Abstract
microRNAs (miRNAs) are an abundant class of ~22 nucleotide (nt) regulatory RNAs that are pervasive in higher eukaryotic genomes. In order to fully understand their prominence in genomes, it is necessary to elucidate the molecular mechanisms that can diversify miRNA activities. In this review, we describe some of the many strategies that allow novel miRNA functions to emerge, with particular emphasis on how miRNA genes evolve in animals. These mechanisms include changes in their sequence, processing, or expression pattern; acquisition of miRNA* functionality or antisense processing; and de novo gene birth. The facility and versatility of miRNAs to evolve and change likely underlies how they have become dominant constituents of higher genomes.
Keywords: microRNA, evolution, subfunctionalization, neofunctionalization
Introduction
microRNAs (miRNAs) are an abundant family of ~21–24 nucleotide (nt) regulatory RNAs derived from hairpin precursor transcripts [1, 2]. They have broad roles in gene regulation during organismal development and adult physiology, in both plants and animals. Although the extent of the miRNA regulatory universe has only been appreciated relatively recently, with hindsight we recognize that the first examples of miRNAs and their targets emerged in the 1990s. In particular, genetic studies of nematode and fruitfly development provided much of the framework for our understanding of miRNA genes and miRNA target identification.
Analysis of the Caenorhabditis elegans heterochronic pathway, which controls the timing of major developmental events, led to the identification of the first two miRNA genes, lin-4 and let-7 [3, 4]. Knowledge of epistatic genetic relationships amongst heterochronic genes fueled the discovery of their key target genes, even before the discovery of miRNAs as a general molecular phenomenon. These include lin-14 and lin-28 as key targets of lin-4 [5, 6], and lin-41 as an essential target of let-7 [4]. lin-4 and let-7 shared the feature of regulating targets via imperfectly complementary sites in target 3′ untranslated regions (3′ UTRs).
Parallel studies of Drosophila revealed that Notch pathway target genes encoding bHLH repressor and Bearded family proteins were repressed post-transcriptionally, via conserved ~7-nt 3′ UTR motifs termed the Brd, K, and GY boxes [7–9]. The motifs are required to restrict Notch signaling during normal developmental patterning, since genomic rescue transgenes bearing specific mutations in “box” motifs (but not wild-type transgenes) induced sensory bristle and eye defects characteristic of Notch target gene gain-of-function. These motifs proved to be amongst the first miRNA target sites known, and it was through them that it was recognized that animal miRNAs primarily identify targets via ~7 nt complements to miRNA 5′ ends [10], also referred to as miRNA “seeds” [11].
Let-7 was the first miRNA known to be well-conserved amongst animal species [12], and the direct cloning and sequencing of small RNAs from various animals resulted in the landmark discovery of ~100 miRNA genes in late 2001 [13–15]. Subsequently, a combination of molecular cloning and computational approaches identified thousands of miRNA genes in animals [16–20], plants [21–23], and even viruses [24, 25], with 600 miRNAs now validated in humans alone [26]. Computational estimates by various groups vary widely, and a consensus on the upper limit of miRNA genes has not been reached. New miRNA genes continue to emerge with the advent of new high-throughput sequencing methods [27–30].
Knowledge of the functional properties of Brd, GY, and K boxes as conserved 7-nt 3′ UTR regulatory motifs enabled forward computational searches for target sites to be performed as early as 1996 ([9] and Christian Burks and Eric C Lai, unpublished data). In addition, while lin-4 was originally characterized as a translational inhibitor [5, 31], studies of Notch pathway targets showed that miRNA binding sites could also destabilize transcripts via a deadenylation-associated mechanism [7, 9]. Many years later, the concept of 7mer seed matching target sites would underlie whole genome computational miRNA target searches [11, 32–35]. In addition, as miRNA targets outside of the Notch pathway also proved regulated at the steady-state mRNA level [36, 37], microarray-based efforts proved efficacious in identifying miRNA targets as transcripts whose levels were inversely correlated with miRNA activity [38–40]. Collectively, these efforts suggest that a majority of animal genes are either under active selection to maintain miRNA binding sites, or actively avoid the acquisition of miRNA binding sites [40, 41]. In addition, the existence of functional non-conserved sites [37, 39, 40] and functional non-seed match sites [4, 42, 43] likely increases the size of the miRNA target network.
In terms of the numbers of genes and regulatory targets, then, miRNAs can be considered amongst the most “successful” gene classes. In order to understand why miRNAs are so successful as genetic entities, it is imperative to understand how their functions and activities can be diversified during evolution. In this review, we highlight several molecular mechanisms by which this can occur. We will focus specifically on “miRNA-centric” mechanisms for the diversification of miRNA functions. Of course, miRNA functions can change through changes in their target genes. Since minimal functional sites are only 7 nt long, virtually all genes are a point mutation away from gaining or losing miRNA binding sites. Such lability of miRNA target sites underlies the wholesale turnover of miRNA target cohorts between animal clades, even though the miRNAs them-selves are often highly or even perfectly conserved [44]. This topic has been extensively reviewed recently [45, 46]. In addition, as other perspectives have presented a detailed discussion of plant miRNA evolution [46–48], we have focused this review on new concepts that have recently been examined in the context of animal genomes. However, at least some of the principles discussed here may apply generally to miRNA evolution across the kingdoms.
