Abstract
The melibiose permease (MelB) from Escherichia coli couples the uptake of melibiose to that of Na+, Li+, or H+. In this work, we applied attenuated total reflection Fourier transform infrared (ATR-FTIR) difference spectroscopy to obtain information about the structural changes involved in substrate interaction with the R141C mutant and with the wild-type MelB reacted with N-ethylmaleimide (NEM). These modified permeases have the ability to bind the substrates but fail to transport them. It is shown that the sugar-induced ATR-FTIR difference spectra of the R141C mutant are different from those corresponding to the Cys-less permease from which it is derived. There are alterations of peaks assigned to turns and β-structures located most likely in loop 4-5. In addition, and quite notably, a peak at 1659 cm−1, assigned to changes at the level of one α-helix subpopulation, disappears in the melibiose-induced difference spectrum in the presence of Na+, suggesting a reduction of the conformational change capacity of the mutated MelB. These helices may involve structural components that couple the cation- and sugar-binding sites. On the other hand, MelB-NEM difference spectra are proportionally less disrupted than the R141C ones. Hence, the transport cycle of these two permeases, modified at two different loops, is most likely impaired at a different stage. It is proposed that the R141C mutant leads to the generation of a partially defective ternary complex that is unable to catalyze the subsequent conformational change necessary for substrate translocation.
Introduction
Melibiose permease (MelB) of Escherichia coli transports the disaccharide melibiose to the cell interior coupled to the downhill electrochemical ion gradient of Na+, Li+, or H+ (1,2). MelB binds and transports melibiose and its coupling ion in a 1:1 ratio (3). The three coupling ions have a common binding site, and Na+ or Li+ ions enhance the cotransporter affinity for melibiose (4,5). Reciprocally, melibiose binding enhances MelB affinity for the coupling ion (6). Biochemical studies, including immunological, PhoA-MelB fusions, and proteolytic mapping analysis (7–9), as well as two-dimensional crystallization (10,11) and Fourier transform infrared (FTIR) studies (12), consistently fit a topological model that includes 12 transmembrane α-helical domains. This secondary structure model is illustrated in Fig. 1. Moreover, genetic and site-directed mutagenesis studies (13–17) suggest that 1), the cation-binding site is principally lined by N-terminal transmembrane domains (light gray helices in Fig. 1) and the sugar-binding site is delimited at least in part by C-terminal domains (darker gray helices in Fig. 1); 2), helix IV may connect the two substrate-binding sites (18), and loops 4-5 and 10-11 are important for MelB function (8,19–21); and 3), Asp and Glu residues are implicated in cation and melibiose binding and/or translocation (16,20,22–24). Other residues, such as Asn (25), Arg (19,26), and Tyr (24), have also been shown to be important in these processes. Analyses of purified MelB reconstituted in proteoliposomes using different biophysical techniques have provided strong evidence for several substrate-induced changes in MelB conformation during the MelB transport cycle. Previous studies reported that the coupling cations induce changes in the fluorescence of Trp located in the N-terminal domain, whereas melibiose interaction preferentially leads to a fluorescence change of Trp located in the C-terminal domain (15,18,27). Moreover, the Na+ (or Li+) dependent change in the fluorescence resonance energy transfer properties of a dansylated sugar derivative suggests that ion binding to MelB promotes a structural change in the sugar-binding site or in its immediate vicinity (13,28). Electrophysiological measurements have revealed fast transient currents (20 ms range) triggered by either Na+ or melibiose binding (6,29). The melibiose-induced charge transfer is best explained by structural changes involving movement of charged amino acids and/or a reorientation of helix dipoles (29). Finally, results from FTIR spectroscopy have not only provided information on the secondary structure components of MelB (12), they also suggest that changes in α-helix tilting occur upon substrate binding (30). In addition, hydrogen/deuterium (H/D) exchange experiments showed that MelB is less accessible to solvent when incubated with sugar and either cation than with the corresponding cation alone (31). The same study also showed that β-structures are protected against H/D exchange by sugar binding.
Figure 1.
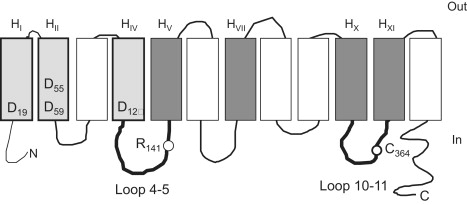
Predicted secondary structural model of MelB. The 12 successive transmembrane helices are represented by boxes and labeled using the H letter and a roman number subscript. Shaded boxes indicate helices putatively lining the cosubstrate binding sites (light gray, Na+ site; dark gray, sugar site). Amino acids of interest for this study are labeled using the single-letter amino acid code and a number subscript corresponding to their position on the primary MelB amino acid sequence. The aspartic acid residues distributed in HI, HII, and HIV are important for Na+ binding. Arg141 is located in loop 4-5. Cys364 in loop 10-11 is the NEM target. The model is from Pourcher et al. (9).
More refined insights into the substrate-induced changes in MelB structural properties, as well as the change in protonation-deprotonation and/or environment of given amino acid side chains, were recently gained with the use of attenuated total reflection (ATR)-FTIR difference spectroscopy applied to MelB proteoliposomes fully equilibrated in either H2O or D2O media (32,33). IR difference spectroscopy is a powerful methodology that can reveal details of changes at the level of only one amino acid (34). Although the difference spectra obtained are of very low intensity, we and other investigators (32,33,35–38) have demonstrated their high repeatability and consistency. The most salient information from these studies on MelB can be summarized as follows (see details in the Results section below): The addition of Na+ (or Li+), and subsequently that of melibiose, gives rise to sets of discrete and sharp peaks that can be considered as specific signatures of the interaction of MelB with each substrate. In the difference ATR-FTIR spectra, peak assignment in the amide I interval suggests that all types of secondary structures (α-helix, β-structures, and turns) participate in the substrate-induced structural variations. In particular, substrate-specific changes (Na+ versus H+, cation/sugar versus cation) at the level of different subtypes of α-helical components with distinct FTIR signatures have been tentatively related in part to helix tilt variations. Finally, the change in the antisymmetric vibration signal of some of the MelB carboxylates has been attributed to (COO−)-Na+ (or Li+) interactions and/or (COO−)-Arg+ (or Lys+) interactions.
Replacing R141 in cytoplasmic loop 4-5 of MelB by a neutral Cys residue leads to selective inactivation of MelB translocation capacity (19). Biochemical analyses have shown that the R141C mutant retains Na+-dependent sugar-binding activity (19), as well as a fluorescence resonance energy transfer signal arising from a bound dansylated sugar-analog (39). In contrast, it does not catalyze active transport or energy-independent, carrier-mediated cosubstrate translocation reactions. It was recently reported that MelB electrical properties resulting from sugar binding (6,29) were also selectively impaired in the mutant (39). Although Na+ (or Li+) addition still elicited a fast electrical transient signal, subsequent addition of melibiose no longer gave rise to any of the fast and slow sugar-induced electrical transients. Also, Na+-dependent sugar-induced variations of MelB tryptophan signal (27) were no longer observed in R141C. Of interest, although acylation of Cys364, localized in loop 10-11, in the wild-type (WT) MeB by N-ethylmaleimide (NEM) also selectively impaired translocation (40), no change in its fast electrical components or Trp fluorescence response was observed (6,27). This suggests that the NEM reaction introduces a defect after sugar binding (39). These data suggest that the extramembranous loop 4-5 has a role in the transport process, and more especially during a step immediately after sugar binding.
