Abstract
The nuclear lamina is part of the nuclear envelope (NE). Lamin filaments provide the nucleus with mechanical stability and are involved in many nuclear activities. The functional importance of these proteins is highlighted by mutations in lamin genes, which cause a variety of human diseases (laminopathies). Here we describe a method that allows one to quantify the contribution of lamin A protein to the mechanical properties of the NE. Lamin A is ectopically expressed in Xenopus oocytes, where it is incorporated into the NE of the oocyte nucleus, giving rise to a prominent lamina layer at the inner nuclear membrane. Nuclei are then isolated and probed by atomic force microscopy. From the resulting force curves, stiffness values are calculated and compared with those of control nuclei. Expression of lamin A significantly increases the stiffness of oocyte nuclei in a concentration-dependent manner. Since chromatin adds negligibly to nuclear mechanics in these giant nuclei, this method allows one to measure the contribution of individual NE components to nuclear mechanics.
Introduction
Eukaryotic nuclei are delimited by the nuclear envelope (NE), which separates genetic material and transcriptional machinery from the cytoplasm. The NE consists of two lipid membranes in which pore complexes are embedded. In metazoans a fourth component, the nuclear lamina, is closely apposed to the inner nuclear membrane. The lamina is an essential component of metazoan cells. It is comprised mainly of type V intermediate filament proteins, the nuclear lamins, and a growing number of lamin-associated polypeptides (1). Lamins recruit and anchor, either directly or indirectly, several NE proteins and interact with chromatin. They are involved in a multitude of cellular functions, including proper chromatin organization, DNA replication, cell cycle regulation, cell differentiation, and apoptosis. The importance of lamina function is highlighted by mutations in genes encoding nuclear lamina proteins that cause a wide range of heritable human diseases called laminopathies (1–4). Two hypotheses have been proposed to understand the mechanisms underlying laminopathies: the gene regulation hypothesis and the structural hypothesis (2). The gene regulation hypothesis proposes that perturbation of the lamina structure will alter gene regulation in a tissue-specific manner. In contrast, the structural hypothesis proposes that mutations in lamina proteins will affect the mechanical properties of the NE, rendering nuclei and cells more fragile and thereby causing diseases, particularly in mechanically stressed tissues. Experimental support is emerging for both hypotheses, which are not mutually exclusive. Given the role of the nuclear lamina as an integral part of the cytoskeleton, the structural hypothesis is particularly intriguing.
Vertebrates express A- and B-type lamins, which share a common structural organization: short N-terminal and larger globular C-terminal domains flank an α-helical rod domain. The rod domains allow lamins to form coiled-coil dimers that assemble via head-to-tail and lateral association into filaments (5). B-type lamins are constitutive components of the NE, whereas A-type lamins are not (6,7). Gene silencing experiments indicate that lamin B1 and lamin B2 are essential for cell survival (8). Mice lacking wild-type lamin B1 die shortly after birth (9). In contrast, cells lacking lamin A are viable and can be propagated in culture. Moreover, lamin A is absent from early embryonic stages but is present in most differentiated cells. Mutations in the lamin A gene give rise to many different tissue-specific diseases, and homozygous lamin A knockout mice are retarded in postnatal growth, suffer from muscular dystrophy, and die a few weeks after birth (10).
The nuclear lamina appears as an electron-dense layer interposed between the inner nuclear membrane and the peripheral chromatin (11). The thickness of this layer varies with cell type from a few nanometers up to 100 nm and might change depending on the physiological or pathological state (12,13). A- and B-type lamins contribute differently to lamina organization and seem to be segregated within the lamina layer (14,15). B-type lamins are permanently isoprenylated (16) and form thin filamentous layers that are closely associated with nuclear membranes. Prelamin A, in contrast, loses its isoprene moiety in the course of proteolytic processing to mature lamin A (17,18) and is probably less closely associated with the nuclear membrane (15). We previously analyzed lamin filaments and their assembly into higher-order structures by means of scanning electron microscopy in amphibian oocytes (14). Upon ectopic expression in oocytes, lamin A assembles into filaments that form a compact layer that covers the endogenous nuclear lamina (19). The lamin-A-containing NEs in oocytes resemble those of somatic cells with particularly thick nuclear laminae, except that chromatin is not associated with the oocyte lamina (14). Nuclei with prominent lamina layers are preferentially found in cells in which the nucleoskeleton significantly adds to cell and tissue architecture, such as certain fibroblast and endothelial cells. Their mechanical properties are therefore of particular interest. However, because they are an integral part of tissues, these cells are not easily accessible to investigation.
