton M.D., Beever R.E., Mackay J.P. Structural analysis of hydrophobins. Micron. 2008;39:773–784. doi: 10.1016/j.micron.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Horne D.S. Casein micelle structure: models and muddles. Curr. Opin. Colloid Interface Sci. 2006;11:148–153. [Google Scholar]
- 10.Adamson A.W., Gast A.P. John Wiley & Sons; New York: 1997. Physical Chemistry of Surfaces. [Google Scholar]
- 11.Weaire D.L., Hutzler S. Oxford University Press; Oxford, UK: 1999. The Physics of Foams. [Google Scholar]
- 12.Rahman P.K.S.M., Gakpe E. Production, characterisation and applications of biosurfactants—review. Biotechnology. 2008;7:360–370. [Google Scholar]
- 13.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Gill S.C., Von Hippel P.H. Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 15.Boucher, W. 2002. Azara suite of programs. http://www.bio.cam.ac.uk/azara/.
- 16.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 17.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Nilges M., Macias M.J., Odonoghue S.I., Oschkinat H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J. Mol. Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 19.Smith B.O., Picken N.C., Westrop G.W., Bromek K., Mottram J.C. The structure of Leishmania mexicana ICP provides evidence for convergent evolution of cysteine peptidase inhibitors. J. Biol. Chem. 2006;281:5821–5828. doi: 10.1074/jbc.M510868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linge J.P., Williams M.A., Spronk C., Bonvin A., Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 21.Flach C.R., Brauner J.W., Taylor J.W., Baldwin R.C., Mendelsohn R. External reflection FTIR of peptide monolayer films in-situ at the air/water interface—experimental-design, spectra-structure correlations, and effects of hydrogen-deuterium exchange. Biophys. J. 1994;67:402–410. doi: 10.1016/S0006-3495(94)80495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maltseva E., Kerth A., Blume A., Mohwald H., Brezesinski G. Adsorption of amyloid β (1–40) peptide at phospholipid monolayers. Chem. Bio. Chem. 2005;6:1817–1824. doi: 10.1002/cbic.200500116. [DOI] [PubMed] [Google Scholar]
- 23.Flach C.R., Gericke A., Mendelsohn R. Quantitative determination of molecular chain tilt angles in monolayer films at the air/water interface: infrared reflection/absorption spectroscopy of behenic acid methyl ester. J. Phys. Chem. B. 1997;101:58–65. [Google Scholar]
- 24.Mendelsohn R., Brauner J.W., Gericke A. External infrared reflection-absorption spectrometry monolayer films at the air-water-interface. Annu. Rev. Phys. Chem. 1995;46:305–334. doi: 10.1146/annurev.pc.46.100195.001513. [DOI] [PubMed] [Google Scholar]
- 25.Lu J.R., Perumal S., Powers E.T., Kelly J.W., Webster J.R.P. Adsorption of β-hairpin peptides on the surface of water: a neutron reflection study. J. Am. Chem. Soc. 2003;125:3751–3757. doi: 10.1021/ja0292290. [DOI] [PubMed] [Google Scholar]
- 26.Lu J.R., Su T.J., Penfold J. Adsorption of serum albumins at the air/water interface. Langmuir. 1999;15:6975–6983. [Google Scholar]
- 27.Lu J.R., Zhao X.B., Yaseen M. Protein adsorption studied by neutron reflection. Curr. Opin. Colloid Interface Sci. 2007;12:9–16. [Google Scholar]
- 28.Lu J.R., Thomas R.K. Neutron reflection from wet interfaces. J. Chem. Soc., Faraday Trans. 1998;94:995–1018. [Google Scholar]
- 29.Lu J.R., Thomas R.K., Penfold J. Surfactant layers at the air/water interface: structure and composition. Adv. Colloid Interface Sci. 2000;84:143–304. doi: 10.1016/s0001-8686(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 30.Jacrot B. Study of biological structures by neutron-scattering from solution. Rep. Prog. Phys. 1976;39:911–953. [Google Scholar]
- 31.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP—a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 32.Hussain H., Kerth A., Blume A., Kressler J. Amphiphilic block copolymers of poly(ethylene oxide) and poly(perfluorohexylethyl methacrylate) at the water surface and their penetration into the lipid monolayer. J. Phys. Chem. B. 2004;108:9962–9969. [Google Scholar]
- 33.Kerth, A. 2003. Infrared reflection absorption spectroscopy at lipid, peptide and liquid crystal films at the air-water interface. PhD thesis. Martin Luther University Halle-Wittenberg, Halle/Saale, Germany.
- 34.Meister A., Kerth A., Blume A. Adsorption kinetics of n-nonyl-β-D-glucopyranoside at the air-water interface studied by infrared reflection absorption spectroscopy. J. Phys. Chem. B. 2005;109:6239–6246. doi: 10.1021/jp045196u. [DOI] [PubMed] [Google Scholar]
- 35.Kerth A., Erbe A., Dathe M., Blume A. Infrared reflection absorption spectroscopy of amphipathic model peptides at the air/water interface. Biophys. J. 2004;86:3750–3758. doi: 10.1529/biophysj.103.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopes D.H.J., Meister A., Gohlke A., Hauser A., Blume A. Mechanism of islet amyloid polypeptide fibrillation at lipid interfaces studied by infrared reflection absorption spectroscopy. Biophys. J. 2007;93:3132–3141. doi: 10.1529/biophysj.107.110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meister A., Nicolini C., Waldmann H., Kuhlmann J., Kerth A. Insertion of lipidated Ras proteins into lipid monolayers studied by infrared reflection absorption spectroscopy (IRRAS) Biophys. J. 2006;91:1388–1401. doi: 10.1529/biophysj.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leon L.J., Idangodage H., Wan C.P.L., Weers P.M.M. Apolipophorin III: Lipopolysaccharide binding requires helix bundle opening. Biochem. Biophys. Res. Commun. 2006;348:1328–1333. doi: 10.1016/j.bbrc.2006.07.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weers P.M.M., Ryan R.O. Apolipophorin III: a lipid-triggered molecular switch. Insect Biochem. Mol. Biol. 2003;33:1249–1260. doi: 10.1016/j.ibmb.2003.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


