Abstract
Phospholamban (PLN) is an integral membrane protein that inhibits the sarcoplasmic reticulum Ca2+-ATPase, thereby regulating muscle contractility. We report a combined electrochemical and theoretical study demonstrating that the pentameric PLN does not possess channel activity for conducting chloride or calcium ions across the lipid membrane. This suggests that the pentameric configuration of PLN primarily serves as a storage form for the regulatory function of muscle relaxation by the PLN monomer.
Main Text
Phospholamban (PLN) is a single-pass membrane protein that reversibly binds and regulates the sarcoplasmic reticulum (SR) Ca2+-ATPase (SERCA) (1). The SERCA/PLN complex is responsible for Ca2+ ion transport from the cytoplasm into the SR lumen, causing muscle relaxation. In the heart, this membrane protein complex contributes to the synchronous beating of cardiac muscle cells (2). PLN exists in equilibrium between the oligomeric (pentameric) and monomeric forms (3). A plethora of biochemical and biological data collected both in vivo and in vitro indicate that the PLN monomer is the active, inhibitory species, regulating Ca2+ transport (4–9). The function of the oligomer, however, has puzzled biochemists and biophysicists for many years. In fact, PLN oligomerizes into well-structured pentamers that are exceptionally resistant to both thermal and chemical denaturations (3). Although there is an ongoing controversy regarding the topology of the cytoplasmic portion of PLN (10), scientists have reached a consensus regarding the helical left-handed coiled-coil configuration of the transmembrane (TM) domain (11–14). The TM helix of PLN can be divided into two domains: a more hydrophilic stretch (domain Ib, residues 22–32), stabilized by inter-protomer hydrogen bonds (15) and a second more hydrophobic one (domain II, residues 33–52), stabilized by a leucine-isoleucine zipper (16). In 1988, Kovacs et al. reported that both full-length and the 26–52 fragment of PLN selectively transport Ca2+ through membranes, suggesting a possible role of PLN in Ca2+ homeostasis (17). To the best of our knowledge, that is the only experimental evidence in the literature for channel activity of PLN. In the past few years, the Ca2+ channel hypothesis has resurfaced several times both in experimental and theoretical works (14,18). More recently, a possible role of pentameric PLN in Cl− ion transport (11) has been hypothesized. The Cl− ion channel hypothesis is exciting for two reasons: first, it would give new insights on ion regulation in the SR; second, it would suggest that completely hydrophobic helical bundles forming pores <4.5 Å can conduct Cl− ions through the membranes by simple diffusion.
To test these hypotheses, we probed Cl− and Ca2+ ion conduction through membranes using electrochemical measurements with PLN reconstituted in tethered bilayer lipid membranes (tBLMs) in concert with molecular dynamics (MD) and potential of mean force (PMF) simulations. As a control for electrochemical measurements, we used sarcolipin (SLN) (10), a PLN homolog that conducts Cl− ions in tBLMs (19). The preparations of tBLMs and the incorporation of both SLN and PLN are reported in the Supporting Material. Fig. 1 shows ΔY′ versus E plots at 0.1 Hz for both SLN and PLN, where ΔY′ is the increase in Y′ (in-phase component of the electrode admittance) at each applied potential after the incorporation of the given protein. The error bars are the standard deviations from four different sets of measurements. Whereas SLN attains a maximum conductivity at −0.600 V, corresponding to an effective transmembrane potential ϕ2=0.72 (−0.600 + 0.460) V ∼−0.1 V, PLN causes no detectable increase in the tBLM conductance. Remarkably, additions of Ca+2 ions of concentration ranging from 1×10−3 to 5×10−3 M to the aqueous solution of 0.1 M KCl have no effect on the conductance of a tBLM incorporating PLN. These results demonstrate that at physiological TM potentials PLN pentamer does not conduct either Cl− or Ca2+ ions.
Figure 1.

Plots of ΔY′ against E at 0.1 Hz obtained at tBLMs bathed by aqueous 0.1M KCl and incorporating SLN from its 0.7 μM solution or PLN from its 0.4 μM solution; ΔY′ is the increase in Y′ at each applied potential after the incorporation of the given protein. The error bars are the standard deviations from four different sets of measurements.
To substantiate our experiments, we performed theoretical calculations on ion conductivity based on two molecular models of PLN pentamer: the first model was the NMR structure of PLN in micelles (Protein Data Bank code: 1ZLL) (11), whereas the second model was built based on the fluorescence anisotropy data (20). Both models were subjected to energy minimization and MD simulations with identical protocols (see the Supporting Material).