Amphibian oocytes are giant cells with nuclei that are ∼100,000-fold larger in volume than those of somatic cells. In contrast to somatic cells, their chromosomes are not in contact with the nuclear periphery (20) and they harbor high amounts of nuclear actin (21). A single lamin, the B-type lamin LIII, is the major lamin of Xenopus oocytes (22,23). Experimentally, the composition of the oocyte lamina can easily be altered by ectopically expressing lamins or lamin-associated proteins, which makes these cells ideal objects for analyses of NE mechanics. Isolated nuclei can be prepared under near-physiological conditions and used for atomic force microscopy (AFM) measurements.
In this study we used AFM to analyze the mechanical properties of isolated amphibian nuclei that ectopically express prelamin A. AFM has been used to study the mechanical properties of mainly eukaryotic cells in physiological conditions (24). In these cells, including fibroblasts, osteoblasts, and others, the mechanical properties are mainly determined by the actin cytoskeleton (25). AFM interrogates the mechanical properties by locally applying a force by the AFM tip and indenting the cell. From this loading curve (indentation versus loading force), the material properties (elastic or Young's modulus) can be inferred when the mechanical model is applied to the experimental situation. A very important point is that AFM allows one to follow processes in cells, such as cell migration (26) and cell division (27). Bacteria have also been investigated by AFM, although adhesion to the support is here an issue (28). The mechanical properties of bacteria are mainly determined by the bacterial cell wall (29). From a mechanical point of view, oocyte nuclei are comparable to bacteria, since the properties of the NE will determine the mechanical response. A previous study investigated the NE by AFM after peeling off the nucleus and spreading NE patches on a suitable support (30), and recently the mechanical properties of these supported membrane patches were accessed (31). Although researchers have used various techniques, such as pipette aspiration (32), to investigate the mechanics of intact oocyte nuclei, this study is the first (to the best of our knowledge) to use AFM for that purpose.
We show here that the elastic properties of oocyte nuclei change concomitantly with the expression of lamin A. Compared with control nuclei, nuclei that contain large amounts of lamin A are much stiffer and resist deformation to a high degree. The method described here can be applied to a wide variety of NE proteins to analyze their effects on the mechanical properties of the NE.
Material and Methods
Oocyte isolation and microinjection
Female Xenopus laevis (Daudin) were purchased from the African Xenopus Facility (Knysna, Republic of South Africa). Oocytes were surgically removed and defolliculated by collagenase treatment as previously described (33).
Plasmid DNA (37 or 110 ng/μL in H2O) was injected into the oocyte nucleus (13.8 nL per nucleus) with a nanoliter injector (Nanoject II; Drummond Scientific, Broomall, PA). DNA was mixed with Blue Dextran (10 mg/mL final concentration; Fluka, Buchs, Switzerland) to ascertain successful nuclear injection. Injected oocytes were incubated for 16–24 h at 18°C to allow expression of lamin proteins. The construction of Flag-epitope-tagged Xenopus laevis prelamin A was described previously (34). Oocyte nuclei were manually isolated in 5:1 buffer (83 mM NaCl, 17 mM KCl, 10 mM Tris-HCl, pH 7.2), cleaned from adhering cytoplasm by repeated pipetting up and down in a hand-drawn glass pipette, and transferred into a small well (∼0.5 mm in diameter) at the bottom of a petri dish.
AFM
An atomic force microscope (MFP-3D; Asylum Research, Santa Barbara, CA) combined with an inverted optical microscope (Zeiss Axiomat 135 microscope; Zeiss, Oberkochen, Germany) equipped with a 10× objective lens was used. The optical microscope allowed positioning of the AFM tip on top of the nucleus. Petri dishes with nuclei were fixed in a custom-built holder that was magnetically attached to the microscope stage. Soft silicon nitride cantilevers (Microlever; Park Scientific, Sunnyvale, CA) with a nominal spring constant of 10 mN/m were used. The setup rested on a granite plate supported by soft rubber bands from the ceiling for vibration isolation.