A brief overview of animal miRNA biogenesis
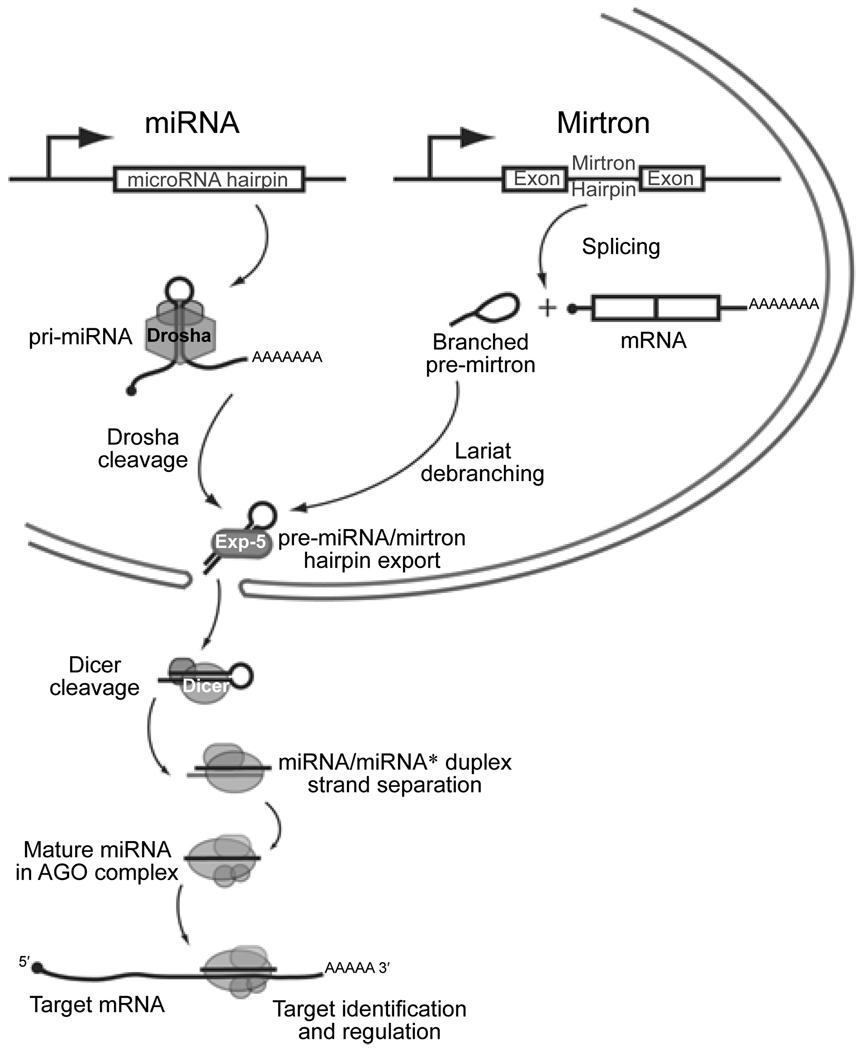
To provide context for the mechanisms discussed in this article, it is appropriate to first review the basic features of animal miRNA biogenesis and function (Figure 1). With only few exceptions [49], animal miRNAs are the products of much longer transcripts generated by RNA Polymerase II [50, 51]. The majority of miRNA genes are processed from the introns or exons of non-coding transcripts, but some 30% of miRNAs are located on the sense strands of introns of protein-encoding genes. It has been widely assumed that intronic miRNAs are processed out of spliced host introns; however, where it has been examined experimentally, it appears that intronic miRNA processing precedes intron splicing [52].
Figure 1.
Features of canonical miRNA and mirtron biogenesis. Canonical miRNAs are transcribed as long primary transcripts whose hairpin structures are cleaved by the nuclear Drosha RNAse III enzyme to release pre-miRNAs. Mirtrons are short hairpin introns that are spliced and then debranched, yielding pre-miRNA mimics. Both types of hairpins are exported from the nucleus by Exportin-5, then cleaved by the cytoplasmic RNAse III enzyme Dicer; this yields a duplex of ~21–24 nt RNAs. One RNA product, termed the mature miRNA, is preferentially loaded into an Argonaute (AGO) protein and guides it to complementary transcripts for regulation. The other duplex strand, termed the miRNA* species, is favored for degradation and accumulates to a lower level than the miRNA. The schematic was adapted from a published model [54].
Canonical miRNA precursors bear a duplex stem of at least ~30 basepairs. The base of the hairpin serves as a binding site for the double-stranded RNA binding protein Pasha/DGCR8, which positions the RNAse III enzyme Drosha to cleave ~1 helical turn away from the hairpin base (Figure 1) [53]. This processed hairpin is referred to as the “pre-miRNA”, and is typically 55–70 nt in length. Recently, it was found that short intron hairpins termed “mirtrons” can bypass Drosha processing and instead use the splicing machinery to directly define their precursor ends [54–56]. Following intron debranching, mirtrons appear as pre-miRNA mimics that gain access to the miRNA pathway.
Both mirtrons and pre-miRNAs are exported to the cytoplasm via Exportin-5, and are then cleaved by the cytoplasmic RNAse III enzyme Dicer (Figure 1) [57]. This yields an ~21–22 nt duplex, of which one strand is preferentially selected for entry into an Argonaute (AGO) silencing complex. The mature miRNA guides the AGO complex to complementary sites on target transcripts usually found in their 3′ UTRs. Although rare examples of atypical, functional miRNA binding sites with non-complementary seeds have been described [4, 5, 42, 43], the vast majority of miRNA targeting in animals appears to involve ~7 nt of Watson-Crick basepairing to positions 2–8 from the 5′ end of the miRNA. Thus, precision of the 5′ end of the mature miRNA is essential for it to select appropriate targets.
Evolutionary fates of duplicated miRNA genes
Just as with protein-encoding genes, a major route for gene diversification begins with aberrant DNA replication resulting in a local gene duplication [58]. It is generally accepted that duplicated genes should acquire distinct functions in order to be maintained over significant evolutionary timescales [59, 60]. Although many genes might appear dispensable or functionally redundant in the laboratory setting, truly redundant genes are not likely to survive during the evolution of “living” genomes.
At least three fates have been proposed for duplicated genes [61–63]. In the case of “nonfunctionalized” duplicates, one of the genes loses its activity through the accumulation of mutations. It eventually becomes a pseudogene or disappears from the genome entirely. “Subfunctionalized” gene pairs lose complementary functions, so that both genes need to be maintained in the genome in order to fulfill the aggregate function of the ancestral gene. Finally, a “neofunctionalized” gene acquires a novel activity that becomes selectively advantageous, leading to its stabilization in the genome. It is often easier to recognize examples of neofunctionalization, although sometimes this occurs hand-in-hand with subfunctionalization [64]. In the next few sections, we consider some of the molecular mechanisms that underlie neofunctionalization or subfunctionalization of duplicated miRNA genes, including via the acquisition of new miRNA sequences or expression patterns. Interestingly, as we shall see, at least some of these mechanisms can also apply to non-duplicated miRNA genes.
