Abstract
Profilin, a small cytoskeletal protein, and phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] have been implicated in cellular events that alter the cell morphology, such as endocytosis, cell motility, and formation of the cleavage furrow during cytokinesis. Profilin has been shown to interact with PI(4,5)P2, but the role of this interaction is still poorly understood. Using giant unilamellar vesicles (GUVs) as a simple model of the cell membrane, we investigated the interaction between profilin and PI(4,5)P2. A number and brightness analysis demonstrated that in the absence of profilin, molar ratios of PI(4,5)P2 above 4% result in lipid demixing and cluster formations. Furthermore, adding profilin to GUVs made with 1% PI(4,5)P2 leads to the formation of clusters of both profilin and PI(4,5)P2. However, due to the self-quenching of the dipyrrometheneboron difluoride-labeled PI(4,5)P2, we were unable to determine the size of these clusters. Finally, we show that the formation of these clusters results in the destabilization and deformation of the GUV membrane.
Introduction
Most investigations of the interaction between membrane lipids and proteins have been performed using either model systems that provide only averaged parameters from bulk data collected from solutions containing many liposomes, or in cellulo systems. The latter are often so complex that primary effects due to protein-lipid interactions are very difficult to observe and analyze. The giant unilamellar vesicle (GUV) has emerged as a model system that can bridge the gap between the bulk data collection (i.e., liposomes) and cells. The low curvature of GUV membranes closely resembles that of native cells, whereas other vesicles have much higher curvatures that could affect the interaction between proteins and lipids. Of importance, one can visualize and image spatially confined events in GUVs at the level of single vesicles in a controlled system that is much simpler than a cell. In the last 3 years, several groups have successfully used the GUV system to study protein phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] interactions (1–8). In this study, we expand on our previous work (7) and further characterize the interaction between profilin and PI(4,5)P2.
PI(4,5)P2 is an important dynamic component of the plasma membrane and constitutes ∼1% of the total membrane lipids in human erythrocytes (9). Despite its relatively low concentration, PI(4,5)P2 plays several important roles in cellular events, including endocytosis/exocytosis (10–12) and normal cytokinesis (13,14).
PI(4,5)P2 interacts with several proteins that are involved in membrane secretion and actin cytoskeleton remodeling. One of these ligands is profilin I, a 14 kDa actin sequestering protein (15). Profilin I is a ubiquitous eukaryotic protein that has been shown to play pivotal roles in many cellular functions.
Profilin gene disruption has led to similar defects in different organisms. In Saccharomyces cerevisiae, the disruption led to the formation of larger (sometimes by up to 50 times) undivided, multinucleated cells due to defective cytokinesis (16). In Dictyostelium discoideum, profilin disruption causes the cell size to increase by up to 10 times, as in the case of Saccharomyces cerevisiae, in which motility was shown to be severely affected and development blocked before fruiting-body formation (17). In normal bovine embryos, profilin was found to be concentrated at the cleavage furrow in 56% of the embryos (18). A similar localization of profilin in cleavage furrows was observed earlier in Tetrahymena (19), suggesting that profilin may facilitate cell division. The suppression of profilin I by RNA interference in human umbilical vein endothelial cells produced a significant reduction of the formation of actin filament and cell adhesion. The absence of profilin was also responsible for a significant reduction in cell protrusions (20).
In human platelets, profilin partitions between the plasma membrane and the cytosol in response to membrane PI(4,5)P2 levels (21). This interaction of profilin with PI(4,5)P2 has been partially assigned to binding of the negatively charged headgroup of the phosphoinositides to basic amino acids (22,23). In a previous study (7), we proposed that the initial interaction of profilin with PI(4,5)P2 lipids may require more than one PI(4,5)P2 and/or that the membrane is an essential component of the profilin interaction with PI(4,5)P2. Our GUV data also suggest that profilin is able to recruit adjacent PI(4,5)P2 lipids. Recent studies on the interaction between PI(4,5)P2 lipids and ezrin (8), PTEN (24), and the basic effector domain of myristoylated alanine-rich C kinase substrate (MARCKS) (25) all indicate clustering of PI(4,5)P2 phospholipids upon protein binding. We demonstrated in our previous work that the short-chain (C6) dipyrrometheneboron difluoride (BODIPY)-labeled polyphosphoinositide (PPI) lipids partition to the membrane only under specific conditions (7). Using the ζ-potential, Carvalho et al. (8) recently demonstrated that the long-chain C16 fluorescent PI(4,5)P2 analogs are effectively incorporated into the GUV membrane. Therefore, we can use C16-labeled PI(4,5)P2 to better characterize the interaction between profilin and the GUV membrane.
In this work, we show that the interaction of profilin with PI(4,5)P2 destabilizes the GUV membrane. This destabilization results in altered GUV morphology. We demonstrate that profilin and PI(4,5)P2 form clusters on the membrane, and that some of these clusters are localized at the sites of morphological alterations. Finally, we show that increasing the concentration of PI(4,5)P2 in the GUV results in lipid demixing and cluster formation, leading to morphological changes and eventually prevention of the electroformation of the GUVs.
Materials and Methods
Preparation of GUVs
GUVs were prepared by the electroformation method first described by Angelova et al. (26), with the GUV chamber described by Fidorra et al. (27). BODIPY TMR PI(4,5)P2 was purchased from Echelon Biosciences (Salt Lake City, UT). Unlabeled L-α-phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] (Brain, Porcine-Triammonium Salt) and 1-palmitoyl 2-oleoyl-phosphatidylcholine (POPC) were purchased from Avanti Lipids (Alabaster, AL). The POPC, BODIPY TMR PI(4,5)P2, and unlabeled PI(4,5)P2 stocks were prepared in chloroform methanol mix 2:1 (v/v). The GUVs used to investigate the profilin interaction with PI(4,5)P2 were made of 99% POPC and 1% BODIPY TMR PI(4,5)P2 (mole fraction). The GUVs used to study the effect of the increased PI(4,5)P2 concentration were made of POPC, BODIPY TMR PI(4,5)P2, and unlabeled PI(4,5)P2 at different molar ratios (99:1:0, 90:1:9, 90:10:0, 80:1:19, 80:20:0, 60:1:39, and 0:100:0) as required. The GUVs were grown as previously described (7) (for details see the Supporting Material).
Site-directed mutagenesis of profilin I
The plasmid containing human profilin I was mutated to introduce cysteine at position 57 by the overlapping polymerase chain reaction technique. The primers (GeneWorks, Hindmarsh, Australia) used were TCCTGGTTGGCAAAGACCGGTGTAGTTTTTACGTG and CCGGTCTTTGCCAACCAGGACACCCACCTC for S57C. The mutations were confirmed by sequencing using the pETBlue Up and Down primers (Novagen 70603 and 70604, respectively; Novagen, Darmstadt, Germany).