The potential of mean force (PMF), or the free energy profile, for Cl− ion transport was determined through a series of umbrella sampling simulations (21). The reaction coordinate, which specifies the ion migration across the lipid membrane, is the difference between the z coordinate of the center of mass of the backbone atoms (N, Cα, C′, and O) from residues 27 to 52 in each monomer and the z coordinate of the Cl− ion, where the z axis is perpendicular to the membrane plane (see the Supporting Material). After initial MD equilibration, the TM domains for both models converge to a similar configuration, which is maintained throughout the entire simulation. The cytoplasmic domains move away from the initial configurations with undefined orientations with respect to the lipid membrane. Since the TM domains from the two models reach a similar configuration, we have carried out the PMF analysis based on the NMR model (1ZLL) alone. Fig. 2 depicts two snapshots of PLN from the initial stage of MD simulations. The central pore of PLN initial configuration is sufficiently large to be filled with water molecules (Fig. 2, left). After 700 ps of equilibration, the water molecules escape from the TM region, leaving domain II dehydrated throughout the entire course of the initial MD simulations (37 ns). Note that during this time we observed no spontaneous Cl− ion transport. Fig. 3 reports the computed PMF profiles for Cl− and Ca2+ translocation across PLN. The error bars corresponding to one standard deviation are estimated using the bootstrap method, with a decorrelation time of 5 ps and 50 Monte Carlo trials. The statistical fluctuations give rise to ∼1 kcal/mol errors near the top of the free energy barrier, suggesting that the free energy profile converged adequately. The convergence of the calculations is also revealed by the nearly equal free energy values for the ions on both sides of the PMF profile.
Figure 2.
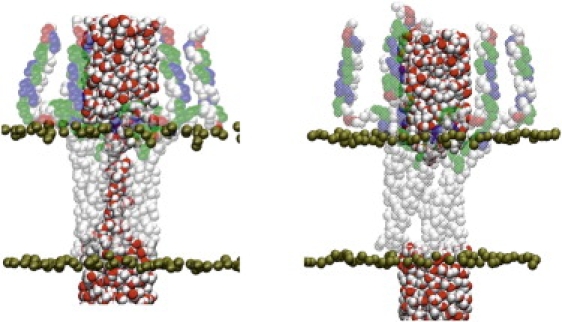
MD snapshots of PLN in lipid membranes. (Left) Initial configuration built from 1ZLL. Water and lipid molecules within 2.5 Å of PLN have been removed for clarity. (Right) PLN configuration after 700 ps of MD simulations.
Figure 3.

(Left) MD snapshot with the Cl− ion approximately at 5 Å with respect to the center of the membrane. (Right) plots of the PMF for Cl− and Ca2+ ions. In the most hydrophobic region of the pore the energy barrier plateaus out at ∼19 and 40 kcal/mol for Cl− and Ca2+ ions, respectively.
Fig. 3 shows a snapshot from the MD simulations with the chloride ion near the center of the bilayer. When the Cl− ion is located in the middle of PLN domain II, a chain of water molecules aligns along the entire pore, with hydrogen atoms pointing toward the ion. The free energy barrier for the passage of the Cl− ion across the pore is 19 kcal/mol. This high free energy barrier indicates that PLN is unable to transport Cl− ions. We repeated the calculations with Ca2+ ions and found that the free energy barrier for the transport is much higher (∼40 kcal/mol). As a comparison, Allen et al. (22) obtained a free energy barrier of ∼15 kcal/mol for Cl− conduction across gramicidin A, a Na+ ion channel, using a similar computational protocol. This is ∼10 kcal/mol greater than that for a Na+ ion. Corrections for finite size and membrane-induced polarization lower the barrier by 4 and 15 kcal/mol for mono and divalent ions, respectively (22). Thus, even with these corrections, the free energy barriers for ion permeation through PLN are still too high. Indeed, the absence of any hydrophilic group in PLN domain II causes strong desolvation effects, resulting in a large free energy barrier for both ion and water transport. We did not observe any evidence of ion stabilization via the Cys sulfur atoms in position 36 and 41. Taken with the electrochemical data, these calculations show that it is highly improbable that the conformations of the TM domain of PLN exemplified by the two models can conduct ions.
In conclusion, both electrochemical measurements under membrane potentials close to physiological conditions and free energy calculations provide strong evidence that both in the bellflower or pinwheel configurations PLN does not conduct Cl− or Ca2+ ion. The high free energy barrier is due to the hydrophobicity and the geometry of the PLN pore, in agreement with the recent computational studies, showing that hydrophobic pores with a diameters <4.5 and 6.5 Å are impermeable to water and ions, respectively (23). Similar conclusions were reached by Kim and co-workers using molecular dynamics calculations (24). These findings reinforce the hypothesis that PLN pentamer is a storage form that depolymerizes upon encountering SERCA and binds with the enzyme in a 1:1 complex (4–9). Cluster formation and oligomerization are a common theme for membrane proteins. Recently, Lang and co-workers have characterized the organization and dynamics of the plasmalemmal protein syntaxin 1, which forms well-defined clusters (25). Giving the crowding of the SR membrane (∼80% is constituted by membrane-embedded proteins), similar mechanisms of self-association/regulation are likely to occur for PLN, SERCA, as well as ryanodine receptors.
Acknowledgements
This work was supported by grants from the National Institutes of Health to G.V. (GM64742, HL80081, GM072701), D.D.T. (GM27906), J.G. (GM46736), J.G. from the Office of Naval Research, and R.G. from Ente Cassa di Risparmio (Firenze, Italy). All computations were performed on computers at PNNL through an EMSL Grand Challenge grant.