Changes in miRNA sequence
As antisense regulators, alteration to the miRNA sequence will have an impact on their targeting capabilities. This is a convenient mechanism for the invention of miRNA gene novelty. One can easily imagine that, following gene duplication, one of the genes may sustain a mutation that changes its targeting activity. Under the right circumstances, this might become favorably selected and eventually fixed in the form of a new miRNA gene. In this scenario, one of the ancestral genes should retain its sequence while its derivatives may drift. This is indeed what one sees amongst conserved animal miRNAs. Many apparent founder genes are identical amongst invertebrate and vertebrate genomes, but each lineage can have a series of related family members [18, 20, 27, 28, 65].
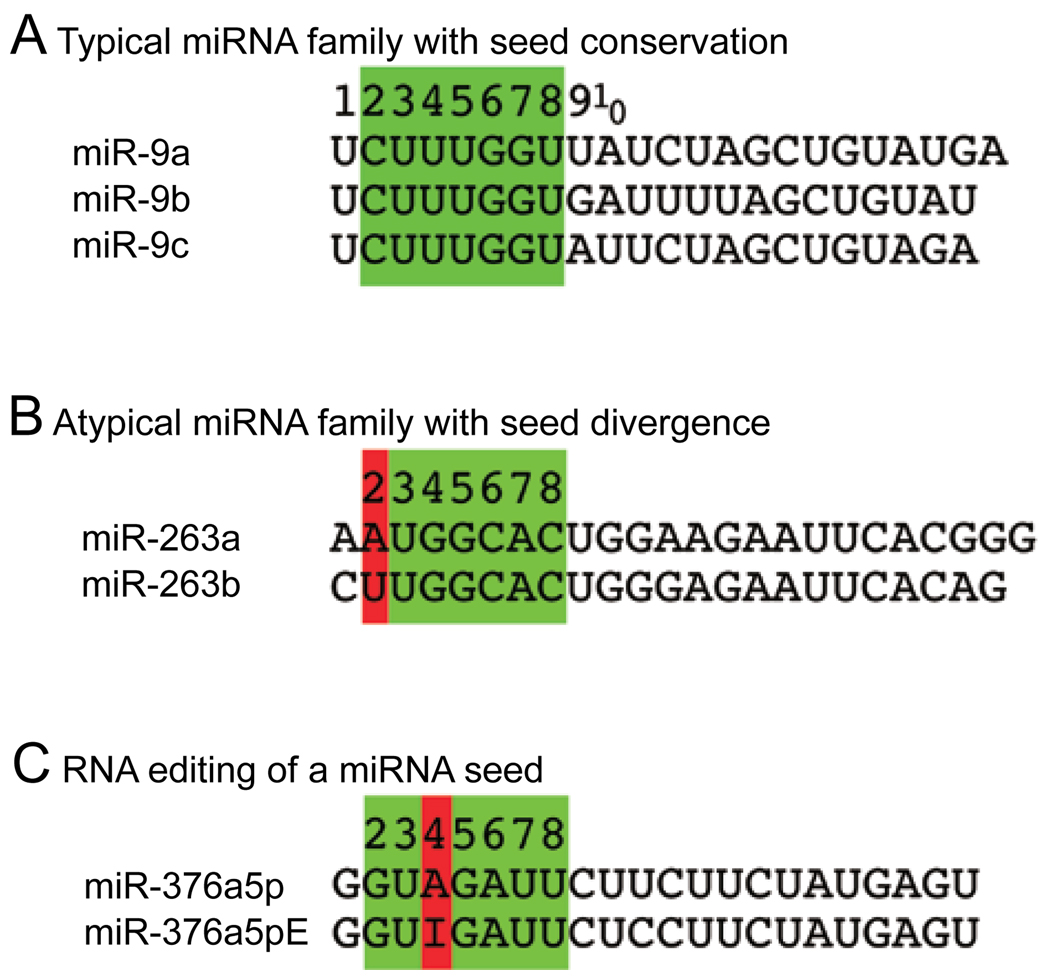
Although changes might occur along the length of the miRNA, changes in the seed region will have particular impact. Inspection of miRNA families reveals a predominant trend in which duplicated miRNA genes are most similar in their seed regions (Figure 2A), perhaps reflecting that miRNA duplicates often shift their target spectra modestly via 3′ pairing [11, 42, 66]. However, there are also examples of related miRNA genes with distinct seed regions. For example, Drosophila miR-263a and miR-263b are highly related miRNAs that nevertheless have different seed regions (Figure 2B). As a consequence of this minor sequence difference, these two miRNAs are expected to have mostly non-overlapping target sets.
Figure 2.
Changes in miRNA sequence can diversify miRNA activity. (A) Typical families of miRNAs can be classified according to shared 7mer motifs located at positions 2–8, also known as the miRNA “seed” (green box). Shown are the three members of the Drosophila miR-9 family; note that their sequences are identical through position 8, but strongly diverge beginning with position 9. The members of this family are inferred to have at least some common targets because of their shared seed, although their target properties are likely somewhat distinct because of their divergent 3′ ends. (B) An atypical family of Drosophila miRNAs which differ in a seed position (red box). These miRNAs are inferred to have mostly non-overlapping target sets. (C) Example of a miRNA that is edited in human and mouse, miR-376-5p. Of several edited positions identified, the A-I conversion at position 4 is significant as it is inferred to redirect its targeting capacity.