The general purpose of this study was to gain a better understanding of the structure-function relationships of the ion-coupled MelB transporter. We sought to determine the extent to which the R141C defect is associated with any structural defect. To that end, we carried out a difference ATR-FTIR analysis of the cosubstrate-induced change recorded on the purified R141C MelB mutant in proteoliposomes. The overall difference ATR-FTIR information previously obtained from WT-MelB was used as a general reference. We also characterized the Cys-less mutant (the quadruple mutant C110S/C235V/C310S/C364S 40) from which R141C was derived, and used it as a control. Finally, the NEM-acylated WT-MelB, which also displays selective inactivation of the translocation capacity (3,5), was analyzed for comparison. Overall, the data show that the R141C mutation introduces a selective defect of the structural properties of MelB during (or at a stage immediately after) sugar binding to the Na+-MelB.
Materials and Methods
Materials
Synthesis of (3-laurylamido)-N,N′-(dimethylamino)propylamine oxide (LAPAO) was performed as previously described (41). Dodecyl maltoside (DM) was obtained from Boehringer Mannheim (Basel, Switzerland), and Ni-NTA resin was obtained from Qiagen (Hilden, Germany). SM-2 Bio-Beads were obtained from Bio-Rad (Hercules, CA). Total E. coli lipids (acetone/ether precipitated) were purchased from Avanti Polar Lipids (Alabaster, AL). High-purity-grade salts or chemicals were used to prepare nominally Na+-free media containing <20 μM sodium salts. All other materials were obtained from commercial sources.
Plasmids and site-directed mutagenesis
A recombinant pK95ΔAHB plasmid with a cassette containing the melB gene (42) encoding a permease devoid of its four native cysteines (Cys-less MelB) was engineered by polymerase chain reaction using the appropriate mutagenesis primers, and subsequently used for control experiments or as background for further permease engineering (19). This Cys-less carrier displayed a valine instead of Cys235, and a serine instead of Cys110, Cys310, and Cys364 (40).
MelB overproduction and purification
A RecA− derivative of E. coli DW2 (Δmel ΔlacZY) (43) was transformed with pK95ΔAHB plasmid to overexpress a WT His-tagged MelB (15). Transformed cells were grown at 30°C in 200 L of M9 medium supplemented with appropriate carbon sources and ampicillin (100 mg/mL) at the Centre de Fermentation, Centre National de la Recherche Scientifique (Marseille, France), and used to prepare inverted membrane vesicles (IMVs), by means of a French press (American Instrument). Purification of the His-tagged MelB was carried out essentially as described previously (42).
Preparation of MelB proteoliposomes
MelB protein (0.5 mg/mL) solubilized in DM (0.1%, w/v) was mixed with E. coli lipids to give a protein/lipid ratio of 1:2 (w/w). DM was removed by an overnight adsorption in SM-2 Bio-Beads at 4°C as described previously (44). The proteoliposomes were then subjected to repeated freeze/thaw-sonication-wash cycles in nominally Na+-free, 0.1 M potassium chloride buffer (pH 6.6) to eliminate NaCl from both the external medium and the internal space. All samples were prepared in H2O buffers.
MelB inhibition with NEM
For MelB inhibition with NEM, the proteoliposome suspension was incubated for 30 min in the presence of 2 mM of NEM and then extensively washed (5).
Data acquisition
The experimental setup was the same as that described in a previous study (32). Briefly, a sample of 20 μL of a proteoliposome suspension (∼150 μg of protein) was spread homogeneously on a germanium ATR crystal (Harrick, Ossining, NY; 50 × 10 × 2 mm, yielding 12 internal reflections at the sample side) and dried under a stream of nitrogen. The substrate-containing buffer and the reference buffer were alternatively perfused over the proteoliposome film at a rate of ≈1.5 mL/min. Spectra were recorded with an FTS6000 Bio-Rad spectrometer equipped with a Mercury-Cadmium-Telluride detector at a resolution of 4 cm−1.
Data corrections and manipulations
Deconvolution by the maximum entropy method was applied to the difference spectra as previously described (45).
Results
Substrate-induced changes of the Cys-less mutant
Given that the MelB mutant was constructed with Cys-less permease used as the molecular background, we checked the similarity of the substrate-induced difference spectra of the Cys-less and the WT permeases (see the Supporting Material). Peaks absorbing in turns, β-structures, α-helices, and carboxylic side-chain regions are observed in the difference spectra of both permeases. All respective difference spectra are similar in shape and share the same main peaks. They are also similar to previously published MelB difference spectra (32,33).
Since the transport characteristics of Cys-less and WT are similar but not identical, one could expect that the difference spectra would also be similar but not identical. In all experiments, the Na+ and/or sugar concentrations used (10 mM) are saturating (3–5). Indeed, the main parts of the difference spectra are very similar, i.e., the amide I region in Na+ versus H+ and melibiose · Na+ versus Na+ difference spectra, the carboxylic peaks around 1400 cm−1 in Na+ versus H+ difference spectrum, etc. (Supporting Material). These facts and the virtual identity between spectra taken from different samples indicate 1), the reliability of the MelB difference spectra despite their low intensity; and 2), the feasibility of the deconvolution applied to the difference spectra (for example, the small peaks at 1515 and 1526 cm−1 are reproduced in the difference spectrum of both permeases).
It can be concluded that the substrate-induced difference spectra of Cys-less and WT are essentially similar, albeit not entirely identical, and yield comparable information on fully active MelB permeases. Hence, the mutation of the four native Cys of MelB has a limited impact on changes due to Na+ binding or to melibiose interaction in the presence of Na+ or H+. This is in keeping with the reported similar functional behavior of the Cys-less mutant compared with the WT (40).
Substrate-induced changes of the R141C mutant
Few alterations are observed when one compares the amide I region of the difference spectra induced by Na+ binding with R141C and Cys-less (Fig. 2 A). The structural changes resulting from the replacement of the coupling H+ by Na+ are therefore alike in both permeases. However, some variations are visible in some parts of the difference spectrum, such as those in the peaks in the 1630–1600 cm−1 region. These changes may be mainly caused by the disappearance of the peak centered at 1614 cm−1, which may be assigned to Arg side chains (34,46) and more specifically to Arg141. The assignment of this peak to Arg side chains is consistent with the fact that this peak is sensitive to H/D exchange (33). In addition, there is a decrease in the intensity of the peaks absorbing in the amide II (around 1550 cm−1). Peaks corresponding to carboxylic acids (33) (at 1598–1600, 1576–1579, 1404, and 1383 cm−1) show no modifications except for a small decrease in the peak at 1600 cm−1, which could also be caused by the disappearance of the nearby peak at 1614 cm−1. A comparison of the deconvoluted spectra (Fig. 2 B) that increase the peak resolution confirms these conclusions.