Labeling of profilin I mutant
Profilin cloning and purification were performed as described by Moens and Bagatolli (7) (for details see the Supporting Material). Then 5-iodoacetamidofluorescein (5-IAF), a thiol-reactive label (catalogue No. I-30451; Molecular Probes, Carlsbad, CA) dissolved in dimethylformamide was added to the purified profilin mutants in 50 mM Tris buffer pH 7.5 at a 1:5 ratio of protein to label with constant stirring. The reaction was allowed to proceed for 1 h at room temperature and free labels were separated from bound using PD-10 columns (catalogue No. 17-0851-01; Amersham Biosciences, Piscataway, NJ). The eluted labeled profilin was dialyzed using 5 kDa cutoff membrane against a large volume of 50 mM Tris pH 7.5 and concentrated on a 5 kDa cutoff Amicon Ultra centrifugal filter (catalogue No. UFC900524; Millipore, Billerica, MA).
Confocal microscopy
The GUVs were observed in a confocal fluorescence microscope (Nikon C1 with a Ti-E motorized inverted microscope) using a Plan Apochromat, VC60x WI, 1.2 NA objective lens). The pinhole was set at 33 μM. Laser lines at 488 and 532 nm were used for excitation of 5-IAF and BODIPY TMR PI(4,5)P2, respectively.
Number and brightness analysis
For the number and brightness analysis, 100 frames of 256 × 256 pixels each were taken. The pixel size was 49.7 nm with a pixel dwell time of 20 μs. These image stacks were analyzed using SimFCS software (Laboratory for Fluorescence Dynamics, University of California, Irvine, CA). The analog detector calibration was obtained from background image stacks taken before and after the experimental image stacks using the exact same settings but with the laser turned off as described previously (28). No differences were found in the detector calibration between the two sets of images. The software allows selection of areas of the images that have the same apparent brightness (B). B = 1 values represent the immobile fraction of the image, and B > 1 values represent the mobile fraction (28). To obtain the molecular brightness in photons/molecule/s, we divide the B − 1 value by the pixel dwell time.
Fluorescence spectroscopy
Excitation and emission spectra of 1% BODIPY TMR PI(4,5)P2 GUV and 100% BODIPY TMR PI(4,5)P2 were recorded on an ISS PC1 spectrofluorimeter (ISS, Champaign, IL) equipped with a 300 W Xenon arc lamp. For the excitation spectrum, the emission wavelength was set at 580 nm and the excitation was scanned from 380 nm to 564 nm. For the emission spectrum, the excitation wavelength was set at the maximum of the excitation spectrum, i.e., 537 nm for the 100% BODIPY TMR PI(4,5)P2 solution and 547 nm for the 1% BODIPY TMR PI(4,5)P2 GUV solution, respectively. The emission was then scanned from 555 nm to 650 nm. Three spectra for each sample were recorded and then normalized, and the average spectrum for each sample was calculated from the three normalized spectra.
Fluorescence lifetime measurements
Fluorescence lifetime measurements were performed with a laser-based fluorometer for time-resolved fluorescence measurements equipped with a supercontinuum fiber laser source SC450-2 from Fianium (Southampton, UK). For a description of the experimental setup, see the Supporting Material. All measurements were performed using excitation and emission polarizers in magic-angle orientations. The sample holder was connected to a water bath, which kept the sample at 20°C. The B&H discriminator threshold level was set to −100 mV during all measurements. After the samples were measured by time-correlated single photon counting, the instrument response function was determined with the use of a scattering solution that directed excitation light onto the photomultiplier tube. The scattering solution consisted of glycogen dissolved in water (glycogen from oyster, Type II, G8751; Sigma-Aldrich, St. Louis, MO). The two lifetime components of the fluorescence decay curve were then obtained by deconvolution of the fluorescence curve from the instrument response function.
Results
PI(4,5)P2 incorporation into GUV
GUVs with 1% BODIPY TMR PI(4,5)P2 and 99% POPC were prepared and observed directly in the growing chamber or in microscopy chambers by exciting the BODIPY TMR PI(4,5)P2 at 532 nm. The emission was detected through a 605/75 nm bandpass barrier filter. As recently reported by Carvalho et al. (8), the long-chain (C16) BODIPY TMR-labeled PI(4,5)P2 are effectively incorporated into the GUV membrane. Most of the GUVs formed ranged in diameter from 10 to 40 μm. As illustrated in Fig. 1, A and B, vesicles were spherical and did not present obvious membrane alterations; <5% of the GUV population presented vesicle inclusions or junctions with other vesicles, or were found to be multilamellar (data not shown).
Figure 1.
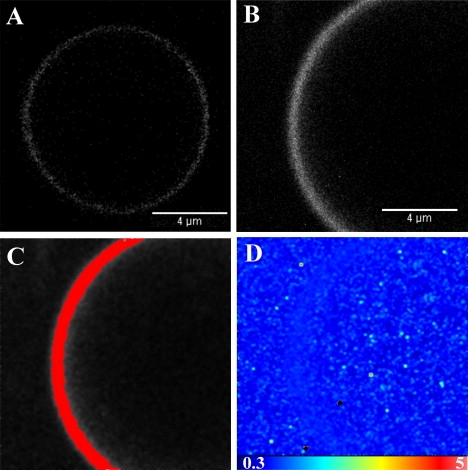
GUV containing 99% POPC and 1% BODIPY TMR-labeled PI(4,5)P2. (A) Confocal microscopy image of a GUV in the absence of profilin. (B) Intensity map of the GUV membrane. (C) The highlighted pixels correspond to a molecular brightness of 1.070. (D) Brightness map of the same GUV membrane.
Brightness analysis demonstrated that the BODIPY TMR PI(4,5)P2 is uniformly distributed in the membrane. The brightness calculated for this GUV (Fig. 1 B) was B = 1.070, giving an average of 3470 photons/molecule/s (pixel dwell time = 20 μs). When the pixels corresponding to that brightness were selected, the entire membrane was highlighted (Fig. 1 C), demonstrating that the brightness is homogeneous (Fig. 1 D). These data show that there is no aggregation or clustering of the BODIPY TMR PI(4,5)P2 at the 1% molar ratio. The GUVs could still be observed after 3 days in the microscope chamber, with no noticeable alteration of their morphology (data not shown).