Data acquisition and analysis
Force curves were acquired at a rate of 1 Hz with a typical scan range of 2–12 μm at 1000 pixels per force curve. Force curves have been analyzed with two different mechanical models. If the nuclear membrane is very stiff compared to the internal three-dimensional nucleoskeleton, the appropriate mechanical model is a thin elastic shell that is deformed by a point force. This model has been found to be appropriate in the case of bacteria being locally indented by an atomic force tip (29). In this case, the loading force versus indentation relation will be linear. From the slopes an effective spring constant kNE of the sample can be calculated as follows:
| (1) |
| (2) |
where d is the deflection of the cantilever, and z is the z height of the sample. If the slope s is equal to one, this formula gives the correct result for an infinitely stiff sample (kNE = ∞); if the spring constant of cantilever and NE are identical, the resulting slope is 0.5, as expected. The slopes were calculated for indentations <2 μm to ensure that only the tip was touching the nuclear membrane.
If the thickness of the shell (i.e., the NE) is known, and it is assumed that the mechanical properties of the shell material are homogenous and isotropic, the elastic modulus can be estimated by:
| (3) |
where h is the thickness of the NE and R is the radius of the oocyte nucleus. This relation is a fit to simulated indentation curves employing finite element models (35). This relation is only valued for small deformations of the shell, which is the case in our experiments, since typical indentations (up to 2 μm) are smaller than the radius of the nucleus, which is on the order of 250 μm.
If the mechanical response of the NE can be neglected, the mechanical response will stem from the underlying three-dimensional actin skeleton. Then the Hertz model (36), which describes the indentation in a infinite elastic half space by a pyramidal indenter, will be appropriate for describing the force curves:
| (4) |
where F is the loading force, α is the half opening angle of the pyramid, E is the elastic modulus, ν is the Poisson ratio, and δ is the indentation. The half opening angle α of the pyramid is quoted by the manufacturer to be 45°, and the Poisson ratio is assumed to be 0.5, corresponding to an incompressible material. This model has been applied in many cases to the mechanical response of eukaryotic cells (for review, see Radmacher (24)). The force indentation relation of this model is quadratic and thus can be easily distinguished from the linear relationship predicted from the shell theory.
Here, we assume that the indentation of the AFM tip (<2 μm) is much smaller than the diameter of the nucleus itself (500 μm). This allows us to use the Hertz formula for a conical indenter pushing on a planar sample of infinite thickness.
Results
To study the contribution of lamin A to the mechanical properties of the NE, we ectopically expressed lamin A in Xenopus oocytes by nuclear injection of a plasmid encoding the cDNA of an N-terminal epitope-tagged version of Xenopus laevis prelamin A. Previous immunohistological and cell fractionation experiments showed that experimentally introduced prelamin A is efficiently targeted to the NE (34,37), where it forms filaments (14). Lamin A filaments are closely associated with the nucleoplasmic side of the inner nuclear membrane. They appear as an electron-dense layer in transmission electron microscopy (TEM) cross sections (compare Fig. 1 A with B and C) that lines the entire inner aspect of the NE but leaves the nuclear pore complexes (NPCs) free (arrows in Fig. 1, B and C). The thickness of the lamin A layer (30 to >100 nm) depends on the expression level but may also vary between different areas of the same nucleus (Fig. 1 B, arrowheads). Fig. 1 A shows a TEM cross section of a control nucleus for comparison. The endogenous lamina, formed by a single layer of lamin LIII filaments (14,38), is inconspicuous and cannot be distinguished as a distinct layer from the inner nuclear membrane to which it is attached (Fig. 1 A).
Figure 1.
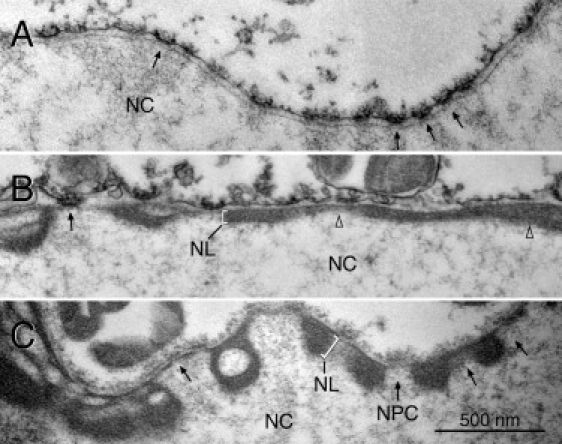
TEM of the NEs of lamin A-expressing oocytes. Prelamin A was expressed in oocytes by nuclear injection of plasmid. TEM sections of isolated oocyte nuclei are shown. The lamina (NL) (brackets in B and C) forms a thick electron-dense layer in oocytes expressing lamin A (B and C), which leaves the NPCs (arrows) free. The lamina in noninjected control oocytes is hardly discernible (A). NC: nuclear content; arrows point to NPCs; open triangles in B point to adjacent lamina layers of different thickness. Images were taken at the same magnification.