Figure 2.
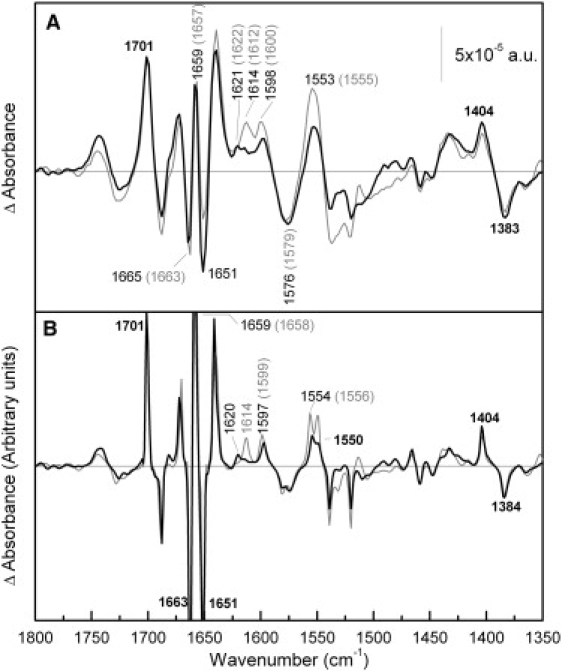
Comparison of Na+-induced difference spectra of R141C and Cys-less. (A) Solid line: difference spectrum of R141C in 20 mM MES, 100 mM KCl, 10 mM NaCl, pH 6.6, minus R141C in 20 mM MES, 110 mM KCl, pH 6.6; shaded line: difference spectrum of MelB Cys-less under the same conditions. In this and subsequent figures, higher substrate concentrations had no further effect on the difference spectra. (B) Deconvoluted difference spectra of panel A.
The R141C difference spectrum induced by melibiose binding in the presence of Na+ (Fig. 3 A) presents an overall shape comparable to that of Cys-less. However, there are some noteworthy dissimilarities in the amide I region that are best seen after deconvolution of the spectra (Fig. 3 B). First, the peaks at 1688, 1682, and 1672 cm−1 of Cys-less, previously assigned to putative β-structures or turns (32,33), are replaced by only two contiguous peaks at 1684 and 1680 cm−1 in R141C. This suggests that the conformational changes of these structures occurring in R141C upon melibiose interaction are different from those of Cys-less. A second and striking modification in the amide I region of R141C (Fig. 3 B) is the absence of the 1659 cm−1 peak, which was previously proposed to arise from one of the two major α-helix subpopulations (α-helix type I) (32,33). Recent FTIR studies on MelB using polarized light suggest that this peak is at least partly due to sugar-induced variation of helix tilting (30). Accordingly, the disappearance of this peak in R141C may indicate the suppression of movements of one or more helices of the α-helix type I subpopulation. Other changes seen in the deconvoluted amide I region include the disappearance of the negative peak at 1632 cm−1 and the appearance of one positive peak at 1636 cm−1 (Fig. 3 B). These two spectral variations are consistent with changes in the contribution of β-structures (12,38) involved in cooperative interaction between the two substrate-binding sites or in the substrate translocation process. A decrease in the intensity of the peaks near 1621 cm−1 in the difference spectra induced by melibiose binding in the presence of Na+ or H+ (Fig. 3; see also Fig. 4) could be interpreted (34) as the disappearance of the signal arising from one or more of the side chains known to be involved in the transport function (Arg52, Arg141, and Arg149).
Figure 3.
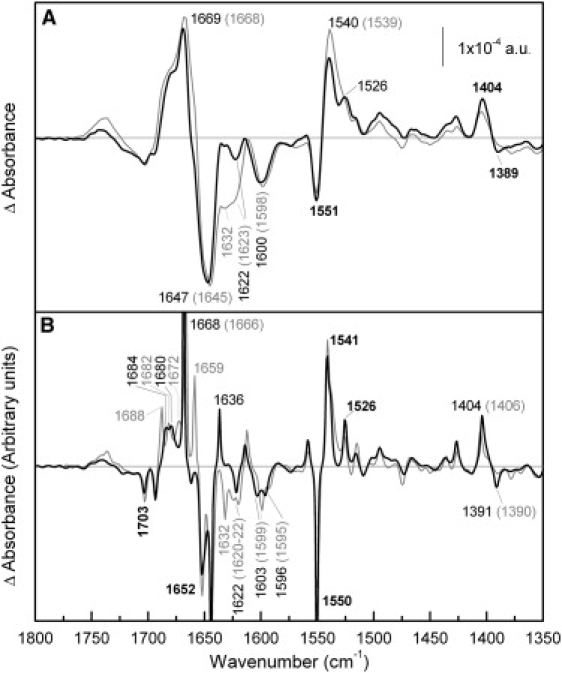
Comparison of melibiose-induced difference spectra of R141C and Cys-less in the presence of Na+. (A) Solid line: Difference spectrum of R141C in 20 mM MES, 100 mM KCl, 10 mM NaCl, 10 mM melibiose, pH 6.6, minus R141C in 20 mM MES, 100 mM KCl, 10 mM NaCl, pH 6.6; shaded line: difference spectrum of MelB Cys-less under the same conditions. (B) Deconvoluted difference spectra of panel A.
Figure 4.
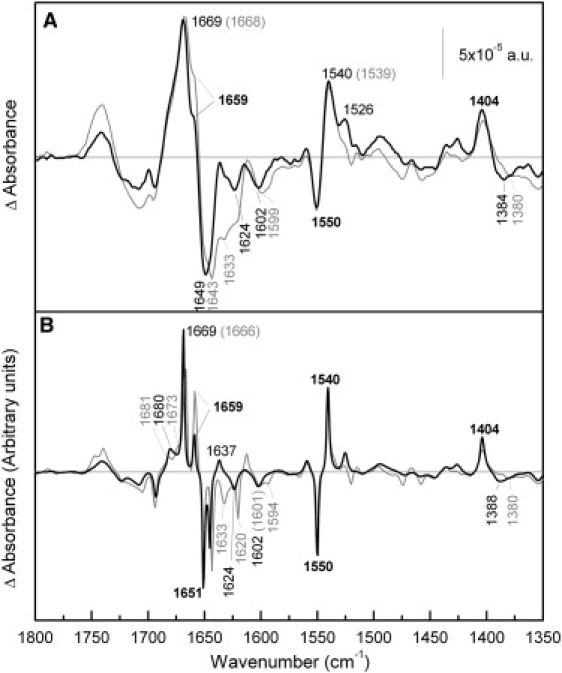
Comparison of melibiose-induced difference spectra of R141C and Cys-less in the presence of H+. (A) Solid line: Difference spectrum of R141C in 20 mM MES, 100 mM KCl, 50 mM melibiose, pH 6.6, minus R141C in 20 mM MES, 100 mM KCl, pH 6.6; shaded line: difference spectrum of MelB Cys-less under the same conditions. (B) Deconvoluted difference spectra of panel A.