Profilin-I binding to PI(4,5)P2 on GUV membrane
In a recent study (7), we indirectly assessed the binding of profilin to the GUV membrane by looking at the resistance to fluorescence resonance energy transfer (FRET) when profilin interacted with the lipids. In the work presented here, we used labeled lipids with a C16 alkyl chain, and expressed and purified a cysteine mutant of profilin that could be specifically labeled with 5-IAF and could therefore be directly visualized on the GUV membrane. Labeled profilin mixed with wild-type profilin in a 1:1500 molar ratio was added to GUVs made of 99% POPC and 1% unlabeled PI(4,5)P2 at a final concentration of ∼55 μM. Fig. 2 clearly confirms that profilin binds to the GUV membrane. It also shows that profilin aggregates in clusters on the membrane.
Figure 2.
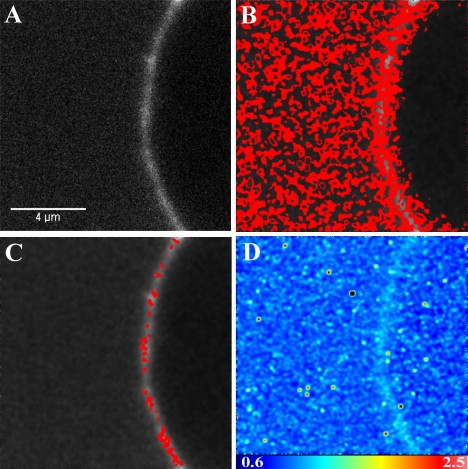
Confocal microscopy image of 5-IAF-labeled profilin binding on a GUV made of 99% POPC and 1% unlabeled PI(4,5)P2. (A) Intensity map of the GUV membrane. (B) The highlighted pixels correspond to a molecular brightness of 1.11. (C) The highlighted pixels are clusters of profilin and correspond to a molecular brightness of 1.23. (D) Brightness map of the same GUV membrane.
The brightness analysis distinguished between two populations of profilin. One population had a brightness of B = 1.11 (Fig. 2 B), which for a pixel dwell time of 20 μs gives an average of 5500 photons/mol/s, and the other population (Fig. 2 C) had a brightness of B = 1.23, giving an average of 11,500 photons/mol/s. Since the brightness depends on the specific fluorophore and the laser power, these values cannot be compared with the brightness of the BODIPY TMR PI(4,5)P2 obtained from a different image, which was calculated to be B = 1.071, giving an average molecular brightness of 3550 photons/mol/s (Fig. 1, C and D). However, we can compare brightness values obtained within the same images. In these cases, the different brightness values correspond to particles of different sizes. The ratio of molecular brightness between the two populations of fluorophores in Fig. 2, B and C, is equal to 2.1, suggesting that profilin forms dimers when interacting with PI(4,5)P2. However, to be able to visualize the binding of profilin to the GUV membrane, we diluted the labeled profilin by a factor of 1500 with unlabeled profilin so that the labeled profilin concentration would be ∼36 nM. Therefore, assuming a stochastic clustering of labeled and unlabeled profilin, the detection of dimers could indeed represent much larger aggregates. To test whether labeled profilin could bind to the GUV through nonspecific interactions, we added labeled profilin to GUVs made of 100% POPC and found that there was no binding of the profilin to the membrane, confirming that the interaction is specific to PI(4,5)P2 (Fig. 3).
Figure 3.

Confocal microscopy images of GUVs. (A) 5-IAF-labeled profilin in the presence of a GUV made of 100% POPC (average of 100 frames). (B–F) GUV containing 99% POPC and 1% BODIPY TMR-labeled PI(4,5)P2 in the presence of profilin. (D) Shmoo-like deformation of the GUV. (B) Budding of the GUV and the presence of lipid inclusions. (C) Altered GUV after several hours in the presence of profilin. (E and F) GUVs grown in the presence of 10 μM profilin.
Profilin 1 destabilizes the membrane of the GUV
To our surprise, we found that although the GUV made of 1% BODIPY TMR PI(4,5)P2 in the absence of profilin could still be observed after several days, when profilin was added, the GUVs could not be seen after 36 h, and only clusters of BODIPY TMR PI(4,5)P2 lipids were detected in the chambers (data not shown). Looking at various intervals after the addition of unlabeled profilin (70–100 μM), we noticed that although not all GUVs were affected simultaneously, a large proportion of GUVs showed altered morphology (Fig. 3), including “shmoo-like” shapes, junctions between GUVs, and budding of GUVs. These morphological changes were accompanied by increased fluorescence intensity at the site of junction or budding. No changes in the GUV morphology were observed when profilin was added to GUV made of POPC labeled with 1% DIiC18 (data not shown), confirming that these changes resulted from the interaction between profilin and PI(4,5)P2.
We further tested this mechanism by growing GUVs in the presence of various amounts of unlabeled profilin (10–100 μM). We found that GUVs were not electroformed when profilin was present at concentrations above 10 μM. Only a few GUVs were formed in the presence of 10 μM profilin, but the shapes of these vesicles were dramatically altered (Fig. 3).
Using brightness analysis, we investigated whether the increased intensity at the site of altered morphology was due to the presence of clusters of profilin or PI(4,5)P2 lipids, or both. Fig. 4 demonstrates that both profilin and PI(4,5)P2 form clusters, and that these clusters are associated with sites of membrane alterations. The cluster sizes determined from the brightness of the labeled profilin range from 3 to 5. However, as mentioned above, the size of these clusters is underestimated due to the presence of unlabeled profilin. The cluster size determined from the brightness of the BODIPY TMR PI(4,5)P2 lipids can be much larger (up to 13). Unfortunately, these do not represent the actual cluster size either. Indeed, although each PI(4,5)P2 has a fluorescent label (unlike profilin), BODIPY TMR PI(4,5)P2 has been shown to undergo aggregation with subsequent self-quenching, and has been used to monitor the interaction of MARCKS (25) and PTEN (24) with PI(4,5)P2 in large vesicles. Such quenching would result in an underestimation of the brightness of the aggregates or clusters. If the BODIPY TMR self-quenching is due to dynamic quenching, by measuring the lifetime of the cluster (29) we could calculate the amount of quenching and thus obtain a corrected value for the brightness of the clusters.
Figure 4.
