Abstract
Mouse mammary epithelial cells (HC-11) and mammary tissues were analyzed for developmental changes in circadian clock, cellular proliferation and differentiation marker genes. Expression of the clock genes, Per1 and Bmal1, were elevated in differentiated HC-11 cells whereas Per2 mRNA levels were higher in undifferentiated cells. This differentiation-dependent profile of clock gene expression was consistent with that observed in mouse mammary glands as Per1 and Bmal1 mRNA levels were elevated in late pregnant and lactating mammary tissues, while Per2 expression was higher in proliferating virgin and early pregnant glands. In both HC-11 cells and mammary glands, elevated Per2 expression was positively correlated with c-Myc and Cyclin D1 mRNA levels while Per1 and Bmal1 expression changed in conjunction with ß-casein mRNA levels. Interestingly, developmental stage had differential effects on rhythms of clock gene expression in the mammary gland. These data suggest that circadian clock genes may play a role in mouse mammary gland development and differentiation.
Keywords: circadian rhythm, oscillation, Period1, Period2, Bmal1, Cryptochrome1, Cryptochrome2, mammary gland, development, differentiation, cell cycle
INTRODUCTION
Biological clocks play a key role in adaptation to daily and seasonal environmental changes. The circadian clock regulates daily oscillations in metabolism, hormone and neurotransmitter levels and a variety of behaviors, including activity, drinking and feeding (Reppert and Weaver, 2001). In mammals, these 24-hour rhythms are controlled centrally by the suprachiasmatic nucleus (SCN), a small region in the anterior hypothalamus that functions as the master circadian clock and receives light input from the retina.
The basic molecular clockworks underlying the generation of circadian rhythms consist of interacting positive and negative transcriptional/translational feedback loops. The dimerization of two basic helix-loop-helix Per-Arnt-Sim (bHLH-PAS) family members, circadian locomotor output cycles kaput (Clock) and brain and muscle ARNT-like protein (Bmal1), drives the rhythmic transcription of three Period genes (Per1, Per2 and Per3) and two Cryptochrome genes (Cry1 and Cry2) at the beginning of each day. By mid-day, CRY and PER proteins begin to accumulate and enter the nucleus and then interact with CLOCK and/or BMAL1 to inhibit their own transcription and enhance Bma1l transactivation. Thus, increased CLOCK:BMAL1 heterodimers activate Per and Cry gene expression and the clock is reset at the beginning of the next day (Reppert and Weaver, 2001).
The expression of these clock genes and their rhythmic regulation are not unique to the SCN, but instead are widely distributed in many cells and tissues. For example, the period genes are expressed and rhythmically regulated in a variety of peripheral tissues including liver, lung and skeletal muscle (Zylka et al., 1998; Yamazaki et al., 2000). These peripheral oscillations in the expression of core clock elements are thought to provide for local coordination of cell populations in specific tissues and the circadian control of “downstream” genes including Cyclin D1 and c-Myc, which regulate cell cycle progression (Nagoshi et al., 2004). It is interesting that the cyclic expression of clock genes does not occur within some tissues such as testis and thymus (Alvarez et al., 2003; Morse et al., 2003). Thus, the local coordination of time between individual cells may be suspended in differentiating tissues or perhaps clock genes may also have non-circadian functions in developing cells.
The mammary gland is a unique tissue because its development mainly occurs post-embryonically during puberty. In addition, the mammary gland undergoes repeated cycles of development that sequentially consist of proliferation, differentiation and regression coinciding respectively with pregnancy, lactation and involution. Numerous hormones and growth factors exert positive or negative effects to regulate these developmental transitions. These regulatory pathways include the hormones estrogen, progesterone and prolactin as well as locally produced signaling molecules including members of the transforming growth factor beta (TGF-β), Wnt, epidermal growth factor (EGF), fibroblast growth factor (FGF) and matrix metalloprotease (MMP) families (Muller and Neville, 2001). Studies have shown that many of these signals governing mammary gland development and differentiation are regulated in a circadian fashion. Ultimately, these orchestrated signals activate critical pathways, which control cell growth, migration, differentiation and apoptosis, in a time-dependent manner to ensure proper branching morphogenesis and regulation of milk production.
The expression and functional implications of clock genes in mammary gland development have not been explored. Recent discoveries have revealed that the circadian clock and disruption of its core molecular components affect fundamental aspects of cellular function including cell cycle progression, apoptosis and differentiation (Matsuo et al., 2003; Xiao et al., 2003; Granda et al., 2005). Many of the genes identified in these studies as targets for circadian clock regulation are known mediators of normal mammary gland development. In addition, animal studies indicate that DNA synthesis and mitosis in the terminal end buds of developing adolescent mouse mammary glands occurs in a rhythmic fashion with a peak in DNA synthesis occurring in the middle of the dark phase (Berger and Daniel, 1982). Therefore, the purpose of these studies was to determine whether developmental changes in clock gene expression occur within mammary epithelial-derived cell lines and mouse mammary tissues. Using quantitative PCR (qt-PCR), the expression of clock genes was compared to genes with distinctive developmental profiles in undifferentiated and differentiated mouse epithelial cells (HC-11) in vitro and in mouse mammary glands at different stages of reproductive development.