In the amide II region, only small differences are observed between both permeases, and they can be assigned mainly to amino acid side chains. One peak that shows some changes is that centered at 1526 cm−1; it is ascribed to Lys side chains, although it could also correspond to amide II vibration (34,47). In the R141C difference spectrum (Fig. 3 A) there is a narrowing of the peak centered at 1540 cm−1 (assigned to amide II). This narrowing allows the observation of the peak at 1526 cm−1 without deconvolution. Finally, carboxylic peaks around 1703, 1600, and 1405 cm−1 are comparable in the difference spectrum (Fig. 3 A), although some changes are seen in the deconvoluted spectrum, especially around 1600 cm−1 (Fig. 3 B).
Fig. 4 compares the R141C and Cys-less difference spectra resulting from interaction of melibiose in the presence of H+. In the amide I region corresponding to turns or β-structures (1690–1670 cm−1, Fig. 4 B), only one peak is resolved by deconvolution at 1680 cm−1 in the R141C difference spectrum, whereas two peaks are observed in the Cys-less spectrum. The positive peak at 1659 cm−1, which is absent in R141C · melibiose · Na+ versus Na+ difference spectrum, is still recorded when the coupling ion is H+, but its intensity is significantly reduced compared to Cys-less. In the amide II absorption region (Fig. 4 A), there is a clear increase in the intensity of the peak centered at 1526 cm−1. As noted above, this peak can be assigned to either amide II vibration or Lys side chain.
Carboxylic antisymmetric vibration presents an upshift to 1602 cm−1, which is best seen in the nondeconvoluted spectra (Fig. 4 A). The negative peak around 1384 cm−1, corresponding to symmetric carboxylic vibrations, also shows changes between both mutants (33).
In summary, the observed variations of sugar-induced FTIR signals at the level of α-helices, as well as from turns and β-structures between the R141C and Cys-less permeases, indicate that the R141C mutation has a significant impact on the MelB structural variations associated with or triggered by sugar binding.
Effect of MelB NEM treatment
MelB acylation by NEM almost completely inhibits its transport ability, whereas Na+-dependent sugar-binding capacity is reduced by only 10% (3,5). Therefore, R141C and NEM-reacted MelB have the same properties with regard to the observed transport: they can bind substrates but have lost the ability to translocate. Fig. 5 shows that the melibiose-induced difference spectrum of MelB-NEM in the presence of Na+ is very similar to that of WT in both the overall shape (Fig. 5 A) and the deconvoluted spectrum (Fig. 5 B).
Figure 5.
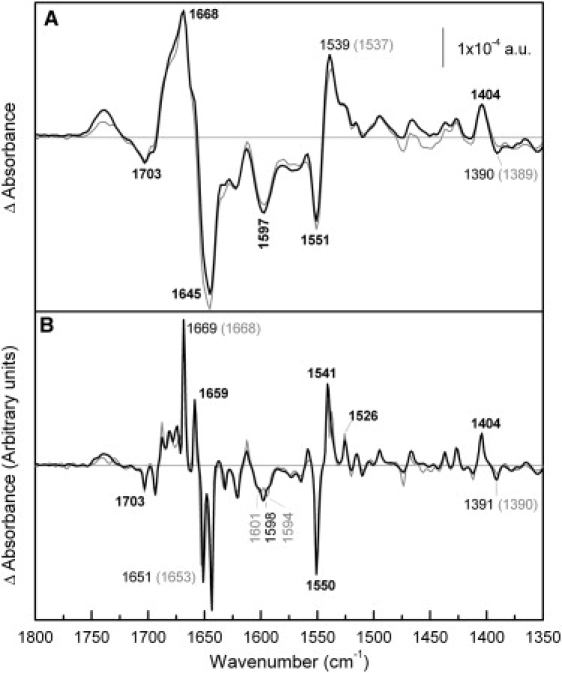
Comparison of melibiose-induced difference spectra of NEM-MelB and WT MelB in the presence of Na+. (A) Solid line: Difference spectrum of NEM-MelB in 20 mM MES, 100 mM KCl, 10 mM NaCl, 10 mM melibiose, pH 6.6, minus NEM-MelB in 20 mM MES, 100 mM KCl, 10 mM NaCl, pH 6.6; shaded line: difference spectrum of WT MelB under the same conditions. (B) Deconvoluted difference spectra of panel A.
Fig. 6 compares the melibiose-induced difference spectrum of MelB-NEM in the presence of H+ with that recorded in WT. The MelB-NEM difference spectrum presents variations in turns and β-structures absorbing above 1670 cm−1. Peaks assigned previously to α-helices (32,33) (at 1669, 1659, and 1551 cm−1) are practically not affected by the NEM reaction, although in the deconvoluted spectrum a new negative peak at 1663 cm−1 that can be assigned to α-helix structure can be appreciated in the MelB-NEM spectrum (Fig. 6 B). Peaks corresponding to vibrations of carboxylic side chains show only small changes. Finally, around 1699 cm−1 there is a groove in the negative broad peak around 1705 cm−1. Variations in this region can be assigned to changes in the C=O vibration of Asp or Glu, although this signal can also be assigned to Asn side chain or turn secondary structure (34,48).
Figure 6.
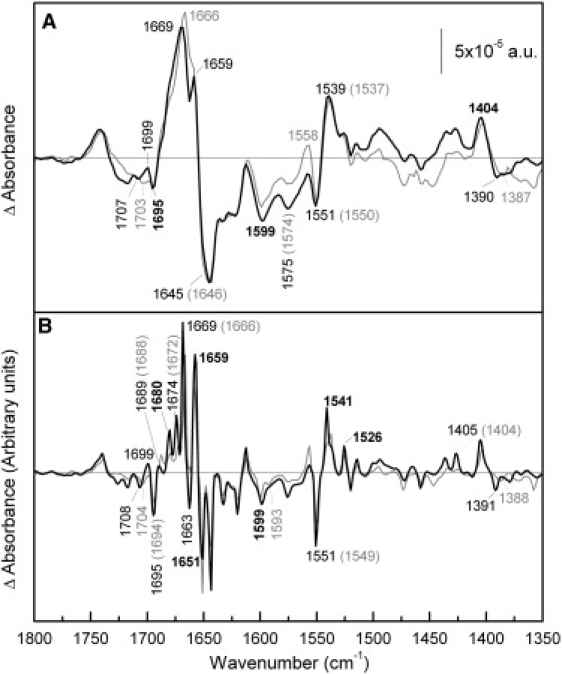
Comparison of melibiose-induced difference spectra of NEM-MelB and WT MelB in the presence of H+. (A) Solid line: Difference spectrum of NEM-MelB in 20 mM MES, 100 mM KCl, 50 mM melibiose, pH 6.6, minus NEM-MelB in 20 mM MES, 100 mM KCl, pH 6.6; shaded line: difference spectrum of MelB WT under the same conditions. (B) Deconvoluted difference spectra of panel A.