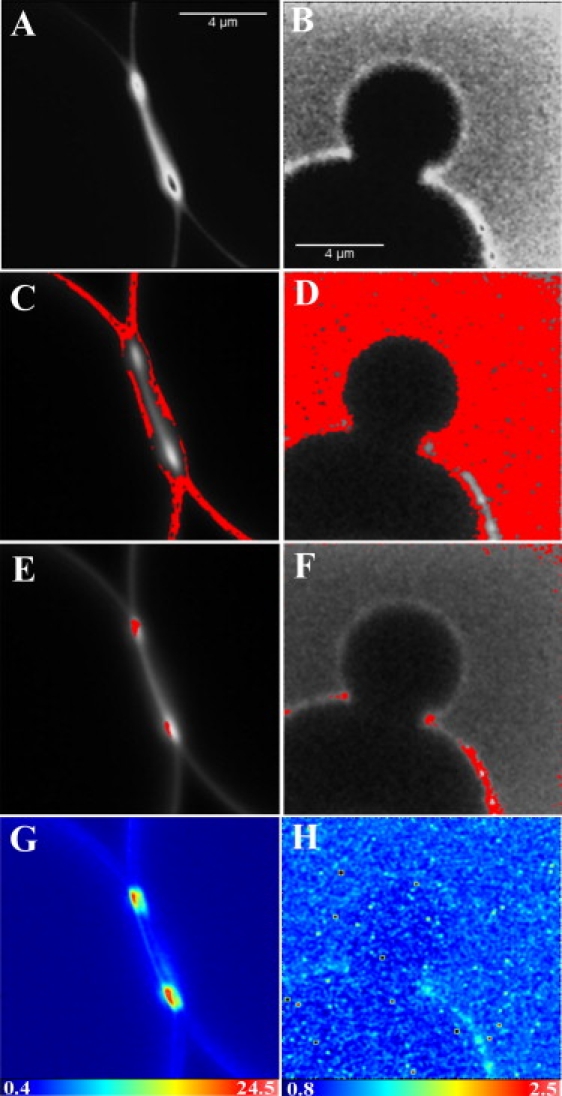
Brightness analysis of a GUV in the presence of ∼55 μM profilin. (A, C, E, and G) GUV containing 99% POPC and 1% BODIPY TMR-labeled PI(4,5)P2. (B, D, F, and H) 5-IAF-labeled profilin binding on a GUV made of 99% POPC and 1% unlabeled PI(4,5)P2. (A and B) Intensity map of the GUV. (C and D) The highlighted pixels correspond to the molecular brightness of monomers. (E and F) The highlighted pixels have a higher brightness and correspond to clusters in the membrane. (G and H) Brightness map of the same GUVs.
Self-quenching of BODIPY TMR PI(4,5)P2 is the result of static quenching
In the presence of dynamic quenching, the quenching rate constant kq can be calculated directly from the lifetime of the fluorophores in the absence (τ0) and presence (τ) of quencher [Q] according to the following equation:
| (1) |
We therefore attempted to determine the lifetime in GUVs in the absence of quenching (25), i.e., in GUVs made of 1% BODIPY TMR PI(4,5)P2 and 99% POPC, and with GUVs formed with 100% BODIPY TMR PI(4,5)P2 when quenching would be maximal. The GUVs were grown simultaneously in different wells of the same chamber. Upon observation with the microscope, we found that whereas GUVs made of 1% BODIPY TMR PI(4,5)P2 and 99% POPC were nicely formed, we could only detect BODIPY TMR PI(4,5)P2 lipid aggregates in the wells of 100% BODIPY TMR PI(4,5)P2. Although we did not produce GUVs when using 100% BODIPY TMR PI(4,5)P2, these clusters could still be used to determine the lifetime in conditions of maximal quenching.
The measured fluorescence decay curves of the two samples can be described by a double exponential decay. The first lifetime component is ascribed to the fluorescence from the probes, whereas the second component is considered to be a scattering signal with a quasi-zero lifetime. This scattering component is not only necessary for the fit, but additional scattering test measurements have confirmed the unavoidable existence of scattered light under the chosen measurement conditions (530 nm excitation and OG550 fluorescence filter). The 100% BODIPY TMR PI(4,5)P2 sample clearly exhibits a lifetime of 5 ns. The GUV made with the 1% BODIPY TMR PI(4,5)P2 sample shows a similar lifetime in the 5–6 ns range, but due to the extremely low fluorescence intensity of this sample (and therefore extremely long data collection times), the data quality does not allow precise lifetime resolution. However, this lifetime is in good agreement with a lifetime of 5.1 ns previously determined for the short-chain (C6) BODIPY TMR PI(4,5)P2 (7). The normalized emission and excitation spectra of these samples (Fig. 5) show that there is a ∼10 nm blue shift of the absorption spectrum for the 100% BODIPY TMR PI(4,5)P2. There is also a slight increase of the intensity in the long-wavelength region. This increase could represent emission from ground-state aggregates as suggested by Bergström et al. (30). However, the fractional intensity is so low for this component that we could not resolve it from the lifetime measurements.
Figure 5.
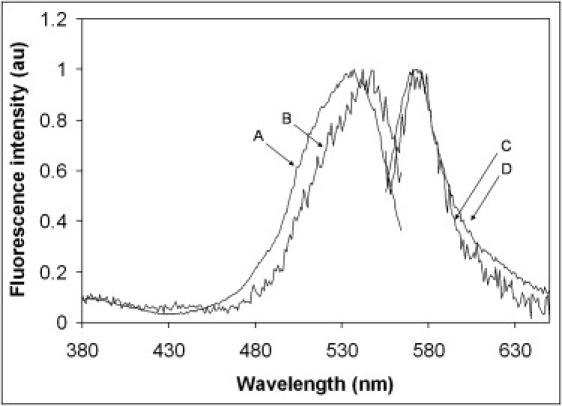
Excitation and emission spectrum of BODIPY TMR-labeled PI(4,5)P2 in GUVs and aggregates. Traces A and D are the normalized excitation and the emission spectrum of the lipid aggregates (100% BODIPY TMR-labeled PI(4,5)P2), respectively. Traces B and C are the normalized excitation and the emission spectrum of GUVs containing 99% POPC and 1% BODIPY TMR-labeled PI(4,5)P2, respectively.
Effect of increasing concentrations of PI(4,5)P2 on the GUV membrane
Since GUVs could not be electroformed with 100% BODIPY TMR PI(4,5)P2, we investigated the effect of increasing concentrations of PI(4,5)P2 on the electroformation of GUVs. To eliminate a possible effect of the fluorescent probe, we kept the concentration of labeled PI(4,5)P2 constant at a 1% molar ratio and increased the ratio of PI(4,5)P2 to POPC using unlabeled PI(4,5)P2. We found that GUVs were not formed above 40% PI(4,5)P2, and at 40% only a few GUVs could be observed in the chamber (Fig. 6, G and H). Below 4% PI(4,5)P2, there are no morphological alterations of the GUVs (Fig. 6, A and B). However, when the molar ratio of PI(4,5)P2 is increased to 8–10%, one starts to detect the formation of lipid aggregates and multivesicles (Fig. 6, C and D). These lipid aggregates and morphological alterations increase with the increased PI(4,5)P2 molar ratio, and at 16–20% PI(4,5)P2, lipid tubes are seen extending from the membrane, and aggregates are numerous. There are also large numbers of small vesicles inside larger GUVs (Fig. 6, E and F). A brightness analysis of GUVs containing 8% BODIPY TMR PI(4,5)P2 shows that there is clustering of PI(4,5)P2 in the membrane. Fig. 7 shows that areas of the membrane with different levels of brightness can be isolated. The lowest brightness was found to be B = 1.071 (3550 photons/mol/s), which probably represents monomers of PI(4,5)P2 (Fig. 7 B). The remainder of the membrane could be separated into two populations with an average brightness of B = 1.376 (18800 photons/mol/s) and B = 2.263 (63150 photons/mol/s), respectively. Because of the self-quenching of the BODIPY TMR moiety, it is not possible to determine the exact size of the aggregate. However, this result provides evidence that at these high PI(4,5)P2 molar ratios, these lipids self-aggregate into large clusters.