However, it should be noted that any change along the length of the mature miRNA is likely to be of some functional impact. miRNAs that differ only in their 3′ regions can select different targets, especially in cases where 3′ compensatory target pairing is in operation. For example, different members of the Drosophila K box miRNA family with identical seed regions nonetheless exhibit distinct capacities to regulate the proapoptotic targets grim and sickle [42]. As another example, nematode let-7 and its related family members miR-48/miR-84/miR-241 have distinct capacity to regulate the lin-41 gene, which is a non-canonical, seed-mispaired target that requires 3′ compensatory pairing unique to let-7 [4, 67].
Post-transcriptional editing of miRNAs
A different strategy to change a miRNA sequence, and thus diversify its function, is through RNA editing. In this process, an adenosine deaminase acts selectively within dsRNA to convert adenosine (A) residues into inosines (I) [68]. Since animal miRNA precursors are necessarily composed of dsRNA, it is conceivable that some miRNAs are edited so as to affect their processing and/or their target spectra. Studies of miR-376 nicely demonstrated this principle (Figure 2C). Since a highly edited site is positioned in the middle of its “seed” region, the genomic and edited versions of miR-376a-5p are predicted to target entirely different sites. Indeed, functional tests demonstrated this to be the case [69]. Systematic analysis has revealed a number of other miRNA genes that are edited at various positions [30, 70]. In addition, editing might theoretically affect processing and/or loading of miRNAs, with subsequent effects on target regulation. For example, editing of the pre-mir-151 hairpin blocks its cleavage by Dicer [71].
Changes in Drosha or Dicer processing
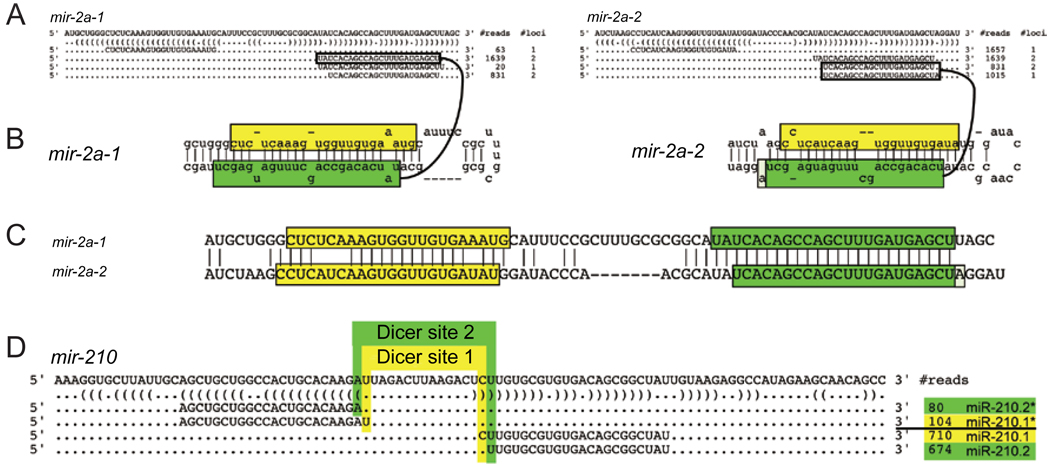
A third mechanism that can change the sequence of mature miRNAs involves shifting their processing to create new small RNAs with different 5′ ends. As mentioned, since the 5′ end of a mature miRNA sets the register for target site recognition, even a single nucleotide shift in its 5′ end will yield a radical alteration in target selection. Some compelling examples of this mechanism emerged following careful re-inspection of long-known miRNAs. For example, “K box” miRNAs constitute the largest family of miRNAs in Drosophila [10, 72]. Because many K box loci have been duplicated, the genomic mapping of several K box miRNAs is ambiguous. In particular, it was earlier presumed that several genes in the miR-2/miR-13 K box subfamily produce identical miRNAs [15, 73]. For example, it was originally thought that mir-2a-1 and mir-2a-2 (Figure 3A) encode the same product, UAUCACAG CCAGCUUUGAUGAGC.
Figure 3.
Changes in Drosha or Dicer processing can diversify miRNA activity. (A) mir-2a-1 and mir-2a-2 share 27 consecutive nucleotides along their miRNA-producing arms; therefore many sequences can be mapped to either genomic locus (# loci=2). However, their unique miRNA* sequences (# loci=1) allow their respective miRNA strands to be deconvolved based on miRNA/miRNA* duplexes with ~2-nt 3′ overhangs (B). This reveals the 5′ ends of miR-2a-1 and miR-2a-2 to be shifted by 2 nt with respect to each other (C). (D) An example of a rare miRNA locus that appears to be subject to alternative Dicer processing, yielding equal numbers of the distinct miRNAs miR-210.1 and miR-210.2. Note that the mir-2 and mir-210 cloning data depict the most abundant isoforms recovered from large-scale sequencing data [28]; less abundant reads mapped to these loci are not shown.
On the contrary, close inspection of larger miRNA sequence datasets, which now included substantial numbers of miRNA* species and even cloned miRNA hairpin loops, permitted the unambiguous assignment of some miRNAs with more than one genomic match to specific miRNA hairpins [28]. This revealed that certain seemingly identical miRNA loci actually generate miRNAs with completely distinct seeds, due to alternative Drosha/Dicer processing. For example, with mir-2a-1 and mir-2a-2, their unique “star” strand sequences allowed their corresponding mature miRNAs to be inferred from small RNA duplexes with 3′ overhangs (Figure 3A–3C). On this basis, one can see that miR-2a-1 and miR-2a-2 are actually distinct miRNAs with a 2-nt offset in their 5′ ends and seeds.
In the case of the miR-2a genes, it seems that a shift in the Drosha cleavage at the hairpin base probably underlies the change in the Dicer cleavage register at the terminal loop side. However, these events can be unlinked since miRNA precursors generated from at least some non-duplicated loci in Drosophila appear to be subject to alternative Dicer processing. A particularly compelling example is miR-210 (Figure 3D), for which approximately equal numbers of miRNAs with different 5′ ends are generated from a single precursor hairpin [28]. Thus, alternate miRNA processing can apparently generate functional diversity without changes to the gene sequence itself.