These results indicate that some changes occur between the difference spectra of MelB reacted with NEM due to melibiose binding in the presence of Na+ or H+, in comparison with those recorded for WT. However, the spectral deviations from the WT behavior observed in the NEM-acylated MelB are less important than the ones observed for the R141C mutant.
Discussion
The R141C mutation causes inhibition of both sugar-induced increases of Na+ affinity and substrate translocation. In contrast, acylation of the WT permease by NEM impairs only substrate translocation. Due to the absence of a high-resolution 3D structure of MelB, the presence of loops or the localization of some amino acid is still tentative and is based on different kinds of studies. Recently obtained 3D structures of transporters such as the lactose permease (LacY) may help in positioning Arg141. Although the sequence similarity of MelB to the lactose permease is only marginal, it is instructive to take into account the role of Arg side chains as deduced from the LacY 3D structure. In LacY, Arg144 forms bidentate hydrogen bonds with the sugar moiety, and is therefore an essential side chain for substrate binding (49). In MelB, Arg141 seems to play a different role, as the mutant R141C retains the capacity to bind the sugar even if transport is inhibited. In contrast, Arg149 of MelB (in helix IV) is more likely equivalent to Arg144 in helix IV of LacY. Indeed, mutation of Arg149 into Cys or Asn (but not Lys) leads to inhibition of sugar binding (19).
Our data indicate that only limited modifications of the substrate-induced difference spectral features take place in R141C or the NEM-reacted WT protein compared with Cys-less or WT. The conclusion emerges that small structural rearrangements occurring in key sites in the permease are sufficient to produce such significant functional effects. In this case, these key sites presumably correspond to loop 4-5 (R141C) or loop 10-11 (NEM acylated MelB). As discussed below, perturbations of loop 4-5 structure may propagate to the helical domains.
As a preliminary remark, it is important to mention that replacing three out of four cysteines of MelB by a serine and the last one by a valine to construct the Cys-less permease has no major impact on the substrate-induced difference spectra of MelB. Thus, Cys-less and WT essentially share the same main peaks. Since these two permeases have comparable kinetic properties of melibiose transport (40), it can be safely concluded that their substrate-induced difference spectra yield equivalent structural information on fully active MelB transporters.
Conformational changes induced by Na+ binding to R141C or Cys-less permease are similar
The R141C mutant cannot transport substrates but retains the ability to bind them (19). The Na+-induced difference spectra for R141C and either Cys-less or WT indicate that replacement of H+ by Na+ as a coupling ion induces very similar conformational changes in the amide I region and the carboxylic side-chain absorption region. This is consistent with previous data showing that, in all three transporters, Na+ activates sugar binding, enhances the fluorescence emission of the bound fluorescent sugar Dns2-S-Gal, and triggers a fast electrical response in a similar fashion (39). Therefore, it can be concluded that the Arg141 mutation modifies neither the Na+-binding site structure nor the associated conformational changes. In view of this, the disappearance of the peak centered at 1614 cm−1 in the difference spectra of the R141C mutant leads to a tentative assignment of this peak to a Na+-induced change of the Arg141 environment.
Secondary structures present in loop 4-5 likely suffer different changes in R141C and Cys-less upon melibiose binding
The difference spectrum induced by melibiose binding to R141C in the presence of Na+ suggests changes in the contribution of reverse turns and β-structures (at 1688, 1682, and 1672 cm−1; Fig. 3 A) to the sugar-induced conformational changes. Because Arg141 is located in loop 4-5 (8), we can tentatively consider that at least part of these conformational alterations occur in this loop. Of interest, the disappearance of the negative peak at 1632 cm−1 and the concomitant appearance of the positive peak at 1636 cm−1 are correlated with perturbation of β-structures by the mutation. Evidence that β-structures are influenced by (or involved in) melibiose interaction was suggested by previous studies of MelB that showed a high degree of protection of some β-structures against H/D exchange when sugar was bound to the permease (31). Furthermore, although loop 4-5 of WT is efficiently protected by Na+ against proteolysis, and even more by Na+ in the presence of melibiose (8), the sugar protection disappears in the R141C mutant (50). All of this structural information is in agreement with the idea that loop 4-5 has a structural role in the transport mechanism. This also agrees with a previous suggestion that loop 4-5 may behave as a functional and mobile reentrant loop that contributes either the stabilization or reorientation of the ternary complex, or participate in the inner gating mechanism (19). In addition, biochemical data and photolabeling experiments suggest that Arg141, and therefore part of loop 4-5, may be close to the sugar-binding site (19,51). Finally, there is growing evidence that loops (reentrant or not) play a functional and/or structural role in several other transporters from different families (1,52–59).
The R141C mutation specifically impacts one of the MelB α-helix subpopulations
One remarkable effect of the mutation at Arg141 on the MelB structural properties is the selective disappearance of the peak at 1659 cm−1 in the sugar-induced spectrum in MelB equilibrated in Na+ medium (Fig. 3), or at least a severe reduction of its intensity in H+ medium (Fig. 4). Dave et al. (12,31) reported that the absorption band at 1660 cm−1 in MelB is one of the two major helix components in the amide I region of FTIR deconvoluted spectra, and assigned it to an α-helix subpopulation (α-helix type I). In contrast, the second major helix component, corresponding to the peak at 1653 cm−1 (α-helix type II), is not modified by the mutation. According to a recent analysis of WT MelB with IR polarized light, these two subpopulations include tilted helices that display substrate-dependent variations of their tilt angle (30). Overall, these data suggest that the mutation preferentially disrupts the conformational properties of the α-helix type I subpopulation.
Although this information is important, in itself it is insufficient to identify which (and how many) of the 12 putative transmembrane domains of MelB become affected by the mutation, or to unravel the catalytic role of MelB. Indeed, IR difference spectroscopy does not reveal whether the 1659 cm−1 signal in WT of Cys-less arises from a single entire α-helix or eventually from part(s) of an interrupted α-helix, or even from a combination of two (or more) α-helices. Despite this complexity, however, a plausible and simple interpretation can be made by combining the absence of the 1659 cm−1 signal in response to sugar binding to R141C with other spectroscopic data and biochemical information collected from WT and R141C MelB. First, the finding that either cation or sugar binding to WT or Cys-less causes a peak variation at 1659 cm−1 in their respective difference spectra (32,33) suggests that the 1659 cm−1 signal arises from the same helix component(s) during the two binding processes. Second, the loss of IR signal at 1659 cm−1 (Fig. 3) in R141C is concomitant with the loss of the cooperative sugar-induced increase of Na+ affinity (39). On the basis of these findings and correlations, we propose that the signal at 1659 cm−1 arises from a set of α-helix type I components lining the cosubstrate-binding pocket.