Figure 6.
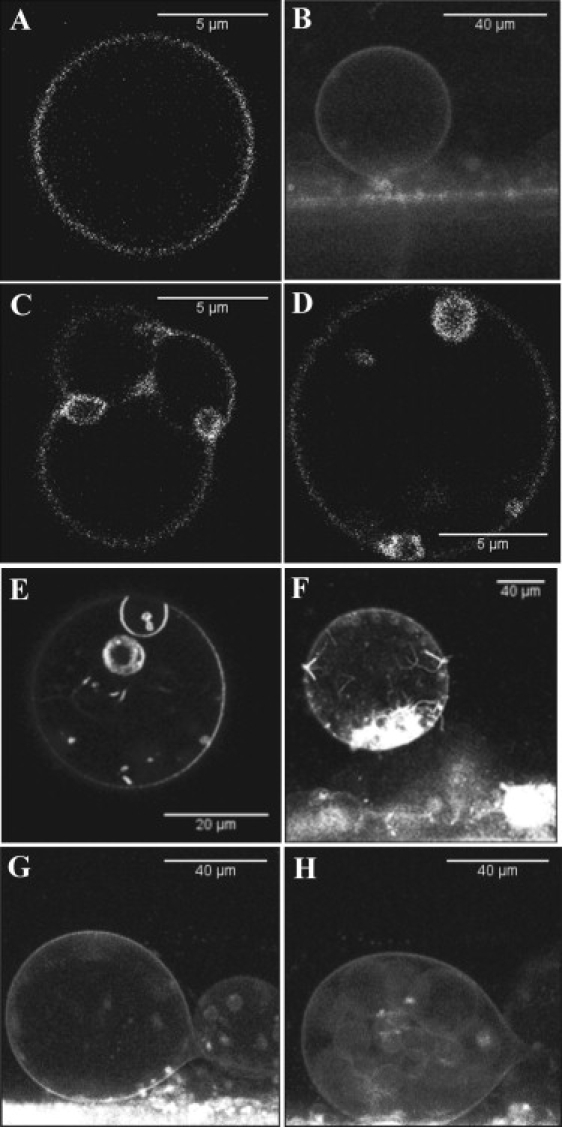
Confocal microscopy images of GUVs made of POPC and an increasing concentration of PI(4,5)P2. The concentration of BODIPY TMR-labeled PI(4,5)P2 is maintained constant at 1% molar ratio, and the increased PI(4,5)P2 concentration is achieved by adding increasing amounts of unlabeled PI(4,5)P2. (A and B) GUVs made with 1% and 2% PI(4,5)P2. (C and D) GUVs containing 8% PI(4,5)P2. (E and F) GUVs containing 20% PI(4,5)P2. (G and H) The only GUVs formed in the chamber when the PI(4,5)P2 concentration was 40% molar ratio.
Figure 7.
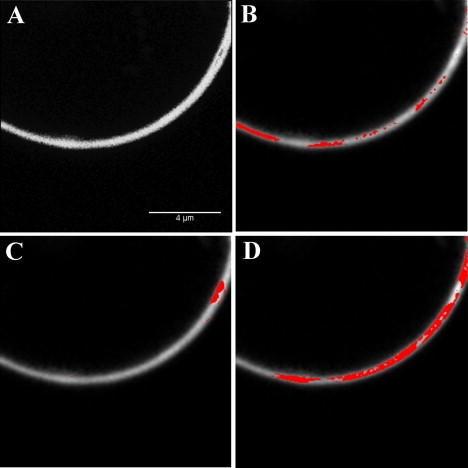
Brightness analysis of GUV made of 92% POPC and 8% PI(4,5)P2. The concentration of BODIPY TMR labeled PI(4,5)P2 was 1% molar ratio. (A) Fluorescence intensity map of the GUV. (B) The pixels highlighted correspond to the molecular brightness of monomers (1.071). (C) The highlighted pixels have an intermediate brightness of 1.376 and correspond to clusters in the membrane. (D) The highlighted pixels have the highest brightness (2.263) and correspond to areas of larger clusters.
Discussion
Self-quenching of BODIPY TMR PI(4,5)P2
To calculate the size of molecular aggregates using brightness analysis, the photon emission rate per molecule in the clusters should be a multiple of the photon emission rate determined for the monomer. Therefore, self-quenching as reported for BODIPY TMR PI(4,5)P2 (24,25) would result in an underestimation of the cluster size. However, if the quenching rate can be determined from the fluorescence lifetime of the clusters, the emission rate in the cluster could be corrected for the quenching process and the number of molecules in the cluster recovered. The mechanism of self-quenching for BODIPY-labeled phosphoinositides is still poorly characterized. Previous works used only steady-state measurements to determine the quenching effect (24,25). The mechanism of self-quenching for the headgroup-labeled lipid N-(lissamine rhodamine B sulfonyl)dipalmitoylphosphatidylcholine was attributed to molecular interactions (31), and the self-quenching mechanism for octadecyl rhodamine B was attributed to Förster resonance energy transfer (32,33). In this work, we demonstrate that there are no changes in the lifetime of the BODIPY TMR PI(4,5)P2 between molecules in the absence of quencher or in aggregates. Moreover, we show that there is a hypsochromic shift of the excitation spectrum of the BODIPY TMR PI(4,5)P2, which shows that these molecules are forming ground-state complexes. However, the aggregates are still fluorescent, which shows that the quenching is only partial. These findings demonstrate that the self-quenching observed is due to static quenching.
Recently, Bergström et al. (30) investigated the aggregation of BODIPY groups in a protein and lipid system. Although they could clearly assign the type of dimers formed in the protein system, it was less straightforward to interpret the aggregation of BODIPY groups in the lipid system. An additional complicating factor in our system is that the position of the BODIPY group is much farther away from the headgroup than in Bergström et al.'s experiments, which would allow the formation of higher aggregates of BODIPY (i.e., trimers, tetramers, etc.). Therefore it is not possible at this stage to describe the type of aggregates present in our system.