Changes in spatial or temporal expression pattern
It is popularly presumed that miRNA genes that produce similar or even identical miRNAs must necessarily have overlapping functions. However, this need not be the case if the miRNA genes are deployed in different places or times. Indeed, the acquisition of distinct expression domains is a common strategy for the functional diversification of duplicated protein-encoding genes, and this seems to occur frequently with miRNA genes as well.
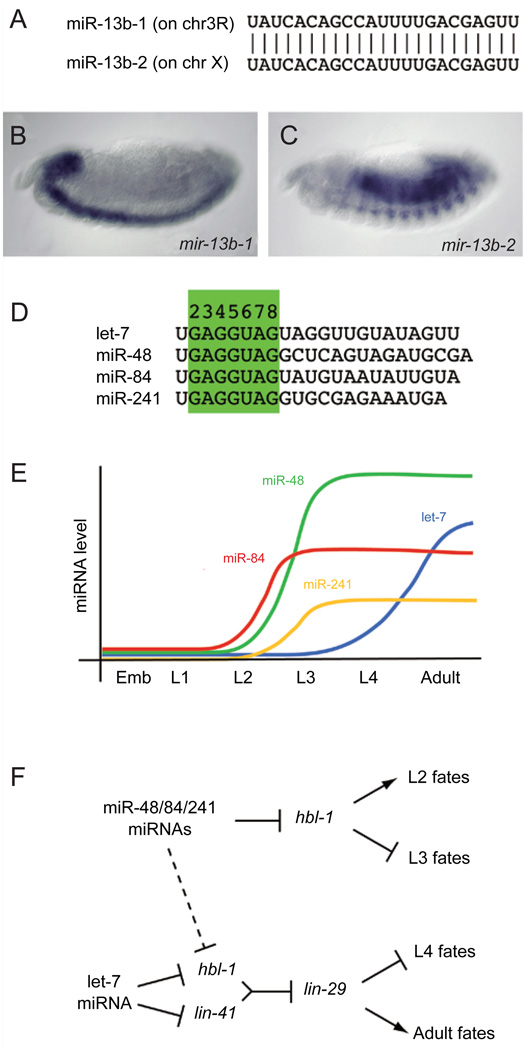
The Drosophila K box family provides a nice illustration of this principle. There are eight genes located in four genomic clusters that comprise the miR-2/miR-13 subfamily [16]. The embryonic expression patterns of three of these loci were determined, and all found to be distinct [74]. In fact, miR-13b-1 and miR-13b-2, which otherwise produce the same mature miRNA (Figure 4A), have nearly non-overlapping expression in the central nervous system and muscles/gut, respectively (Figure 4B and 4C). Therefore, it is undoubtedly the case that the diversification of the K box family has been accomplished, at least in part, by deploying them in distinct spatial domains.
Figure 4.
Changes in spatial or temporal expression can diversify miRNA activity. (A) miR-13b-1 and miR-13b-2 are identical miRNAs produced from loci on different chromosomes. Their non-redundant activity is evidenced by the distinct expression of their primary transcripts in the central nervous system (B) and the gut and musculature (C). (D) The let-7 sisters comprise related miRNAs with distinct temporal expression. (E) The levels of miR-48/miR-84/ miR-241 peak during the transition from L2 to L3, while the level of let-7 peaks during the transition from L4 to the adult. (F) Genetic hierarchy of the control of L2-L3 transition by miR-48/miR-84/miR-241 and control of the L4-adult transition by let-7. Note that let-7 is a unique regulator of lin-41 at this developmental stage since it requires 3′ compensatory pairing that is specific to let-7; mir-48/miR-84/miR-241 may repress hbl-1 at both stages.
A similar concept applies to members of the nematode let-7 family, not at the spatial level but instead at the temporal level. Members of the let-7 family have similar seeds and thus at least some overlapping target specificity (Figure 4D). However, they are deployed at different times in development [4, 75], and their particular temporal expression underlies their essential roles in specifying stage-specific developmental events (Figure 4E and 4F). Genetic analyses revealed that three related let-7 “sisters” termed miR-48, miR-84, and miR-241 are activated between the second (L2) and third (L3) larval stages, and they promote L3 development by repressing hbl-1. Slightly later in development, the let-7 miRNA is activated between the L4 and adult stages, and it promotes adult development by repressing lin-41 and hbl-1 (Figure 4F). Inspection of large-scale small RNA cloning efforts reveals a variety of other miRNA families in which individual members are expressed at different times and/or places in invertebrate or mammalian organisms [28, 30].
Acquistion of miRNA* functionality
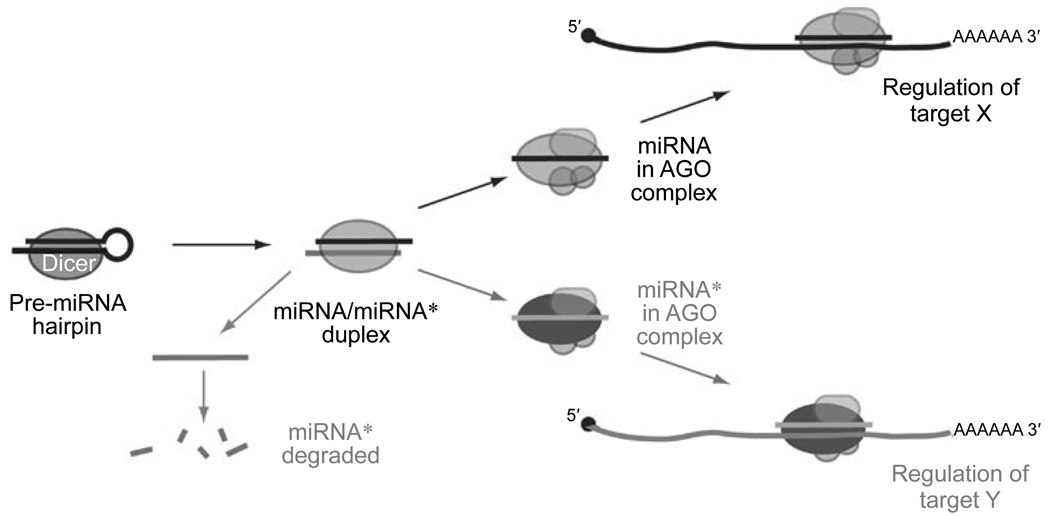
miRNA biogenesis proceeds via an obligate small RNA duplex step (Figure 1), and the small RNAs are necessarily produced initially at a 1:1 ratio by transcription. However, the mature miRNA/miRNA* ratio is asymmetric at steady-state, sometimes at a discrepancy of >10 000:1 [27, 28]. The asymmetry of miRNA strand selection has been taken as evidence that miRNA* species are merely carrier strands that are needed only to promote accurate processing of their partner miRNA strands. In support of this, thermodynamic rules can rationalize which miRNA strand is favored for selection into a mature regulatory complex [76, 77]. Nevertheless, the notion that miRNA* species are “junk” strands has come about mostly through lack of study.