Results from several mutagenesis studies may be used to consider the predicted transmembrane segments HII and HIV (42) as potential components of the 1659 cm−1 helix subpopulation. Indeed, both HII and HIV carry Asp residues (Asp55 and Asp59 in HII; Asp124 in HIV) that are essential for Na+ recognition and are expected to be part of the ion coordination network (16,17). Proximity between these two transmembrane segments in the MelB structure is suggested, in particular, by the observation that the introduction of an aspartate residue substituting Gly117 (in HIV) can restore Na+-dependent transport activity to the Na+-insensitive D55C (in HII) mutant (60). Similarly, the introduction of an Arg residue in HIV (W116R) compensates for the lack of an Arg at position 52 in HII of the R52S mutant (26). Other data suggest that HII and HIV are close to and interact with structural components of the sugar-binding site, or may even be part of them. On the one hand, alkylation of the D59C mutant with NEM prevents sugar binding, and melibiose affords protection against this inactivation (22). A similar effect of the SH-reagent p-chloromercuribenzene sulfonate was observed in permeases carrying individual Cys substitution of other residues of HII (61). Lastly, studies of single point mutations introduced in HIV led to the suggestion that this transmembrane segment may act as a hinge between the two putatively overlapping substrate-binding sites (18,24).
It should be stressed that HIV is one of the two flanking helices of loop 4-5. Thus it is possible that the conformational defect in loop 4-5 could directly perturb the properties of this helix component (as well as the HII helix interacting with HIV), perhaps by interfering with its (or their) substrate-dependent tilt capacity.
Conclusions
One of the most interesting results of this work is the absence of the peak at 1659 cm−1, ascribed to α-helix subpopulations, in the difference spectrum of R141C caused by melibiose interaction in the presence of Na+. Following the arguments given above, this helix most likely forms part of the structures that couple the cation-binding site to the sugar-binding site. This coupling may be disabled by the R141C mutation. The NEM acylation does not affect this peak, which means that the coupling mechanism is still operating in the NEM-reacted permease even when transport is blocked.
A comparison of the difference spectra of R141C with those of MelB-NEM and WT suggests that the sugar-induced conformational changes in the NEM-acylated protein are more similar to those observed in WT than to those in R141C. This may be explained by considering that the transport cycle of the R141C mutant is blocked at an earlier stage than that of the NEM-reacted MelB. To examine the possible relationships between conformational and kinetic transition defects occurring in the R141C or NEM-reacted permeases during MelB cycling, we used the kinetic model of the MelB transport cycle shown in Fig. S4. This model is derived from a six-state model based on alternating-access principles, to which was added an occluded-state intermediary (C'Namel) during the carrier translocation process (CONamel↔CiNamel) (39). The more extensive disruption of the structural and functional properties in the R141C mutant than in NEM-WT is in agreement with previous studies demonstrating that even if both transporters still bind the two cosubstrates, R141C does not exhibit sugar activation of Na+ binding or generation of a sugar-induced fast transient electrical signal, or any sugar-induced tryptophan signal variation (19,39). All of these properties are retained by the MelB-NEM permease, as is the coupling between the cation and the sugar sites. The suggestion that R141C transport is blocked before reaching a putative “occluded” state of MelB, whereas MelB-NEM permease transport is blocked in the “occluded” state (39), could account for the observed effects. On the other hand, the different effects of modification at loops 4-5 (R141C) and 10-11 (NEM) presumably reflect their involvement in different steps of the transport reaction. In this regard, the use of time-resolved spectroscopic techniques to follow the time course of the structural changes that occur during MelB cycling should enable a better understanding of the structural basis of the Na+-coupled sugar transport mechanism of this symporter. This was illustrated in a recent study showing that the sugar-induced fast electrical transient and Trp fluorescence variations in WT MelB have comparable rate constants, which suggests that they monitor highly correlated structural events during cycling (62).
Supporting Material
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00768-1.
Supporting Material
Acknowledgments
We thank Víctor Lórenz-Fonfría for a critical reading of the manuscript; Raymonde Lemonnier, Elodia Serrano, and Neus Ontiveros for technical assistance; and Meritxell Granell for help in the Cys-less purification.
This work was supported by Dirección General de Investigación grant BFU2006-04656/BMC and in part by a grant from the Commissariat à l'Energie Atomique (CEA-Saclay).
Footnotes
Xavier León's present address is Centre de Biotecnologia Animal i Teràpia Gènica, Universitat Autònoma de Barcelona, Barcelona, Spain.
References
- 1.Poolman B., Knol J., van der Does C., Henderson P.J., Liang W.J. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 2.Pourcher T., Bassilana M., Sarkar H.K., Kaback H.R., Leblanc G. The melibiose/Na+ symporter of Escherichia coli: kinetic and molecular properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990;326:411–423. doi: 10.1098/rstb.1990.0021. [DOI] [PubMed] [Google Scholar]
- 3.Bassilana M., Pourcher T., Leblanc G. Facilitated diffusion properties of melibiose permease in Escherichia coli membrane vesicles. Release of co-substrates is rate limiting for permease cycling. J. Biol. Chem. 1987;262:16865–16870. [PubMed] [Google Scholar]
- 4.Bassilana M., Pourcher T., Leblanc G. Melibiose permease of Escherichia coli. Characteristics of co-substrates release during facilitated diffusion reactions. J. Biol. Chem. 1988;263:9663–9667. [PubMed] [Google Scholar]
- 5.Damiano-Forano E., Bassilana M., Leblanc G. Sugar binding properties of the melibiose permease in Escherichia coli membrane vesicles. Effects of Na+ and H+ concentrations. J. Biol. Chem. 1986;261:6893–6899. [PubMed] [Google Scholar]
- 6.Ganea C., Pourcher T., Leblanc G., Fendler K. Evidence for intraprotein charge transfer during the transport activity of the melibiose permease from Escherichia coli. Biochemistry. 2001;40:13744–13752. doi: 10.1021/bi011223k. [DOI] [PubMed] [Google Scholar]
- 7.Botfield M.C., Wilson T.H. Peptide-specific antibody for the melibiose carrier of Escherichia coli localizes the carboxyl terminus to the cytoplasmic face of the membrane. J. Biol. Chem. 1989;264:11649–11652. [PubMed] [Google Scholar]