Since there were no changes in the lifetime of the BODIPY TMR PI(4,5)P2 in the clusters, we could not determine the quenching rates in particular aggregates, and thus could not establish a relationship between brightness and cluster size. Nevertheless, we were able to demonstrate the presence or absence of clusters and obtain the lowest possible number of molecules forming these clusters.
Effect of an increased PI(4,5)P2 molar ratio on the GUV membrane
Using lifetime measurements of diphenylhexatriene as a function of increasing concentration of NBD-labeled PI(4,5)P2 in liposomes, Fernandes et al. (34) showed that below a 5% molar ratio, the NBD-labeled PI(4,5)P2 is homogeneously distributed in the POPC lipid matrix. In our experiments, brightness analysis demonstrates that the labeled PI(4,5)P2 are uniformly distributed in the membrane, confirming that there is no aggregation of PI(4,5)P2 in GUVs made with 1% PI(4,5)P2. These results are also supported by the fact that there is no self-quenching of the BODIPY-labeled PI(4,5)P2 below a 1% molar ratio (24,25). However, when the molar ratio of the polyphosphoinositol lipids is increased to ≥8%, dramatic changes in the membrane occur. Clear modifications in the distribution of the fluorescence intensity within the GUV membrane can be qualitatively observed. Also, for molar ratios of PI(4,5)P2 of ≥20%, we noticed alterations of the membrane morphology resulting in regions of high fluorescence intensity and small lipid tubes or vesicles. Brightness analysis was used to ascertain whether the regions of increased intensity were due to an increased number of molecules per pixel or to the aggregation of several PI(4,5)P2 molecules. This analysis demonstrated that the increased intensity was the result of molecular aggregation of the PI(4,5)P2 lipids forming clusters within the GUV membrane. These results suggest that there is a threshold of the PI(4,5)P2 molar ratio between 5% and 8%, above which these lipids start to aggregate and form clusters. The notion of such lipid demixing in the fluid phase is supported by previous work. In 1981, Knoll et al. (35) reported immiscibility in the fluid state. These authors reported that deprotonated mixtures of distearoylphosphatidylcholine and dimyristoylphosphatidylcholine-d54 above the liquidus line showed a nonrandom lipid distribution in the fluid phase. They attributed this lipid demixing to critical concentration fluctuation. Also, using FRET between NBD-phosphatidylcholine and BODIPY TMR PI(4,5)P2 in vesicles formed from a PI(4,5)P2/POPC mixture, Redfern and Gericke (36) showed a decrease in the acceptor/donor emission ratio as a function of pH, suggesting the formation of fluid-type domains.
We also show that GUVs could not be formed when the PI(4,5)P2 molar ratio exceeded 40%. Taken together, these data suggest that the clustering of PI(4,5)P2 destabilizes the GUV membrane. This result is in agreement with the work of Carvalho et al. (8), who reported that the ζ-potential of GUVs containing 15% cholesterol and 10% PI(4,5)P2 increased after five measurements, suggesting that GUVs made of high levels of PI(4,5)P2 are more fragile. They also reported that this effect was not observed for GUVs containing 5% PI(4,5)P2. These data suggest that in cells, a 10-fold local increase in PI(4,5)P2 concentration after, for instance, protein translocation (37) or increased PI(4,5)P2 synthesis (38) could result in self-aggregation of PI(4,5)P2. Since we have shown that monodispersed PI(4,5)P2 has a low affinity for profilin (7), PI(4,5)P2 clustering could dramatically increase the chances of protein-lipid interaction by increasing the association constant for such proteins severalfold. In addition, local increases in PI(4,5)P2 synthesis have been shown to be essential for cell events that require membrane morphological changes, such as membrane ruffling formation (38) and endocytosis/exocytosis (10). Our findings suggest that increased PI(4,5)P2 concentration and PI(4,5)P2 clustering could destabilize the GUV membrane and contribute to the local membrane deformation.
Profilin destabilizes the GUV membrane
We demonstrate in this work that in the absence of PI(4,5)P2, profilin does not bind to the GUV membrane. In the presence of PI(4,5)P2, however, profilin is found to interact with the membrane. In our experiments, profilin concentrations of ∼55 μM were used, corresponding to the range of profilin concentrations found in human platelets (30–50 μM) (39). In the cell, these profilins would be associated with other proteins, such as actin, and therefore free concentrations would be much lower. Nevertheless, such concentrations could be easily attained during local redistribution of proteins. For example, Rawe et al. (18) found that profilin is observed throughout the cytoplasm at all stages of early bovine development through blastocyst formation, but it concentrates at the cleavage furrow of bovine embryos. The association-dissociation constant for the interaction between profilin and PI(4,5)P2 micelles or liposomes has been reported to be between 0.13 μM (40) and 35 μM (41). Therefore, by using ∼55 μM profilin, we should maximize the interaction with the membrane. The downside of using such concentrations of profilin is that to image the binding of profilin to the membrane and analyze the brightness of the particles, we have to dilute the labeled profilin with unlabeled ones to prevent image saturation. Because of the presence of unlabeled profilin in the clusters, the number of profilin molecules forming clusters could not be determined from the brightness analysis. However, we show that profilin, similarly to PI(4,5)P2, forms clusters on the membrane, and that these clusters are also localized on the GUV membrane at the site of morphological alterations. Furthermore, we show that although GUVs made of 1% PI(4,5)P2 and 99% POPC are stable, and PI(4,5)P2 does not form aggregates in these vesicles, the addition of profilin induced the clustering of PI(4,5)P2 and destabilized the GUV membrane, resulting in the formation of shmoo, budding, and fusing vesicles, and eventually the destruction of the GUVs. This destabilization of the membrane is also supported by the profilin concentration dependence on the electroformation of the GUVs. It is possible that the destabilization of the membrane leads to the formation of multilamellar vesicles or multilamellar regions in a vesicle, and that clusters form between bilayers. However, this is unlikely since a brightness analysis of multilamellar GUVs containing 1% PI(4,5)P2 did not show any aggregation or clusters of PI(4,5)P2 (data not shown).
It is tempting to speculate that the membrane destabilization effect seen upon self-clustering of PI(4,5)P2 is the same as that seen in PI(4,5)P2 clustering due to profilin interaction; however, this might not be the case, and further studies to characterize the destabilization of the membrane by PI(4,5)P2 in the presence and absence of profilin are under way.