RESULTS AND DISCUSSION
Expression of circadian clock genes and differentiation markers in mouse mammary epithelial-derived cells
Because tissues composed of differentiating cells show developmental changes in the regulation of molecular components of the circadian clockworks (Alvarez et al., 2003), the present studies were conducted to determine whether development affects clock gene expression in the mammary gland, which is capable of repeated cycles of differentiation. We first examined the influence of development on clock gene expression in the HC-11 mammary epithelial cell line. HC-11 cells are an immortalized, but non-transformed, line isolated from the mid-pregnant mouse mammary gland (Ball et al., 1988). Similar to the mammary gland in vivo, HC-11 cells stop dividing and differentiate into epithelial cells that show high expression of the milk protein ß-casein following stimulation with prolactin and hydrocortisone.
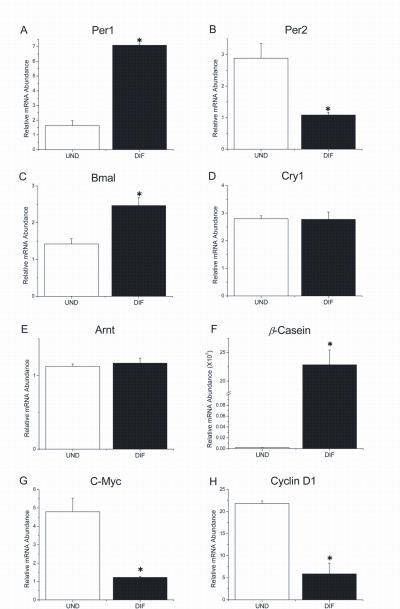
Thus, HC-11 cells differentiated by exposure to these lactogenic hormones were examined in relation to undifferentiated cells for differences in clock gene expression by qt-PCR. Interestingly, differentiated HC-11 cells were found to express significantly higher levels of Per1 but lower levels of Per2 (p<0.05) mRNA than undifferentiated cells (Figs. 1A and 1B). Levels of Bmal1 expression in differentiated HC-11 cells were also significantly elevated (p<0.05) relative to those found in undifferentiated cells (Fig. 1C). These differentiation-induced increases in Per1 and Bmal1 mRNA levels were 4.7-fold and 1.7-fold, respectively whereas Per2 expression was decreased by 3-fold in differentiated cells. However, undifferentiated and differentiated HC-11 cells exhibited no significant differences in Cry1 expression. Further analysis of the temporal profiles of clock gene expression revealed no evidence of circadian or regular rhythmic variations in serum-induced HC-11 cells that were either undifferentiated or differentiated by exposure to lactogenic hormones (data not shown). To examine the effect of differentiation on expression of a non-circadian clock member of the bHLH-PAS family, levels of Arnt mRNA were also analyzed in undifferentiated and differentiated HC-11 cells. Although abundant Arnt expression was observed in HC-11 cells, relative mRNA levels were not altered by differentiation (Fig. 1E).
Figure 1. Expression of circadian clock, cellular proliferation and mammary differentiation marker genes in undifferentiated and differentiated mouse mammary epithelial-derived HC-11 cells.
Per1 (A), Per2 (B), Bmal1 (C), Cry1 (D), Arnt (E), ß-casein (F), c-Myc (G) and Cyclin D1 (H) mRNA levels were analyzed in undifferentiated (UND) and differentiated (DIF) HC-11 cells. Total RNA was isolated from three plates per treatment and analyzed by quantitative real-time RT-PCR. Data are expressed as the mean (±SEM) for each differentiation state. Asterisks indicate significant differences in gene expression levels between UND and DIF HC-11 cells.
To provide an index of developmental state, expression of mammary differentiation and proliferation markers was also analyzed in the same HC-11 cultures. Expression of ß-casein, a major milk protein and widely-used marker of mammary epithelial cell differentiation, was significantly elevated (p<0.05) in differentiated HC-11 cells (Fig. 1F). In contrast, expression of two principal markers of mammary epithelial cell proliferation, c-Myc and Cyclin D1, were significantly higher (p<0.05) in undifferentiated HC-11 cells (Figs. 1G and 1H). Thus, growth and proliferation in undifferentiated HC-11 cells were associated with increased c-Myc and Cyclin D1 expression, which coincided with high levels of Per2 mRNA. Conversely, differentiation of HC-11 cells, which is delineated by high levels of ß-casein expression, occurred in conjunction with high levels of Per1 and Bmal1 mRNA. Collectively, these findings indicate that the clock genes, Per1, Per2 and Bmal1, in the HC-11 mammary epithelial cell line are differentially expressed during development in association with expression of specific differentiation markers.