Recently, this premise was put to experimental test in the Drosophila system [66]. In the null hypothesis, if miRNA* strands were truly irrelevant as regulatory RNAs, they would not exhibit any particular sequence constraint, would not associate with AGO proteins, would not be capable of repressing target transcripts, and would not exhibit preferential conservation of seed-matching sites. In fact, every single one of these premises turns out to be false: miRNA* species are often highly conserved small RNAs that are actively sorted to AGO proteins to regulate conserved targets (Figure 5). Although it is true that miRNA* species mediate much smaller regulatory networks than their sister miRNA species, ~40% of all Drosophila miRNA* species are under stringent sequence selection, implying their function as regulatory RNAs, and about half of these have confident evidence for the selective conservation of complementary seed sites in 3′ UTRs [66].
Figure 5.
Acquisition of miRNA* functionality can diversify miRNA activity. While bulk miRNA* species are preferentially degraded, a substantial fraction of miRNA* species are actively sorted into AGO complexes and are used to repress endogenous targets. The schematic was adapted from a published model [66].
The implications of these findings are substantial. Every pre-miRNA hairpin has the inherent property of being able to generate two distinct regulatory RNAs. Although one of these is always dominant, in that it accumulates to a higher level and typically has more conserved seed matches, pre-miRNA hairpins are nevertheless evolutionarily selected so as to produce a specific amount of functional miRNA* species. This means that individual miRNA genes may constantly be in a state of neofunctionalization, in which the two strands essentially compete for incorporation into a regulatory complex.
One might imagine that a trend to switch the effective miRNA strand might be favored amongst duplicated miRNA genes, in which a gene copy is now freed from functional constraint to choose a particular arm for regulatory purposes. Indeed, careful analysis of miRNA cloning patterns revealed members of at least four different miRNA families that have switched their dominant arm, and many more in which the miRNA/miRNA* strand bias is widely divergent [66, 78].
Antisense miRNA transcription and processing
A perfect genomic inverted repeat can adopt a hairpin structure that is identical on sense and antisense strands. On the other hand, as animal pre-miRNAs are imperfect hairpins containing bulged nucleotides, internal loops, and non-canonical G:U basepairs, miRNA hairpins are generally dissimilar as sense vs antisense transcripts [16]. Typically, one of the two strands adopts a hairpin with significantly better hairpin characteristics, so that computational methods can often accurately predict the transcribed strand of a candidate miRNA locus. However, in many cases, the sense and antisense hairpins are insufficiently different to permit a confident strand prediction. In fact, some confirmed miRNA hairpins have less duplex structure than their antisense counterparts. However, as early small RNA cloning efforts yielded miRNAs only from a single strand of a given genomic hairpin, it was widely assumed that antisense strands of miRNA hairpins were not relevant.
This assumption was challenged by recent studies of the Drosophila Hox clusters, which are physically broken into the Bithorax Complex (BX-C) and the Antennapedia Complex (ANTP-C) (Figure 6). Canonical Hox genes encode homeodomain transcription factors that are central to the appropriate assignment of segmental identities along the anterior-posterior axis of all animals [79]. Misregulation of canonical Hox gene activity results in striking transformations of body segments; thus, it is essential to control their activity with great precision.
Figure 6.
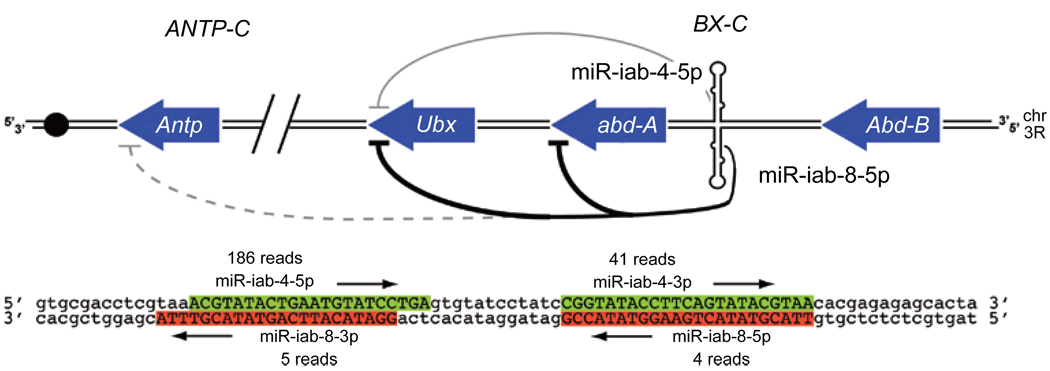
Antisense miRNA transcription and processing can diversify miRNA activity. The mir-iab-4 hairpin in the Drosophila Bithorax Complex (BX-C) is transcribed on its antisense strand as the mir-iab-8 hairpin. Thus, four different small RNAs are produced from a single hairpin locus in embryos. The 5p miRNAs of mir-iab-4 and mir-iab-8 directly regulate other Hox genes; stronger regulatory interactions are depicted with darker lines. The schematic was adapted from a published model [86].