It has been demonstrated that clustering or segregation of PI(4,5)P2 in GUVs can be induced by proteins other than profilin. Bagatolli et al. (42) showed that dynamin II binds to GUV composed of POPC/PI(4,5)P2 10:1 mol/mol. They demonstrated that the protein distribution on the membrane was not uniform, and the fluorescence was concentrated in patches on the surface of the GUV. However, it is not clear whether these patches were the result of self-aggregation of PI(4,5)P2 before dynamin binding or were induced by dynamin, since clusters of PI(4,5)P2 are already present at an 8% mole ratio (Fig. 7). Nevertheless, clusters resulting from the interaction of proteins with PI(4,5)P2 have been clearly demonstrated for annexin A2 (1) and ezrin (8). Of interest, none of these proteins seem to have a destabilization effect on the GUV membrane. Because the destabilization is a slow process, a possible explanation for this difference could be the different timeframes of observation used in the studies. Another possibility is that the interaction of the proteins with PI(4,5)P2 form different types of clusters that could lead to different functions. This is supported by the fact that the interaction of ezrin with PIP2 reduces ezrin's hole-opening activity (4).
A possible explanation for the effect of profilin relies on the binding stoichiometry of PI(4,5)P2 for profilin, and the small size of the protein. Indeed, it has been shown that profilin can bind up to five PI(4,5)P2 lipids (43) and that there are multiple binding sites for PI(4,5)P2 on profilin (23,44). Therefore, when multiple PI(4,5)P2 molecules bind to profilin, they wrap around it, creating a local change in the membrane curvature. The formation and aggregation of several of these complexes could trigger a local destabilization of the membrane. Of interest, both PI(4,5)P2 and profilin have now been independently reported to accumulate at sites of membrane deformation, such as cleavage furrow (13,14,18) and endocytosis (10–12,45,46). The effects of profilin and PI(4,5)P2 on the cell membrane suggest that these molecules may work cooperatively and facilitate these cellular processes.
Supporting Material
Additional materials and methods are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00779-6.
Supporting Material
Acknowledgments
We thank Dr. S. Sanchez for her help at the Laboratory for Fluorescence Dynamics (LFD; University of California, Irvine, CA), and Prof. D.M. Jameson, Prof. L.A. Bagatolli, and Dr. J. Coumans for critical readings of the manuscript.
This work was supported by a National Health and Medical Research Council (NHMRC) research project grant (568301), an internal research grant (RE209781) from the University of New England (Australia), a Linkage Infrastructure Equipment and Facilities Grant (LE0561041) from the Australian Research Council (ARC), and a grant from the ARC/NHMRC Fluorescence Application in Biotechnology and Life Sciences network (RN0460002). K. Krishnan was supported by a University of New England (Australia) Research Award international scholarship. A. Clayton was supported by an NHMRC R. D. Wright Career Development Award grant (280920). Some of the experiments reported in this work were performed at the LFD. The LFD is supported jointly by the National Center for Research Resources, National Institutes of Health (PHS 5 P41-RR003155) and the University of California, Irvine, CA.
References
- 1.Gokhale N.A., Abraham A., Digman M.A., Gratton E., Cho W. Phosphoinositide specificity of and mechanism of lipid domain formation by annexin A2-p11 heterotetramer. J. Biol. Chem. 2005;280:42831–42840. doi: 10.1074/jbc.M508129200. [DOI] [PubMed] [Google Scholar]
- 2.Golebiewska U., Gambhir A., Hangyás-Mihályné G., Zaitseva I., Rädler J. Membrane-bound basic peptides sequester multivalent (PIP2), but not monovalent (PS), acidic lipids. Biophys. J. 2006;91:588–599. doi: 10.1529/biophysj.106.081562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu A.P., Fletcher D.A. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys. J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda S., Saitoh A., Furuta M., Satomi N., Ishino A. Opening of holes in liposomal membranes is induced by proteins possessing the FERM domain. J. Mol. Biol. 2006;362:403–413. doi: 10.1016/j.jmb.2006.07.071. [DOI] [PubMed] [Google Scholar]
- 5.Heuvingh J., Franco M., Chavrier P., Sykes C. ARF1-mediated actin polymerization produces movement of artificial vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:16928–16933. doi: 10.1073/pnas.0704749104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong J., Nguyen L., Vidal A., Simon S.A., Skene J.H.P. Role of GAP-43 in sequestering phosphatidylinositol 4,5-bisphosphate to Raft bilayers. Biophys. J. 2008;94:125–133. doi: 10.1529/biophysj.107.110536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moens P.D.J., Bagatolli L.A. Profilin binding to sub-micellar concentrations of phosphatidylinositol (4,5) bisphosphate and phosphatidylinositol (3,4,5) trisphosphate. Biochim. Biophys. Acta. 2007;1768:439–449. doi: 10.1016/j.bbamem.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho K., Ramos L., Roy C., Picart C. Giant unilamellar vesicles containing phosphatidylinositol(4,5)bisphosphate: characterization and functionality. Biophys. J. 2008;95:4348–4360. doi: 10.1529/biophysj.107.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrell J.E., Huestis W.H. Phosphoinositide metabolism and the morphology of human erythrocytes. J. Cell Biol. 1984;98:1992–1998. doi: 10.1083/jcb.98.6.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aikawa Y., Martin T.F.J. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J. Cell Biol. 2003;162:647–659. doi: 10.1083/jcb.200212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyagi K., Sugaya T., Umeda M., Yamamoto S., Terakawa S. The activation of exocytotic sites by the formation of phosphatidylinositol 4,5-bisphosphate microdomains at syntaxin clusters. J. Biol. Chem. 2005;280:17346–17352. doi: 10.1074/jbc.M413307200. [DOI] [PubMed] [Google Scholar]
- 12.Gong L., Di Paolo G., Diaz E., Cestra G., Diaz M. Phosphatidylinositol phosphate kinase type I γ regulates dynamics of large dense-core vesicle fusion. Proc. Natl. Acad. Sci. USA. 2005;102:5204–5209. doi: 10.1073/pnas.0501412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emoto K., Inadome H., Kanaho Y., Narumiya S., Umeda M. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 2005;280:37901–37907. doi: 10.1074/jbc.M504282200. [DOI] [PubMed] [Google Scholar]