- 8.Gwizdek C., Leblanc G., Bassilana M. Proteolytic mapping and substrate protection of the Escherichia coli melibiose permease. Biochemistry. 1997;36:8522–8529. doi: 10.1021/bi970312n. [DOI] [PubMed] [Google Scholar]
- 9.Pourcher T., Bibi E., Kaback H.R., Leblanc G. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusion analysis. Biochemistry. 1996;35:4161–4168. doi: 10.1021/bi9527496. [DOI] [PubMed] [Google Scholar]
- 10.Hacksell I., Rigaud J.L., Purhonen P., Pourcher T., Hebert H. Projection structure at 8 A resolution of the melibiose permease, an Na-sugar co-transporter from Escherichia coli. EMBO J. 2002;21:3569–3574. doi: 10.1093/emboj/cdf378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purhonen P., Lundback A.K., Lemonnier R., Leblanc G., Hebert H. Three-dimensional structure of the sugar symporter melibiose permease from cryo-electron microscopy. J. Struct. Biol. 2005;152:76–83. doi: 10.1016/j.jsb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Dave N., Troullier A., Mus-Veteau I., Duñach M., Leblanc G. Secondary structure components and properties of the melibiose permease from Escherichia coli: a Fourier transform infrared spectroscopy analysis. Biophys. J. 2000;79:747–755. doi: 10.1016/S0006-3495(00)76332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordat E., Mus-Veteau I., Leblanc G. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. II. Identification of the tryptophan residues acting as energy donors. J. Biol. Chem. 1998;273:33198–33202. doi: 10.1074/jbc.273.50.33198. [DOI] [PubMed] [Google Scholar]
- 14.Hama H., Wilson T.H. Cation-coupling in chimeric melibiose carriers derived from Escherichia coli and Klebsiella pneumoniae. The amino-terminal portion is crucial for Na+ recognition in melibiose transport. J. Biol. Chem. 1993;268:10060–10065. [PubMed] [Google Scholar]
- 15.Mus-Veteau I., Leblanc G. Melibiose permease of Escherichia coli: structural organization of cosubstrate binding sites as deduced from tryptophan fluorescence analyses. Biochemistry. 1996;35:12053–12060. doi: 10.1021/bi961372g. [DOI] [PubMed] [Google Scholar]
- 16.Pourcher T., Zani M.L., Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na(+)-dependent transport and binding properties. J. Biol. Chem. 1993;268:3209–3215. [PubMed] [Google Scholar]
- 17.Wilson D.M., Wilson T.H. Asp-51 and Asp-120 are important for the transport function of the Escherichia coli melibiose carrier. J. Bacteriol. 1992;174:3083–3086. doi: 10.1128/jb.174.9.3083-3086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordat E., Leblanc G., Mus-Veteau I. Evidence for a role of helix IV in connecting cation- and sugar-binding sites of Escherichia coli melibiose permease. Biochemistry. 2000;39:4493–4499. doi: 10.1021/bi991852i. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Dayem M., Basquin C., Pourcher T., Cordat E., Leblanc G. Cytoplasmic loop connecting helices IV and V of the melibiose permease from Escherichia coli is involved in the process of Na+-coupled sugar translocation. J. Biol. Chem. 2003;278:1518–1524. doi: 10.1074/jbc.M210053200. [DOI] [PubMed] [Google Scholar]
- 20.Ding P.Z. An investigation of cysteine mutants on the cytoplasmic loop X/XI in the melibiose transporter of Escherichia coli by using thiol reagents: implication of structural conservation of charged residues. Biochem. Biophys. Res. Commun. 2003;307:864–869. doi: 10.1016/s0006-291x(03)01290-7. [DOI] [PubMed] [Google Scholar]
- 21.Ding P.Z. Loop X/XI, the largest cytoplasmic loop in the membrane-bound melibiose carrier of Escherichia coli, is a functional re-entrant loop. Biochim. Biophys. Acta. 2004;1660:106–117. doi: 10.1016/j.bbamem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Pourcher T., Deckert M., Bassilana M., Leblanc G. Melibiose permease of Escherichia coli: mutation of aspartic acid 55 in putative helix II abolishes activation of sugar binding by Na+ ions. Biochem. Biophys. Res. Commun. 1991;178:1176–1181. doi: 10.1016/0006-291x(91)91016-6. [DOI] [PubMed] [Google Scholar]
- 23.Zani M.L., Pourcher T., Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. II. Effect on cationic selectivity and coupling properties. J. Biol. Chem. 1993;268:3216–3221. [PubMed] [Google Scholar]
- 24.Zani M.L., Pourcher T., Leblanc G. Mutation of polar and charged residues in the hydrophobic NH2-terminal domains of the melibiose permease of Escherichia coli. J. Biol. Chem. 1994;269:24883–24889. [PubMed] [Google Scholar]
- 25.Franco P.J., Wilson T.H. Alteration of Na(+)-coupled transport in site-directed mutants of the melibiose carrier of Escherichia coli. Biochim. Biophys. Acta. 1996;1282:240–248. doi: 10.1016/0005-2736(96)00062-4. [DOI] [PubMed] [Google Scholar]
- 26.Franco P.J., Wilson T.H. Arg-52 in the melibiose carrier of Escherichia coli is important for cation-coupled sugar transport and participates in an intrahelical salt bridge. J. Bacteriol. 1999;181:6377–6386. doi: 10.1128/jb.181.20.6377-6386.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mus-Veteau I., Pourcher T., Leblanc G. Melibiose permease of Escherichia coli: substrate-induced conformational changes monitored by tryptophan fluorescence spectroscopy. Biochemistry. 1995;34:6775–6783. doi: 10.1021/bi00020a024. [DOI] [PubMed] [Google Scholar]
- 28.Maehrel C., Cordat E., Mus-Veteau I., Leblanc G. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. I. Evidence for ion-induced conformational change. J. Biol. Chem. 1998;273:33192–33197. doi: 10.1074/jbc.273.50.33192. [DOI] [PubMed] [Google Scholar]
- 29.Meyer-Lipp K., Ganea C., Pourcher T., Leblanc G., Fendler K. Sugar binding induced charge translocation in the melibiose permease from Escherichia coli. Biochemistry. 2004;43:12606–12613. doi: 10.1021/bi0489053. [DOI] [PubMed] [Google Scholar]
- 30.Dave N., Lórenz-Fonfría V.A., Leblanc G., Padrós E. FTIR spectroscopy of secondary-structure reorientation of melibiose permease modulated by substrate binding. Biophys. J. 2008;94:3659–3670. doi: 10.1529/biophysj.107.115550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dave N., Lórenz-Fonfría V.A., Villaverde J., Lemonnier R., Leblanc G. Study of amide-proton exchange of Escherichia coli melibiose permease by attenuated total reflection-Fourier transform infrared spectroscopy: evidence of structure modulation by substrate binding. J. Biol. Chem. 2002;277:3380–3387. doi: 10.1074/jbc.M105466200. [DOI] [PubMed] [Google Scholar]
- 32.León X., Lórenz-Fonfría V.A., Lemonnier R., Leblanc G., Padrós E. Substrate-induced conformational changes of melibiose permease from Escherichia coli studied by infrared difference spectroscopy. Biochemistry. 2005;44:3506–3514. doi: 10.1021/bi048301z. [DOI] [PubMed] [Google Scholar]