Expression of circadian clock genes in the developing mouse mammary gland
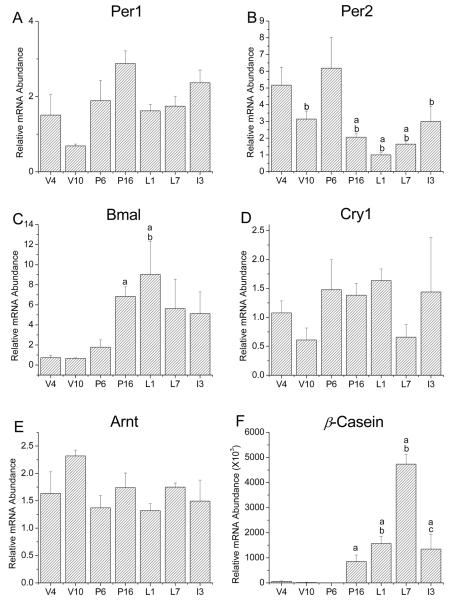
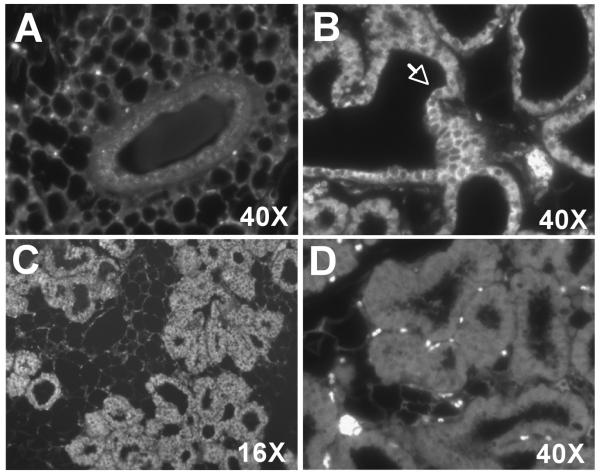
Because our in vitro studies suggest that clock gene expression may be developmentally regulated in mouse mammary epithelial cells, we extended this analysis to determine whether clock gene mRNA levels are similarly altered during mammary gland development in vivo. Using qt-PCR, clock gene expression was analyzed in mouse mammary tissues collected between Zeitgeber Time (ZT)10-12 at seven different stages of development. Our results indicate that clock gene expression in the mammary gland is developmentally regulated (Fig.2). Analysis with one-way ANOVA revealed a main effect of developmental stage on the levels of Per2 and Bmal1 [FPer2(6, 16)=4.551, FBaml1(6,16)=3.721, p<0.05], but not Per1 and Cry1, mRNA in the mammary gland. Of these developmentally-regulated clock genes, Per2 and Bmal1 exhibited the most distinctive patterns of expression and the largest differences between stages. Per2 expression in the mammary gland was relatively high during virgin stages and early pregnancy, decreased during the latter stage of pregnancy, remained low throughout lactation and increased slightly during involution. The developmental pattern of Bmal1 expression in the mammary gland was inversely related to that for Per2; mammary content of Bmal1 mRNA was low in virgin and P6 animals, increased during late pregnancy and remained high throughout lactation and involution. Although morphological changes occur throughout virgin, pregnancy, lactation and involution stages of mammary gland development, we focused on critical comparisons of clock gene expression at V10 and L1 because these stages serve to distinguish fully mature, but undifferentiated, mammary gland from differentiated tissues that resemble the developmental status of HC-11 cells stimulated with lactogenic hormones. Consistent with our in vitro observations, Per1 expression in the mammary gland at L1 was approximately 2-fold higher than that at V10. To determine if this increase in Per1 mRNA expression is due to expansion of the epithelial compartment during pregnancy and lactation, the spatial expression of PER1 protein in the mouse mammary glands from virgin and lactating mice was analyzed using indirect immunofluorescence methods. Similar to qt-PCR results, PER1 protein was not detected in the mammary glands of virgin mice, but was robustly expressed in ductal epithelial cells during lactation (Fig. 3). These results confirm that mammary epithelial cell differentiation is associated with increased Per1 expression. In contrast, Per2 expression was highest in early adolescent (V4) and pregnant (P6) glands, which are periods associated with rapid expansion of the mammary epithelial compartment through active proliferation (Fig. 2B). Post-hoc analysis showed that Per2 mRNA levels in the mammary gland were significantly higher (p<0.05) at V4 than at P16, L1 and L7. In addition, Per2 expression was also significantly greater (p<0.05) in P6 than in V10, L1, L7 and I3 glands. Changes in Bmal1 expression also occurred during the mammary gland development (Fig. 2C) such that mRNA levels were significantly higher (p<0.05) at P16 and L1 than those observed in V4 mice. Although Cry1 showed a trend for increased expression during pregnancy and early lactation (P6, P16 and L1), no significant differences in Cry1 mRNA levels were detected over the course of mammary gland development (Fig. 2D). Similar to in vitro observations in HC-11 cells, the non-clock bHLH-PAS gene Arnt exhibited no significant differences in expression between developmental stages (Fig. 2E). In contrast, expression of the milk protein and mammary differentiation marker ß-casein was significantly increased [F(6, 14)=22.16, p<0.05] in late pregnancy and throughout lactation relative to levels found in virgin and early pregnancy glands (Fig. 1F).