- 14.Field S.J., Madson N., Kerr M.L., Galbraith K.A.A., Kennedy C.E. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr. Biol. 2005;15:1407–1412. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson L., Nystrom L.E., Sundkvist I., Markey F., Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 16.Haarer B.K., Lillie S.H., Adams A.E., Magdolen V., Bandlow W. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J. Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haugwitz M., Noegel A.A., Karakesisoglou J., Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79:303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 18.Rawe V.Y., Payne C., Schatten G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Hum. Reprod. 2006;21:1143–1153. doi: 10.1093/humrep/dei480. [DOI] [PubMed] [Google Scholar]
- 19.Edamatsu M., Hirono M., Watanabe Y. Tetrahymena profilin is localized in the division furrow. J. Biochem. 1992;112:637–642. doi: 10.1093/oxfordjournals.jbchem.a123952. [DOI] [PubMed] [Google Scholar]
- 20.Ding Z., Lambrechts A., Parepally M., Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 2006;119:4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- 21.Hartwig J.H., Chambers K.A., Hopcia K.L., Kwiatkowski D.J. Association of profilin with filament-free regions of human leukocyte and platelet membranes and reversible membrane binding during platelet activation. J. Cell Biol. 1989;109:1571–1579. doi: 10.1083/jcb.109.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machesky L.M., Goldschmidt-Clermont P.J., Pollard T.D. The affinities of human platelet and Acanthamoeba profilin isoforms for polyphosphoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1990;1:937–950. doi: 10.1091/mbc.1.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambrechts A., Jonckheere V., Dewitte D., Vandekerckhove J., Ampe C. Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem. 2002;3:1–12. doi: 10.1186/1471-2091-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redfern R.E., Redfern D., Furgason M.L.M., Munson M., Ross A.H. PTEN phosphatase selectively binds phosphoinositides and undergoes structural changes. Biochemistry. 2008;47:2162–2171. doi: 10.1021/bi702114w. [DOI] [PubMed] [Google Scholar]
- 25.Gambhir A., Hangyas-Mihalyne G., Zaitseva I., Cafiso D.S., Wang J. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelova M.I., Soleau S., Meleard P., Faucon J.F., Bothorel P. Preparation of giant vesicles by external AC fields. Kinetics and application. Prog. Colloid Polym. Sci. 1992;89:127–131. [Google Scholar]
- 27.Fidorra M., Duelund L., Leidy C., Simonsen A., Bagatolli L.A. Absence of fluid-ordered/fluid-disordered phase coexistence in ceramide/POPC mixtures containing cholesterol. Biophys. J. 2006;90:4437–4451. doi: 10.1529/biophysj.105.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalal R.B., Digman M.A., Horwitz A.F., Vetri V., Gratton E. Determination of particle number and brightness using a laser scanning confocal microscope operating in the analog mode. Microsc. Res. Tech. 2008;71:69–81. doi: 10.1002/jemt.20526. [DOI] [PubMed] [Google Scholar]
- 29.Valeur B. Wiley-VCH; Weinheim, Germany: 2002. Molecular Fluorescence: Principles and Applications. [Google Scholar]
- 30.Bergström F., Mikhalyov I., Hagglof P., Wortmann R., Ny T. Dimers of dipyrrometheneboron difluoride (BODIPY) with light spectroscopic applications in chemistry and biology. J. Am. Chem. Soc. 2002;124:196–204. doi: 10.1021/ja010983f. [DOI] [PubMed] [Google Scholar]
- 31.Massari S., Colonna R., Folena E. Interaction of the fluorescent probe N-(lissamine rhodamine B sulfonyl)dipalmitoylphosphatidylethanolamine with phosphatidylcholine bilayers. Biochim. Biophys. Acta. 1988;940:149–157. doi: 10.1016/0005-2736(88)90019-3. [DOI] [PubMed] [Google Scholar]
- 32.Johansson L.B.A., Niemi A. Electronic energy transfer in anisotropic systems. 1. Octadecylrhodamine B in vesicles. J. Phys. Chem. 1987;91:3020–3023. [Google Scholar]
- 33.MacDonald R. Characteristics of self-quenching of the fluorescence of lipid-conjugated rhodamine in membranes. J. Biol. Chem. 1990;265:13533–13539. [PubMed] [Google Scholar]
- 34.Fernandes F., Loura L.M.S., Fedorov A., Prieto M. Absence of clustering of phosphatidylinositol-(4,5)-bisphosphate in fluid phosphatidylcholine. J. Lipid Res. 2006;47:1521–1525. doi: 10.1194/jlr.M600121-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Knoll W., Ibel K., Sackmann E. Small-angle neutron scattering study of lipid phase diagrams by the contrast variation method. Biochemistry. 1981;20:6379–6383. doi: 10.1021/bi00525a015. [DOI] [PubMed] [Google Scholar]
- 36.Redfern D.A., Gericke A. pH-dependent domain formation in phosphatidylinositol polyphosphate/phosphatidylcholine mixed vesicles. J. Lipid Res. 2005;46:504–515. doi: 10.1194/jlr.M400367-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Golebiewska U., Nyako M., Woturski W., Zaitseva I., McLaughlin S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol. Biol. Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda A., Nogami M., Yokozeki T., Yamazaki M., Nakamura H. Phosphatidylinositol 4-phosphate 5-kinase [α] is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 39.Goldschmidt-Clermont P.J., Machesky L.M., Doberstein S.K., Pollard T.D. Mechanism of the interaction of human platelet profilin with actin. J. Cell Biol. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostrander D.B., Gorman J.A., Carman G.M. Regulation of profilin localization in Saccharomyces cerevisiae by phosphoinositide metabolism. J. Biol. Chem. 1995;270:27045–27050. doi: 10.1074/jbc.270.45.27045. [DOI] [PubMed] [Google Scholar]
- 41.Lu P.J., Shieh W.R., Rhee S.G., Yin H.L., Chen C.S. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry. 1996;35:14027–14034. doi: 10.1021/bi961878z. [DOI] [PubMed] [Google Scholar]
- 42.Bagatolli L.A., Binns D.D., Jameson D.M., Albanesi J.P. Activation of dynamin II by POPC in giant unilamellar vesicles: a two-photon fluorescence microscopy study. J. Protein Chem. 2002;21:383–391. doi: 10.1023/a:1021126415320. [DOI] [PubMed] [Google Scholar]
- 43.Goldschmidt-Clermont P.J., Kim J.W., Machesky L.M., Rhee S.G., Pollard T.D. Regulation of phospholipase C-γ 1 by profilin and tyrosine phosphorylation. Science. 1991;251:1231–1233. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- 44.Skare P., Karlsson R. Evidence for two interaction regions for phosphatidylinositol(4,5)- bisphosphate on mammalian profilin I. FEBS Lett. 2002;522:119–124. doi: 10.1016/s0014-5793(02)02913-7. [DOI] [PubMed] [Google Scholar]
- 45.Gareus R., Di Nardo A., Rybin V., Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J. Biol. Chem. 2006;281:2803–2811. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- 46.Witke W., Podtelejnikov A.V., Di Nardo A., Sutherland J.D., Gurniak C.B. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