- 33.León X., Lemonnier R., Leblanc G., Padrós E. Changes in secondary structures and acidic side chains of melibiose permease upon cosubstrates binding. Biophys. J. 2006;91:4440–4449. doi: 10.1529/biophysj.106.090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth A., Zscherp C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 35.Baenziger J.E., Miller K.W., Rothschild K.J. Fourier transform infrared difference spectroscopy of the nicotinic acetylcholine receptor: evidence for specific protein structural changes upon desensitization. Biochemistry. 1993;32:5448–5454. doi: 10.1021/bi00071a022. [DOI] [PubMed] [Google Scholar]
- 36.Ryan S.E., Blanton M.P., Baenziger J.E. A conformational intermediate between the resting and desensitized states of the nicotinic acetylcholine receptor. J. Biol. Chem. 2001;276:4796–4803. doi: 10.1074/jbc.M007063200. [DOI] [PubMed] [Google Scholar]
- 37.Nyquist R.M., Heitbrink D., Bolwien C., Wells T.A., Gennis R.B. Perfusion-induced redox differences in cytochrome c oxidase: ATR/FT-IR spectroscopy. FEBS Lett. 2001;505:63–67. doi: 10.1016/s0014-5793(01)02769-7. [DOI] [PubMed] [Google Scholar]
- 38.Barth A., Mantele W. ATP-Induced phosphorylation of the sarcoplasmic reticulum Ca2+ ATPase: molecular interpretation of infrared difference spectra. Biophys. J. 1998;75:538–544. doi: 10.1016/S0006-3495(98)77543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer-Lipp K., Sery N., Ganea C., Basquin C., Fendler K. The inner interhelix loop 4–5 of the melibiose permease from Escherichia coli takes part in conformational changes after sugar binding. J. Biol. Chem. 2006;281:25882–25892. doi: 10.1074/jbc.M601259200. [DOI] [PubMed] [Google Scholar]
- 40.Weissborn A.C., Botfield M.C., Kuroda M., Tsuchiya T., Wilson T.H. The construction of a cysteine-less melibiose carrier from. E. coli. Biochim. Biophys. Acta. 1997;1329:237–244. doi: 10.1016/s0005-2736(97)00116-8. [DOI] [PubMed] [Google Scholar]
- 41.Brandolin G., Doussiere J., Gulik A., Gulik-Krzywicki T., Lauquin G.J. Kinetic, binding and ultrastructural properties of the beef heart adenine nucleotide carrier protein after incorporation into phospholipid vesicles. Biochim. Biophys. Acta. 1980;592:592–614. doi: 10.1016/0005-2728(80)90103-6. [DOI] [PubMed] [Google Scholar]
- 42.Pourcher T., Leclercq S., Brandolin G., Leblanc G. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry. 1995;34:4412–4420. doi: 10.1021/bi00013a033. [DOI] [PubMed] [Google Scholar]
- 43.Botfield M.C., Wilson T.H. Mutations that simultaneously alter both sugar and cation specificity in the melibiose carrier of Escherichia coli. J. Biol. Chem. 1988;263:12909–12915. [PubMed] [Google Scholar]
- 44.Rigaud J.L., Paternostre M.T., Bluzat A. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry. 1988;27:2677–2688. doi: 10.1021/bi00408a007. [DOI] [PubMed] [Google Scholar]
- 45.Lórenz-Fonfría V.A., Padrós E. Maximum entropy deconvolution of infrared spectra: use of a novel entropy expression without sign restriction. Appl. Spectrosc. 2005;59:474–486. doi: 10.1366/0003702053641504. [DOI] [PubMed] [Google Scholar]
- 46.Ryan S.E., Hill D.G., Baenziger J.E. Dissecting the chemistry of nicotinic receptor-ligand interactions with infrared difference spectroscopy. J. Biol. Chem. 2002;277:10420–10426. doi: 10.1074/jbc.M107412200. [DOI] [PubMed] [Google Scholar]
- 47.von Germar F., Barth A., Mantele W. Structural changes of the sarcoplasmic reticulum Ca(2+)-ATPase upon nucleotide binding studied by Fourier transform infrared spectroscopy. Biophys. J. 2000;78:1531–1540. doi: 10.1016/S0006-3495(00)76705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 49.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H.R. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 50.Meyer-Lipp, K. 2005. Time-resolved measurements of sugar-binding-induced conformational changes in the melibiose permease from Escherichia coli. PhD thesis. Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany.
- 51.Ambroise Y., Leblanc G., Rousseau B. Active-site-directed photolabeling of the melibiose permease of Escherichia coli. Biochemistry. 2000;39:1338–1345. doi: 10.1021/bi9916224. [DOI] [PubMed] [Google Scholar]
- 52.Boudker O., Ryan R.M., Yernool D., Shimamoto K., Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 53.Grunewald M., Kanner B.I. The accessibility of a novel reentrant loop of the glutamate transporter GLT-1 is restricted by its substrate. J. Biol. Chem. 2000;275:9684–9689. doi: 10.1074/jbc.275.13.9684. [DOI] [PubMed] [Google Scholar]
- 54.Lambert G., Forster I.C., Stange G., Kohler K., Biber J. Cysteine mutagenesis reveals novel structure-function features within the predicted third extracellular loop of the type IIa Na+/Pi cotransporter. J. Gen. Physiol. 2001;117:533–546. doi: 10.1085/jgp.117.6.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.López-Corcuera B., Nuñez E., Martínez-Maza R., Geerlings A., Aragón C. Substrate-induced conformational changes of extracellular loop 1 in the glycine transporter GLYT2. J. Biol. Chem. 2001;276:43463–43470. doi: 10.1074/jbc.M107438200. [DOI] [PubMed] [Google Scholar]
- 56.Slotboom D.J., Konings W.N., Lolkema J.S. Cysteine-scanning mutagenesis reveals a highly amphipathic, pore-lining membrane-spanning helix in the glutamate transporter GltT. J. Biol. Chem. 2001;276:10775–10781. doi: 10.1074/jbc.M011064200. [DOI] [PubMed] [Google Scholar]
- 57.Stephan M.M., Chen M.A., Penado K.M., Rudnick G. An extracellular loop region of the serotonin transporter may be involved in the translocation mechanism. Biochemistry. 1997;36:1322–1328. doi: 10.1021/bi962150l. [DOI] [PubMed] [Google Scholar]
- 58.Wakabayashi S., Pang T., Su X., Shigekawa M. A novel topology model of the human Na(+)/H(+) exchanger isoform 1. J. Biol. Chem. 2000;275:7942–7949. doi: 10.1074/jbc.275.11.7942. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita A., Singh S.K., Kawate T., Jin Y., Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 60.Wilson D.M., Hama H., Wilson T.H. GLY113→ASP can restore activity to the ASP51→SER mutant in the melibiose carrier of Escherichia coli. Biochem. Biophys. Res. Commun. 1995;209:242–249. doi: 10.1006/bbrc.1995.1495. [DOI] [PubMed] [Google Scholar]
- 61.Matsuzaki S., Weissborn A.C., Tamai E., Tsuchiya T., Wilson T.H. Melibiose carrier of Escherichia coli: use of cysteine mutagenesis to identify the amino acids on the hydrophilic face of transmembrane helix 2. Biochim. Biophys. Acta. 1999;1420:63–72. doi: 10.1016/s0005-2736(99)00087-5. [DOI] [PubMed] [Google Scholar]
- 62.Garcia-Celma J.J., Dueck B., Stein M., Schlueter M., Meyer-Lipp K. Rapid activation of the melibiose permease MelB immobilized on a solid-supported membrane. Langmuir. 2008;24:8119–8126. doi: 10.1021/la800428h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