Figure 2. Developmental regulation of circadian clock gene expression in the mouse mammary gland.
Per1 (A), Per2 (B), Bmal1 (C), Cry1 (D), Arnt (E) and ß-casein (F) mRNA levels were analyzed in mouse mammary glands during different stages of development. Total RNA was isolated between ZT10-12 from mammary tissue obtained from at least three individual mice per time point and analyzed by quantitative real-time RT-PCR. Time points were chosen to represent significant developmental stages including early (4 week old virgin, V4) and late (10 week old virgin, V10) adolescence, early (pregnancy day 6, P6) and late (pregnancy day 16, P16) pregnancy, early (lactation day 1, L1) and late (lactation day 7, L7) lactation and mid-involution (day 3 involution, I3). Mean values that were significantly different (p<0.05) from virgin week 4, pregnancy day 6 and lactation day 1 are denoted by a, b and c, respectively.
Figure 3. PER1 expression in differentiated mammary ductal epithelial cells.
Immunofluorescent localization of PER1 in 10 week virgin (A) and lactation day 1 mouse mammary glands (B, C). Staining is specific for ductal epithelial cells (see arrow). Negative control analysis of lactation day 1 mammary gland in which the primary antibody was omitted (D).
Expression of cell cycle regulation genes in the developing mouse mammary gland
Similar to the circadian clock, cell division is cyclically regulated involving precise transitions between component processes. Specific regulatory proteins gate transitions between resting state, DNA synthesis and mitosis. Control checkpoints at these transitions assure that progression into the cell cycle is timed and tightly regulated. Defects in these checkpoint proteins are responsible for uncontrolled proliferation that can lead to mutations, genomic instability and ultimately cancer. Several studies have shown that the central circadian clock regulates cell-cycle progression in numerous tissues including the intestine, oral epithelium, skin and bone marrow progenitor cells (Scheving et al., 1983; Buchi et al., 1991; Garcia et al., 2001; Reddy et al., 2005). Because Per2 has been implicated in the regulation of Cyclin D1 and c-Myc expression in the liver (Fu et al., 2002; Fu and Lee, 2003), we examined mammary gland expression of these two cell cycle genes and compared their developmental profiles to those observed for clock genes. c-Myc and Cyclin D1 expression were similarly altered during mammary gland development (Figs. 3A and 3B) such that mRNA levels for both genes were low in adult virgin mice (V10) and increase during early (P6) and late pregnancy (P16) when the epithelial cells proliferate in preparation for milk production. Decreases in both c-Myc and Cyclin D1 mRNA levels were observed during lactation and early involution. However, analysis with one-way ANOVA revealed a main effect of developmental stage on c-Myc expression [F (6,14)=9.414, p<0.05], but not Cyclin D1. Mammary gland levels of c-Myc mRNA during pregnancy (P6 and P16) were significantly greater (p<0.05) than those observed at V4 and during lactation and early involution. The lack of significant differences in Cyclin D1 expression among stages is probably related to the high degree of variation of Cyclin D1 mRNA levels in P6, P16 and L1 glands (Fig. 4B). Comparison of c-Myc and Per2 expression in the mammary gland during pregnancy is especially interesting. When mammary gland levels of c-Myc mRNA were relatively low on P6, peak expression of Per2 was observed whereas elevated c-Myc expression on P16 coincided with low Per2 mRNA abundance. This inverse relationship between c-Myc and Per2 expression during pregnancy may have implications for interactions between these genes in mammary gland development and carcinogenesis.
Figure 4. Expression of c-Myc and Cyclin D1 in the developing mouse mammary gland.
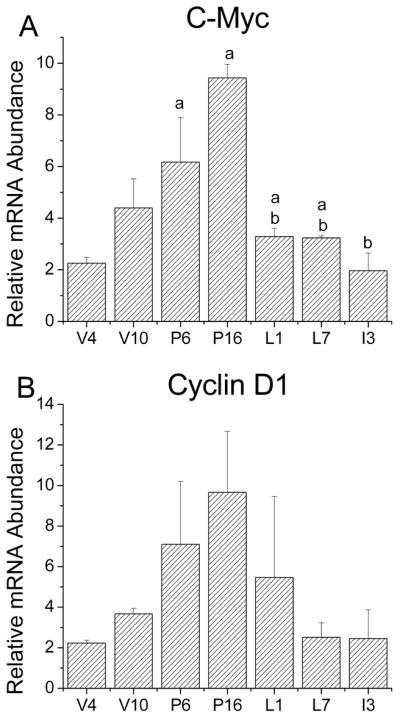
Quantitative real-time PCR analysis of c-Myc (A) and Cyclin D1 (B) expression in mouse mammary glands (n = 3-5) at virgin week 4 (V4) and 10 (V10), pregnancy day 6 (P6) and 16 (P16), lactation day 1 (L1) and 7 (L7), and involution day 3 (I3). Data are expressed as the mean (±SEM) at each developmental time point. Mean values that were significantly different (p<0.05) from virgin week 4 (V4) and pregnancy day 6 (P6) are denoted by a, and b, respectively.