Xenopus oocyte nuclei can easily be isolated in physiological buffer. Handling of isolated nuclei (e.g., by sucking up and down with a narrow-bore pipette or by pushing the nucleus down with a pipette tip) revealed that nuclei of oocytes expressing lamin A are much stiffer and resist deformations much more than nuclei of control oocytes, indicating that lamin A filaments contribute to the mechanical properties of the nucleus. To quantify these effects, we used AFM analysis. Isolated nuclei of lamin A-expressing or control oocytes were transferred into a small well at the bottom of a petri dish mounted under the AFM. The AFM tip was positioned on top of the nucleus guided by the optical microscope. Several force curves were then recorded at this position, and data often were recorded for different locations of the same nucleus. Typical force curves of lamin A-expressing nuclei and control nuclei are shown in Fig. 2 A. Nuclei expressing lamin A are generally stiffer than control nuclei, as demonstrated by the increased slope value. For the high-expression nucleus (110 μg/mL DNA) the force curve is basically linear, which is an indication that the mechanical response of the nucleus is mainly determined by the NE including the lamin A layer, whereas the actin nucleoskeleton can be neglected. The other curves, especially control curves with no DNA injected, exhibit a significant curvature, which indicates that the shell model is not appropriate any more. Here the underlying three-dimensional actin skeleton and, more generally, the nucleoplasm contribute to the mechanical response. If the NE could be neglected, from a mechanical point of view, the Hertz model would be the appropriate model for data analysis. However, since we are probing the mechanical properties of the (upper) nuclear membrane of an intact oocyte nucleus of diameter on the order of 250 μm, we do not need to worry about “feeling” the underlying support, as is often the case in AFM studies of live cells, particularly thin lamellipodial regions in cells (39,40). To test the applicability of the Hertz model, we tried to fit the data of the softest force curve from Fig. 2 A (control) with this model. However, Fig. 2 B shows that the Hertz model fit does not fit the measured data very well. Therefore, we have to conclude that even with noninjected nuclei the mechanical response of the NE cannot be neglected. As a matter of fact, a linear fit leads to a much better fit to the data (Fig. 2 B). Thus we determined for all data sets the slope of the force curve and calculated from the slope the stiffness values using Eq. 2.
Figure 2.
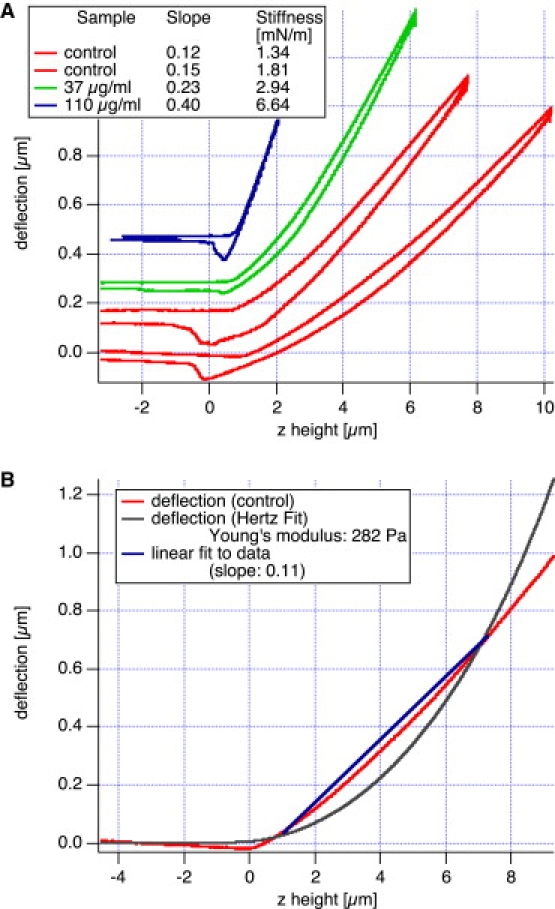
Typical force curve on a NE. The deflection (i.e., loading force) is proportional to the sample height z, as expected from the mechanical response of a thin elastic shell. (A) Force curves are shown for control nuclei (no DNA injected) and nuclei with DNA injected at 37 μg/mL and 110 μg/mL DNA, respectively. Depending on the amount of DNA injected, lamin A is expressed and the nucleus stiffens. The lamin A nuclei mostly exhibit a linear force curve, which shows that the mechanical response is mainly determined by the NE. However, in the softer samples (control), the nonlinearity of the force curve indicates that the underlying nucleoplasm, including its actin cytoskeleton, also contributes to sample stiffness. In B the softest force curve of A (control) has been fitted to a line (shell model) or to the Hertz fit. As can be seen from these two fits, the Hertz fit does not adequately describe our data.