Earlier cloning efforts showed that miRNAs are conserved components of animal Hox clusters [15, 73, 80, 81], and the BX-C locus mir-iab-4 regulates the canonical BX-C Hox gene Ultrabithorax [82]. Curiously, earlier in situ hybridization studies showed that the sense and antisense strands of the mir-iab-4 locus are transcriptionally active in distinct spatial domains of the developing embryo [83, 84]. In fact, transcription and processing of the antisense strand was recently shown to yield novel miRNAs from the mir-iab-8 hairpin (Figure 6) [85–87]. Interestingly, the mature miRNAs of the antisense hairpin are functionally distinct from their sense counterparts, and have both overlapping and distinct roles in BX-C and potentially ANTP-C gene regulation [85–87].
Antisense miRNAs are not an oddity of the Drosophila Hox complex. The recent availability of large small RNA catalogs permitted additional antisense miRNAs to be found in flies and mammals [85, 86]. This demonstrates antisense processing as a general principle that can diversify miRNA function in animals. When combined with miRNA* function, one can imagine that four distinct regulatory RNAs might be produced from a single locus. Indeed, tests in both cultured cells and transgenic animals showed that both miRNA and miRNA* of sense and anti-sense mir-iab-4/mir-iab-8 are functional inhibitory RNAs [66, 85, 86].
It remains the case that the quantity of antisense miRNAs in any organism is quite low. Nevertheless, the study of mir-iab-4/mir-iab-8 clearly shows that this principle has been utilized by highly conserved regulatory circuits. The general phenomenon of broad euchromatic transcription, coupled with the observation that a substantial fraction of miRNA hairpins may plausibly be processed as antisense hairpins, therefore raises this as an economical method to generate new miRNA genes via pre-existing miRNA hairpins.
De novo generation of miRNA hairpins
In addition to EST evidence, global analyses of genomic transcription using tiling microarrays have yielded the picture that a majority of the animal euchromatin is converted into RNA, usually in a temporally and/or defined manner [88, 89]. As with the concept of broad antisense transcription [90], the biological significance of broad transcription of the genome is controversial at present. However, one can easily imagine that this might have tangible consequences for the birth of miRNA genes.
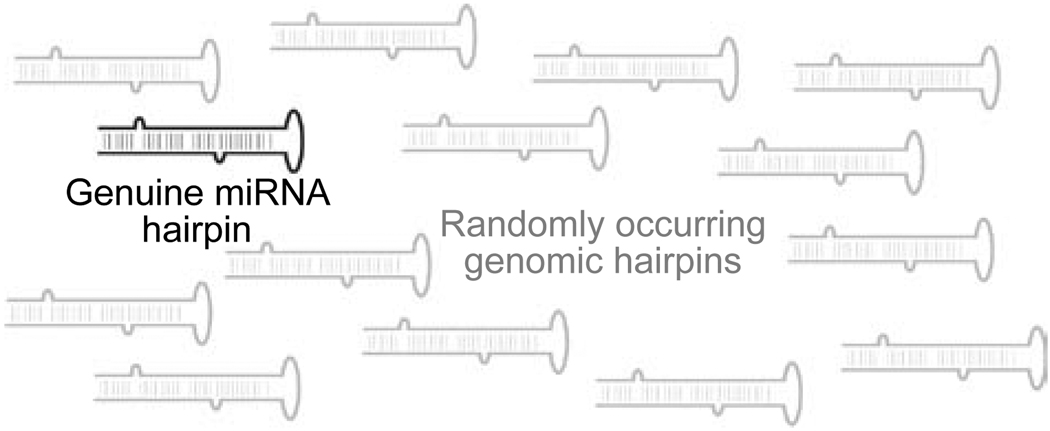
Animal genomes encode very large numbers of candidate hairpins that are plausible as miRNA precursors. Analysis of Drosophilid genomes yields 100 000s of candidate hairpins [16, 91], while mammalian genomes have 1000 000s of such hairpins [20]. While it cannot be formally excluded that there are a million human miRNAs, extensive cloning evidence strongly suggests that the real number is perhaps three orders of magnitude smaller. Because of the large number of raw hairpins, all effective computational methods for miRNA genefinding in animal genomes rely upon the usage of evolutionary conservation as an essential filter to distinguish between bulk genomic hairpins and ones with possible regulatory activity. Even the existence of a conserved hairpin is insufficient to categorize miRNA loci with confidence, since at least some conserved hairpins are fortuitous or represent conserved structures that are not processed into ~21–24 nt regulatory RNAs. Conversely, the mere finding of a cloned small RNA that maps to an arm of a genomic hairpin [92] is today no longer sufficient to warrant classification as a miRNA gene. Large-scale cloning efforts yield a surplus of singleton hairpin-matching reads, for which one cannot confidently infer that the small RNA was produced by Drosha/Dicer cleavage (as opposed to representing a degradation RNA fragment that fortuitously matches a genomic hairpin).
Although this sea of hairpins might be seen as an annoyance for computational miRNA genefinding efforts, they conversely have very compelling evolutionary implications. Such incidental hairpins may be routed at low levels through the miRNA processing pathway to generate small regulatory RNAs lacking beneficial targets (Figure 7). Likely, if they do find targets, their regulation would be detrimental. On the other hand, their regulatory capacity would serve as a general proving ground for natural selection, by which “useful” target interactions would be stabilized in the genome [45, 93, 94].
Figure 7.
Typical genomes encode many miRNA-like hairpins but only a limited set of genuine miRNA hairpins that are specifically processed by the canonical miRNA or mirtron machinery (Figure 1). The large surplus of genomic hairpins that occur frequently throughout the genome may serve as a breeding ground of nascent miRNAs that may accidentally enter the miRNA processing pathway at some low frequency. If they provide a useful function to the organism, these may eventually be stabilized as genuine miRNA genes.