Circadian rhythmicity of mammary clock gene expression is altered during differentiation
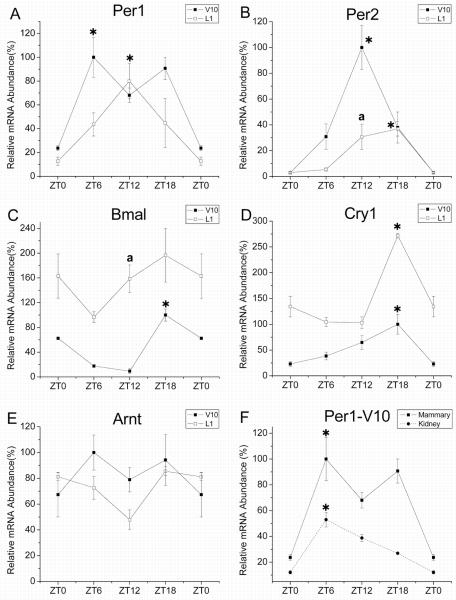
Clock gene expression is rhythmically regulated in many peripheral tissues (Reppert and Weaver, 2001; Reddy et al., 2005). Because the mammary tissue used in our developmental analysis was obtained around the same time of day (between ZT10-12) for all stages, it is possible that the observed changes in clock gene expression during development may reflect alterations in the circadian phase of clock gene oscillations, rather than their relative expression. To explore this possibility, we analyzed the daily patterns of clock gene expression in V10 and L1 mice because these two stages showed the greatest differences in our developmental panel. As shown in Figure 5, V10 mammary tissues were characterized by rhythmic expression of Per1, Per2, Bmal1 and Cry1. In the mammary gland of V10 animals, the rhythmic peak in Per1 expression occurred during the day at ZT 6 (Fig. 5A). This rhythmic pattern of Per1 expression is similar to that observed in the kidney (Fig. 5F) and that reported in other peripheral tissues (Tong et al., 2004). Maximal levels of Per2 mRNA in V10 mammary tissues were observed at ZT 12 (Fig. 5B), which coincides with the peak in DNA synthesis and mitosis found in developing mouse mammary glands (Berger and Daniel, 1982). Bmal1 and Cry1 mRNA levels in the virgin mammary gland oscillated in an anti-phase relationship to the rhythm of Per2 expression with a nocturnal peak occurring at ZT 18 (Fig. 5C-D). During lactation, mammary gland levels of Per1, Per2, and Cry1, but not Bmal1, mRNA fluctuated rhythmically. Further comparison between developmental stages revealed differences in the phase of these diurnal oscillations such that Per1 and Per2 gene expression levels peaked 6 hours later in L1 mice than in V10 animals. However, the implications of these developmental differences in rhythm phase are somewhat restricted by the limited temporal resolution associated with the 6-hour sampling interval used in this experiment. During lactation, the amplitude of the mammary gland rhythm in Per2 expression was altered such that peak-to-trough differences were greater at V10 (≈100-fold) than at L1 (≈40-fold). It is also noteworthy that Cry1 mRNA levels were consistently higher in L1 than V10 animals at all time points examined (Fig. 5D) although rhythm amplitude remained similar during lactation. In contrast to the clock genes, expression of the bHLH-PAS gene Arnt was similar, but not rhythmic in V10 and L1 mammary glands (Fig. 5E). The relative differences observed between V10 and L1 mice in this analysis are compatible with similar comparisons between these stages in our developmental panel (see Fig. 2) and suggest that the observed differences in circadian clock gene expression during mammary gland development occur largely through alterations in the amplitude, not in the phase, of the mammary gland oscillations.
Figure 5. Daily expression of circadian clock genes in 10 week virgin and day 1 lactating mouse mammary glands.