We examined a total of 45 nuclei. Some of these nuclei were injected with DNA at two different concentrations (37 μg/mL or 110 μg/mL, respectively), and some were uninjected and used as controls. The total number of force curves was 1112. Some force data could not be analyzed because the cantilever was not pulled far enough to get out of contact. In total, 1076 force curves from 43 nuclei were analyzed. Fig. 3 shows the stiffness values that were calculated from the slopes of the force curves grouped for each nucleus. We recorded several force curves for each nucleus (three to 125 curves per nucleus, 25 on average) and observed some variations in the stiffness values for each nucleus, which were typically smaller than ±10% but occasionally up to ±10%. As can be seen from this figure, the variation within one nucleus is usually small; however, the differences between different nuclei, even within the same group, are large. In all cases we recorded several force curves at the same location on a nucleus. We never observed any drift in the mechanical response, which would result from destroying or altering the cell locally. Since the number of force curves taken on a single nucleus varied, we calculated for each nucleus the average stiffness value and the standard deviation (SD), respectively (Fig. 4). The SD is shown as an error bar. The data are grouped by experimental day, and the amount of DNA injected is encoded by colors and symbols for clarity (blue symbols denote high concentration (110 μg/mL), green symbols indicate low concentration (37 μg/mL), and red data points denote control nuclei with no DNA injected). As a general trend, the nuclei got stiffer by a factor of 3–4 when DNA was injected. However, the variation among different nuclei was very large, presumably due to different degrees of expression of lamin A. Nevertheless, the differences between control and DNA-injected nuclei is highly significant (see Table 1). Even the difference between low and high DNA concentrations is highly significant, as judged by an analysis of variance (ANOVA) and a t-test (with p < 5% considered significant).
Figure 3.
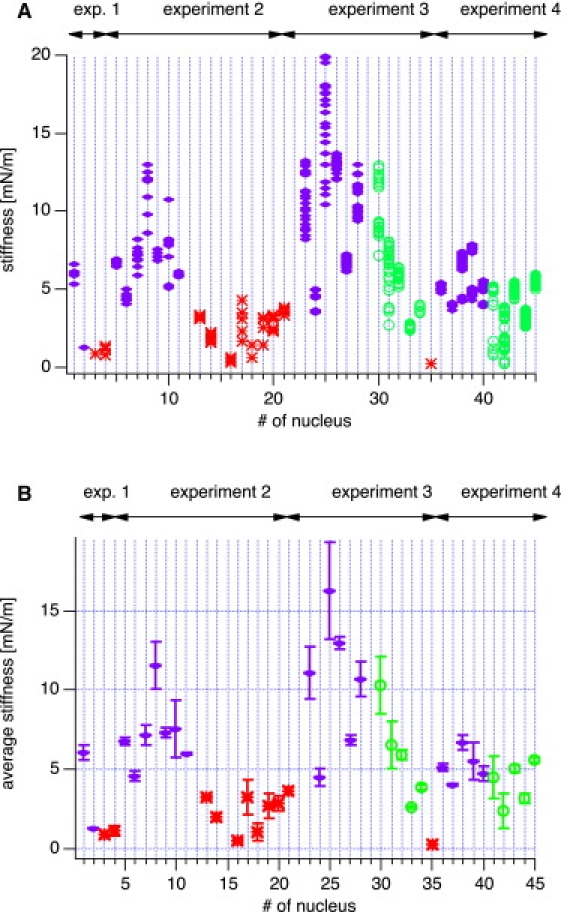
(A) Calculated stiffnesses from AFM force curves on isolated nuclei. The data are grouped for each nucleus. The color code refers to the concentration of injected DNA. Red (asterisk) corresponds to control (no DNA injected), green (circles) to 37 μg/mL, and blue (diamonds) to 110 μg/mL of injected DNA, respectively. (B) Although there is some variability for subsequent measurements of one nucleus, the differences between the different DNA concentrations are obvious and become more pronounced when all measurements of each individual nucleus are averaged. Here the error bars correspond to the SD of each nucleus. There is still some variability between different experiments corresponding to different batches of oocytes. Oocytes within one experiment are all from the same frog. Nevertheless, the degree of expression of lamin A seems to vary to some degree from experiment to experiment and within experiments.