In this scenario, newly born miRNA genes might be part of a general mechanism by which speciation might occur. It is well-accepted that speciation can occur by changes in the cis-regulatory control of existing genes. However, the idea that newly evolved genes might play a substantial role has been less studied to date [95–97]. Although these types of evolutionary questions provide a great challenge for informative experimental tests, small RNA cloning efforts have revealed miRNA genes in every animal and plant subclade that are highly species-specific [27–29]. Cloning of primate brain miRNAs revealed a rich assortment of miRNAs specific to individual primates [98], leading to the proposition that human-specific brain miRNAs might theoretically impart human-specific traits. In addition, it has been observed that the mirtron subclass of miRNA genes evolves more rapidly than canonical miRNAs [54–56]. Therefore, the potential contribution of newly evolved mirtrons to speciation may be greater than that of canonical miRNAs.
Transposon-assisted miRNA gene birth
In addition to the de novo emergence of miRNA hairpins from randomly occurring hairpins, the birth of miRNA genes may also be assisted by pre-existing genetic elements. A substantial proportion of animal genomes is composed of transposons or their defunct remnants. In fact, the human genome may in some sense be considered to be composed of more of these parasitic genetic elements than actual “human” DNA. This of course has substantial consequences for endogenous gene regulation and gene evolution [99–102]. The transposition of selfish genetic elements can mutate genes by directly inserting into them, or by disrupting cis-regulatory elements. Proximity to transcriptional regulatory elements carried on transposons can alter the expression of host genes. Transposons can even become incorporated into host genes, contributing exons and protein-coding potential.
Transposons often carry terminal inverted repeats, but even ones with direct terminal repeats can insert near or into each other, resulting in inverted repeat gene arrangements between once distinct elements. Transcription across such elements might be a substantial source of hairpins that comprise “accidental” miRNA substrates. Their possible regulatory activities might favor their mutation and loss if deleterious, but could also be subject to positive selection if beneficial to the organism. For example, a number of mammalian miRNAs derive from LINE transposable elements or other repetitive elements [103]. Another perhaps more direct route might be through the fortuitous entry of miniature inverted-repeat transposable element (MITE) transcripts, which can be in the range of typical pre-miRNA hairpins, through the miRNA biogenesis pathway [104]. A recent systematic analysis revealed at least 55 human miRNAs derived from LINE elements, SINE elements, or MITEs, with some 85 additional novel TE-derived miRNA candidates predicted [105].
Conclusions
Although a major conclusion of evolutionary-developmental biology has been that fundamental molecular circuits remain the same over the eons, even the most conserved of pathways is subject to change, adaption, and functional diversification in different settings. In this perspective, we have examined some of the many ways that miRNAs have been able to diversify their functions. We argue that their extraordinary flexibility and versatility explain their success in the genomic landscape.
First, miRNAs are easily able to acquire new functions-even a single nucleotide change in their seed region will completely alter their target spectra. Conversely, it is relatively trivial for a given target gene to be captured by a miRNA, since virtually every gene is a point mutation away from acquiring a high-affinity binding site for one or more miRNAs. Second, miRNA genes are frequently duplicated, creating abundant possibilities for their subfunctionalization or neofunctionalization. Third, mechanisms for neofunctionalization can apply even to single miRNAs. We have described examples in which alternate Drosha/Dicer cleavages can diversify the output of a major miRNA species. In addition, inherent miRNA* functionality frequently allows a second functional species to emerge from any given miRNA hairpin. If that were not enough, antisense transcription and processing of miRNA hairpins is yet another genuine mechanism to diversity a single miRNA locus. Finally, miRNAs are easily born de novo. Unlike protein-encoding genes, they do not need to acquire translational initiation context, nor evolve an open reading frame of significant length, nor produce a polypeptide that should fortuitously resist unfolding and degradation. Truly, miRNAs are an evolutionary wonder, constantly capable of reinventing themselves, and even of making something out of thin air.
Acknowledgments
ECL was supported by grants from the Burroughs Well-come Foundation, the V Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research, and the NIH, USA (R01-GM083300 and U01-HG004261).
References
- 1.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack F, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 6.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 7.Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- 8.Lai EC, Posakony JW. Regulation of Drosophila neurogenesis by RNA:RNA duplexes? Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- 9.Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 14.Lau N, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 16.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42.41–R42.20. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 18.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 19.Lim LP, Lau NC, Weinstein EG, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 21.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miR-Base: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruby JG, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 28.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahlgren N, Howell MD, Kasschau KD, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression Atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen P, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 32.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 35.Grun D, Wang Y-L, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comp Biol. 2005;1:0051–0063. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 37.Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 38.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 39.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 41.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′ UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 42.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 44.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′ UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 45.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 46.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 47.Willmann MR, Poethig RS. Conservation and evolution of miRNA regulatory programs in plant development. Curr Opin Plant Biol. 2007;10:503–511. doi: 10.1016/j.pbi.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li A, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 49.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YK, Kim VN. Processing of intronic microRNAs. Embo J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 54.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 58.Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- 59.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 60.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 61.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacCarthy T, Bergman A. The limits of subfunctionalization. BMC Evol Biol. 2007;7:213. doi: 10.1186/1471-2148-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berezikov E, van Tetering G, Verheul M, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bass BL. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb Symp Quant Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blow MJ, Grocock RJ, van Dongen S, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aravin A, Lagos-Quintana M, Yalcin A, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 74.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abbott AL, Alvarez-Saavedra E, Miska EA, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 77.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 78.Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 80.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 82.Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cumberledge S, Zaratzian A, Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc Natl Acad Sci USA. 1990;87:3259–3263. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stark A, Bushati N, Jan CH, et al. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tyler DM, Okamura K, Chung WJ, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 89.Manak JR, Dike S, Sementchenko V, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- 90.Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 91.Lu J, Shen Y, Wu Q, et al. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 92.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu J, Fu Y, Kumar S, et al. Adaptive evolution of newly-emerged microRNA genes in Drosophila. Mol Biol Evol. 2008;25:929–938. doi: 10.1093/molbev/msn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 95.Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 2007;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. PLoS Biol. 2007;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- 98.Berezikov E, Thuemmler F, van Laake LW, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 99.Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- 100.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 101.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 102.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 103.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 104.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]