Temporal patterns of Per1 (A), Per2 (B), Bmal1 (C), Cry1 (D) and Arnt (E) expression in 10 week old virgin (V10; ■, solid line) and day 1 lactating (L1; □, solid line) mouse mammary glands. Symbols denote PCR determinations of mRNA levels (mean ± SEM) in V10 and L1 mammary glands (@n = 3) collected at 6hr intervals for 24 hours. The plotted values correspond to the ratios of mouse Per2, Bmal1 or Cry1/CypA or β-actin mRNA signal and are represented as a percentage of the maximal value obtained in the V10 group. Daily pattern of Per1 expression of in kidney tissues from V10 animals (λ, dashed line) is also depicted in comparison with its temporal profile in the mammary gland (F). Asterisks indicate time points during which peak values for gene expression were significantly greater (P<0.05) than those observed during preceding or succeeding minima of the same developmental stage. Because all tissues in Figure 2 were collected around ZT 10-12, V10 and L1 were compared further for differences in clock gene expression at ZT 12. Mean values at ZT12 that were significantly different (p<0.05) between V10 and L1 animals are denoted by a.
Using an in vitro mouse mammary epithelial cell model, we have found that mammary epithelial cell differentiation is associated with increases in Per1 and Bmal1 mRNA levels (Fig. 1). These results were corroborated in vivo by analyzing a mouse mammary gland developmental panel (Fig. 2) and indicate that Per1 and Bmal1 expression are associated with periods of increased cellular differentiation. Normally, BMAL1 and CLOCK induce Per1, Per2 and Cry1 expression. However, our results show that during times when Bmal1 expression is upregulated when Per1, but not Per2, mRNA levels are increased. This is consistent with the testes and thymus, where normal interactions between core elements of the transcription-translation feedback loop are suspended or restricted to specific developmental stages (Alvarez et al., 2003; Morse et al., 2003). However, unlike the testes and thymus, cycling of the peripheral clockworks appears to continue at least during critical stages of mouse mammary gland development and changes in clock gene expression during differentiation are due to modulation of rhythm amplitude or perhaps some alteration of phase. Similar to oscillations found in other peripheral tissues (Tong et al., 2004), mammary expression of the circadian clock genes Per2, Bmal1 and Cry1 was rhythmic in virgin animals (Fig. 5). In addition, Bmal1 was expressed arrhythmically and at higher levels during lactation (Fig. 5C). Collectively, these data suggest that the regulation of clock gene expression during cellular differentiation of the mammary gland may differ from the normal feedback mechanisms comprising the clockworks in the SCN and other peripheral tissues. In the testes where a similar disruption of normal mechanisms for clock gene regulation has been observed, CREMτ may represent a potential regulator of Per1 expression because a putative CRE has been identified in the Per1 promoter, and the pattern of CREMτ expression resembles that for Per1 during spermiogenesis (Morse et al., 2003). CREMτ-mediated expression of Per1 though its CRE enhancer is further supported by the observation that Per1 expression is attenuated in the testes of CREMτ null mice. It remains to be determined whether Per1 expression in the mammary gland is controlled by a similar mechanism. However, the application of specific clock knockout models may provide an opportunity to determine how individual clock components are involved in normal and aberrant mammary gland development. We are currently using such models to specifically examine the effects of Per1 and Per2 on mammary development and differentiation.
EXPERIMENTAL PROCEDURES
HC-11 Cell culture
HC-11 cells are prolactin-responsive cells derived from the COMMA-1D mouse mammary epithelial cell line (Ball, 1988). Without hormone stimulation, these cells divide and do not express markers of the differentiated mammary gland. When treated with lactogenic hormones hydrocortisone and prolactin, cell division is arrested and induction of ß-casein expression, which is the marker of differentiation, is observed.
HC-11 cells were maintained in RPMI 1640 medium (Life Technologies) containing 10% fetal bovine serum (FBS), 50μg/ml gentamicin (Life Technologies), 5μg/ml insulin (Sigma) and EGF (JRH Biosciences) in a humidified incubator in 95% air, 5% CO2 at 37°C. For induction of ß-casein expression, cells were grown to confluence and growth medium was replaced on a daily basis for three consecutive days. On the fourth day, cells were washed three times with PBS and priming medium (RPMI 1640 medium containing 10% charcoal-stripped serum, 50 μg/ml gentamicin, 5 μg/ml insulin, and 1 μg/ml hydrocortisone [Sigma]) was added. Twenty-four hours later cells were exposed to induction medium (priming medium containing 1μg/ml ovine prolactin [National Hormone and Pituitary Program]). Cells were maintained in induction medium for four days with daily replacement. Total RNA was isolated and DNA was removed by on-column digestion using the RNeasy mini extraction kit (Qiagen).
Animals
Female C57Bl/6J mice were obtained from Charles River (Wilmington, MA). All animals were housed 3/cage and maintained under a standard 12-hour photoperiod (LD 12:12; lights-on at 0600hr or Zeitgeber Time [ZT] 0). Animals were provided with access to food and water ad libitum. The procedures used in this study were approved by the University Laboratory Animal Care Committee at Texas A&M University.