Footnotes
Lucia Becucci and Alessandro Cembran contributed equally to this work.
Supporting Material
References and Footnotes
- 1.Mac Lennan D.H., Kranias E.G. Phospholamban: a crucial regulator of cardiac contractility. Natl. Rev. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 2.Bers D.M. Calcium cycling and signaling in cardiac myocytes. Annu. Rev. Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 3.Thomas D.D., Reddy L.G., Karim C.B., Li M., Cornea R. Direct spectroscopic detection of molecular dynamics and interactions of the calcium pump and phospholamban. Ann. N. Y. Acad. Sci. 1998;853:186–194. doi: 10.1111/j.1749-6632.1998.tb08266.x. [DOI] [PubMed] [Google Scholar]
- 4.Robia S.L., Campbell K.S., Kelly E.M., Hou Z., Winters D.L. Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ. Res. 2007;101:1123–1129. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou Z., Kelly E.M., Robia S.L. Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J. Biol. Chem. 2008;283:28996–29003. doi: 10.1074/jbc.M804782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly E.M., Hou Z., Bossuyt J., Bers D.M., Robia S.L. Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J. Biol. Chem. 2008;283:12202–12211. doi: 10.1074/jbc.M707590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy L.G., Jones L.R., Thomas D.D. Depolymerization of phospholamban in the presence of calcium pump: A fluorescence energy transfer study. Biochemistry. 1999;38:3954–3962. doi: 10.1021/bi981795d. [DOI] [PubMed] [Google Scholar]
- 8.MacLennan D.H., Kimura Y., Toyofuku T. Sites of regulatory interaction between calcium ATPases and phospholamban. Ann. N. Y. Acad. Sci. 1998;853:31–42. doi: 10.1111/j.1749-6632.1998.tb08254.x. [DOI] [PubMed] [Google Scholar]
- 9.Young H.S., Jones L.R., Stokes D.L. Locating phospholamban in co-crystals with ca(2+)-ATPase by cryoelectron microscopy. Biophys. J. 2001;81:884–894. doi: 10.1016/S0006-3495(01)75748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traaseth N.J., Ha K.N., Verardi R., Shi L., Buffy J.J. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca(2+)-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oxenoid K., Chou J.J. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc. Natl. Acad. Sci. USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traaseth N.J., Verardi R., Torgersen K.D., Karim C.B., Thomas D.D. Spectroscopic validation of the pentameric structure of phospholamban. Proc. Natl. Acad. Sci. USA. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith S.O., Kawakami T., Liu W., Ziliox M., Aimoto S. Helical structure of phospholamban in membrane bilayers. J. Mol. Biol. 2001;313:1139–1148. doi: 10.1006/jmbi.2001.5101. [DOI] [PubMed] [Google Scholar]
- 14.Arkin I.T., Adams P.D., Brunger A.T., Smith S.O., Engelman D.M. Structural perspectives of phospholamban, a helical transmembrane pentamer. Annu. Rev. Biophys. Biomol. Struct. 1997;26:157–179. doi: 10.1146/annurev.biophys.26.1.157. [DOI] [PubMed] [Google Scholar]
- 15.Liu W., Fei J.Z., Kawakami T., Smith S.O. Structural constraints on the transmembrane and juxtamembrane regions of the phospholamban pentamer in membrane bilayers: Gln29 and Leu52. Biochim. Biophys. Acta. 2007;1768:2971–2978. doi: 10.1016/j.bbamem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmerman H.K., Kobayashi Y.M., Autry J.M., Jones L.R. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs R.J., Nelson M.T., Simmerman H.K., Jones L.R. Phospholamban forms Ca2+-selective channels in lipid bilayers. J. Biol. Chem. 1988;263:18364–18368. [PubMed] [Google Scholar]
- 18.Sansom M.S., Smith G.R., Smart O.S., Smith S.O. Channels formed by the transmembrane helix of phospholamban: A simulation study. Biophys. Chem. 1997;69:269–281. doi: 10.1016/s0301-4622(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 19.Becucci L., Guidelli R., Karim C.B., Thomas D.D., Veglia G. An electrochemical investigation of sarcolipin reconstituted into a mercury-supported lipid bilayer. Biophys. J. 2007;93:2678–2687. doi: 10.1529/biophysj.107.109280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robia S.L., Flohr N.C., Thomas D.D. Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry. 2005;44:4302–4311. doi: 10.1021/bi0478446. [DOI] [PubMed] [Google Scholar]
- 21.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in monte carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 22.Allen T.W., Andersen O.S., Roux B. Molecular dynamics - potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels. Biophys. Chem. 2006;124:251–267. doi: 10.1016/j.bpc.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Beckstein O., Sansom M.S. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys. Biol. 2004;1:42–52. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 24.Kim T., Lee J., Im W. Molecular dynamics studies on structure and dynamics of phospholamban monomer and pentamer in membranes. Proteins. 2008 doi: 10.1002/prot.22322. [DOI] [PubMed] [Google Scholar]
- 25.Sieber J.J., Willig K.I., Kutzner C., Gerding-Reimers C., Harke B. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