For characterization of gene expression during mammary gland development, female mice were analyzed at seven developmental stages: virgin week 4 and 10 (V4 and V10), pregnancy day 6 and 16 (P6 and P16), lactation day 1 and 7 (L1 and L7), and involution day 3 (i3). At each stage of development, animals (n = 3-5) were sacrificed by cervical dislocation around 11 hours after lights-on (ZT 11) and mammary gland tissues were collected for later extraction of total RNA. To profile differences in the daily pattern of clock gene expression during development, mammary gland tissues were collected from mice at V10 and L1 (@n = 3) sacrificed by cervical dislocation at 6-hour intervals for 24 hours beginning at ZT0.For each animal, approximately 30 mg of mammary tissue was homogenized in 1 ml Trizol reagent (Invitrogen) and total RNA was extracted from the aqueous phase. The final RNA pellet was suspended in 100 μl RNase-free water, and subjected to purification and on-column DNase digestion using the RNeasy mini extraction kit (Qiagen).
Real-time PCR
Quantification of relative mRNA abundance was performed using TaqMan or SYBR-Green real-time PCR technology (Applied Biosystems, Inc. [ABI], Foster City, CA) as described previously (Allen et al., 2004). To generate single-strand cDNAs, total RNA (≈1μg) was reverse transcribed using Superscript II (Invitrogen) and random hexamers or oligo-dTs. For each sample, the cDNA equivalent of 50ng of total RNA was amplified in an ABI PRISM 7700 sequence detection system. Because limited information is available on clock genes expression in mouse mammary gland tissue or cell lines, each sample was analyzed in triplicate to ensure the accuracy of our data. To control for differences in sample RNA content, either Cyclophilin A (CypA) or β-actin mRNA was amplified with the cDNA equivalent of 1ng total RNA from the same samples. Results generated from the same experiments using different internal controls were consistent with each other.
The comparative CT method described in the ABI Prism 7700 Sequence Detection System User Bulletin #2 (PE-ABI) was utilized to calculate the relative mRNA abundance for a given target gene. Using this method, the amount of target gene mRNA in each sample was normalized first to corresponding CypA or β-actin mRNA levels, and then relative to a calibrator consisting of pooled cDNA from multiple samples that was analyzed on each reaction plate. For comparison of the daily patterns of clock gene expression in V10 and L1 mice, relative abundance of target mRNA was represented as a percentage of the maximal value obtained in the V10 group. Primer sequences for PCR amplification are listed in Table 1.
Table 1.
Primers Used in Real-time RT PCR Analyses
| Gene | Primers Sequence |
|---|---|
| Cyclin D1 | Forward: 5′-TCCGCAAGCATGCACAGA-3′ Reverse: 5′-GGTGGGTTGGAAATGAACTTCA-3′ |
| C-Myc | Forward: 5′-ACAGCAGCTCGCCCAAATC-3′ Reverse: 5′-CGAGTCCGAGGAAGGAGAGA-3′ |
| Arnt | Forward: 5′-GCCAGCCTGAGGTCTTTCAA-3′ Reverse: 5′-AATTCTTCATTGTTGTAGGTGTTGCT-3′ |
| β-Casein | Forward: 5′-TGTGCTCCAGGCTAAAGT TCACT-3′ Reverse: 5′-GGTTTGAGCCTGAGCATATGG-3′ |
| Per1 | Forward: 5′-AAACCTCTGGCTGTTCCTACCA-3′ Reverse: 5′-AATGTTGCAGCTCTCCAAATACC-3′ |
| Per2 | Forward: 5′-ATGCTCGCCATCCACAAGA-3′ Reverse: 5′-GCGGAATCGAATGGGAGAAT-3′ |
| Bmal1 | Forward: 5′-CCAAGAAAGTATGGACACAGACAAA-3′ Reverse: 5′-GCATTCTTGATCCTTCCTTGGT-3′ |
| Cry1 | Forward: 5′-AGTTCCCCTCCCCTTTCTCTT-3′ Reverse: 5′-GGGTTCCCTTCCATTTTGTCA-3′ |
| β-Actin | Forward: 5′-GCAACGAGCGGTTCCG-3′ Reverse: 5′-CCCAAGAAGGAAGGCTGGA-3′ |
| Cyclophilin | Forward: 5′-TGTGCCAGGGTGGTGACTT-3′ Reverse: 5′-TCAAATTTCTCTCCGTAGATGGACTT-3′ |
Indirect Immunofluorescence
Mammary glands were harvested from 10 week virgin and lactation day 1 mice that were sacrificed by cervical dislocation during the middle of the 12-hour photoperiod (1200-1400 h). Tissue samples were then fixed in 4% paraformaldehyde in PBS for two hours at 4°C and subsequently stored in 70% ethanol. The mammary glands were then processed, embedded in paraffin and five-micron sections were cut and mounted on slides. For indirect immunofluorescence, the sections were rehydrated by sequential washes in xylene and a series of graded ethanol washes. Antigen retrieval was performed for 20 minutes at 98°C in 0.01 mol/L sodium citrate buffer, pH 6.4, in a microwave oven. After the antigen retrieval step, slides were blocked and incubated for 1 hour at room temperature with PER1 antibody (Affinity Bioreagents). Slides were then washed in PBS-tween and incubated with Alexa-488 conjugated goat anti-rabbit (Covance, Princeton, NJ) at room temperature for 1 hour. Slides were then washed, coverslipped with Vectashield with DAPI (Vector Labs) and visualized by fluorescent microscopy.
Statistical Analyses
For in vitro observations, statistical analysis was performed using independent sample t-tests to determine whether the mRNA levels for a target gene were significantly different between undifferentiated and differentiated HC-11. For analysis of the mammary gland in vivo, statistical analyses were performed using one-way analysis of variance (ANOVA) to determine whether mRNA levels for a given gene were significantly different across different stages of development (V4, V10, P6, P16, L1, L7 and I3). Fisher’s LSD post-hoc analyses were used if significant main effects of development were obtained. The daily patterns of gene expression in mammary gland were assayed using two-way ANOVAs with developmental stage (V10 vs. L1) and time (ZT 0, 6, 12, 18) as two independent variables. To identify rhythmic variation in mammary gland patterns of gene expression, time-point determinations within V10 and L1 mice were evaluated separately using one-way ANOVAs. Paired comparisons between determinations at specific time points were analyzed post-hoc for statistical differences using Fisher’s LSD tests. Further statistical analyses were performed using t-tests to compare V10 and L1 animals for differences in gene expression at ZT 12 because mammary tissues were collected around this time for developmental comparisons. The α value was set at 0.05 for all statistical analyses.
Acknowledgments
This study was supported by NIH grants RO1CA111551 (W.W.P.), NIH Program Project grant P01 NS39546 (D.J.E.) and P30ES09106
REFERENCES
- Allen GC, Farnell Y, Bell-Pedersen D, Cassone VM, Earnest DJ. Effects of altered Clock gene expression on the pacemaker properties of SCN2.2 cells and oscillatory properties of NIH/3T3 cells. Neuroscience. 2004;127:989–999. doi: 10.1016/j.neuroscience.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Chen D, Storer E, Sehgal A. Non-cyclic and developmental stage-specific expression of circadian clock proteins during murine spermatogenesis. Biol Reprod. 2003;69:81–91. doi: 10.1095/biolreprod.102.011833. [DOI] [PubMed] [Google Scholar]
- Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. Embo J. 1988;7:2089–2095. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JJ, Daniel CW. Diurnal rhythms in developing ducts of the mouse mammary gland. J Exp Zool. 1982;224:115–118. doi: 10.1002/jez.1402240113. [DOI] [PubMed] [Google Scholar]
- Buchi KN, Moore JG, Hrushesky WJ, Sothern RB, Rubin NH. Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology. 1991;101:410–415. doi: 10.1016/0016-5085(91)90019-h. [DOI] [PubMed] [Google Scholar]
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Garcia MN, Barbeito CG, Andrini LA, Badran AF. Circadian rhythm of DNA synthesis and mitotic activity in tongue keratinocytes. Cell Biol Int. 2001;25:179–183. doi: 10.1006/cbir.2000.0585. [DOI] [PubMed] [Google Scholar]
- Granda TG, Liu XH, Smaaland R, Cermakian N, Filipski E, Sassone-Corsi P, Levi F. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. Faseb J. 2005;19:304–306. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Morse D, Cermakian N, Brancorsini S, Parvinen M, Sassone-Corsi P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol Endocrinol. 2003;17:141–151. doi: 10.1210/me.2002-0184. [DOI] [PubMed] [Google Scholar]
- Muller WJ, Neville MC. Introduction: Signaling in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:1–5. doi: 10.1023/a:1009540813521. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Wong GK, O’Neill J, Maywood ES, Hastings MH. Circadian clocks: neural and peripheral pacemakers that impact upon the cell division cycle. Mutat Res. 2005;574:76–91. doi: 10.1016/j.mrfmmm.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Scheving LE, Tsai TH, Pauly JE, Halberg F. Circadian effect of ACTH 1-17 on mitotic index of the corneal epithelium of BALB/C mice. Peptides. 1983;4:183–190. doi: 10.1016/0196-9781(83)90111-0. [DOI] [PubMed] [Google Scholar]
- Tong Y, Guo H, Brewer JM, Lee H, Lehman MN, Bittman EL. Expression of haPer1 and haBmal1 in Syrian hamsters: heterogeneity of transcripts and oscillations in the periphery. J Biol Rhythms. 2004;19:113–125. doi: 10.1177/0748730403262871. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li C, Zhu NL, Borok Z, Minoo P. Timeless in lung morphogenesis. Dev Dyn. 2003;228:82–94. doi: 10.1002/dvdy.10346. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Levine JD, Jin X, Weaver DR, Reppert SM. Molecular analysis of mammalian timeless. Neuron. 1998;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]