Figure 4.
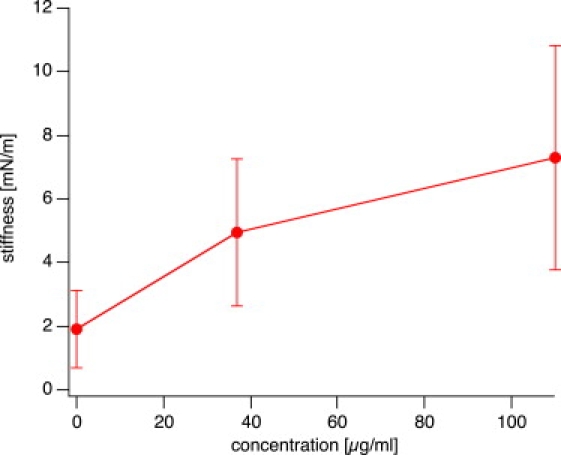
Average stiffness from all measurements as a function of concentration of injected DNA. The difference in stiffness at the three concentrations is highly significant between control and injected samples, and significant between low and high concentrations of injected DNA (see Table 1 for data).
Table 1.
| Average stiffness [mN/m] | SD [mN/m] | No. of nuclei | Noninjected | 37 | 110 | |
|---|---|---|---|---|---|---|
| non injected | 1,90 | 1,22 | 13 | — | 0.0055 | 0.034 |
| 37 | 4,52 | 2,27 | 11 | 0.0031 | — | 0.034 |
| 110 | 6,99 | 3,12 | 21 | 6,8e-7 | 0.020 | — |
The statistical significance of whether the stiffness values of each nucleus (average of the stiffness for this particular nucleus) differed depending on the amount of DNA injected was tested. For comparison, the significance values for the hypothesis of identical distributions for a Student's t-test (lower half of the table) and a one-dimensional ANOVA (upper half) were computed. The differences between all three groups are highly significant.
Discussion
Several experimental methods have been developed to probe the mechanical properties of cell nuclei, including micropipette aspiration, cell strain, cell compression, particle tracking, and AFM (24,41,42). With these methods, measurements are done with either intact cells or isolated nuclei. In somatic cells, both the NE lamina and chromatin contribute to the mechanical properties of nuclei (32,43). The nuclear lamina is a filamentous meshwork that in vertebrates is composed of A- and B-type lamin proteins. The contribution of A-type lamins to the mechanical properties of nuclei is of particular interest because 1), in contrast to B-type lamins, the level of lamin A expression varies greatly among cells of different tissues (6,44); and 2), mutations in A-type lamins cause at least 10 distinct human laminopathies, including Emery-Dreifuss muscular dystrophy, dilated cardiomyopathy, Dunningham-type familiar partial lipodystrophy, and Hutchinson-Gilford progeria syndrome (1–4). Previous studies analyzed the role of lamin A in cell mechanics by applying strain or compression on cells lacking expression of the LMNA gene, and then monitoring for nuclear deformation and damage (45,46). These analyses showed that A-type lamins are important contributors to the mechanical stiffness of nuclei. Cells deficient in lamin A have also been used to study the effect of lamin A on the micromechanical properties of the cytoplasm (47). Moreover, using micropipette aspiration methods, Pajerowski et al. (48) showed that the physical plasticity of stem cell nuclei decreases during terminal differentiation in parallel with an up-regulation of A-type lamins.
Alterations in the mechanical properties of Hutchinson-Gilford progeria cells were observed with the use of micropipette aspiration as well as photobleaching and nuclear swelling (49). Unexpectedly, it was found that although mutations in the lamin A gene may alter the structural properties of the nuclear lamina, such alterations do not necessarily result in altered resistance to mechanical stress when intact Hutchinson-Gilford progeria cells are challenged, e.g., by pressure.
In somatic cells, alterations in the lamina not only influence the mechanical properties directly, they may also contribute indirectly by altering chromatin organization. Therefore, to analyze the contribution of particular NE components to nuclear mechanics, experimental systems are needed in which chromatin adds minimally to the mechanical properties of the nuclei. Xenopus oocyte nuclei meet that criterion. They are up to 200 times larger in diameter than a typical vertebrate somatic cell nucleus, yet both types of nuclei contain approximately the same amount of DNA. As a consequence, the DNA (chromatin) concentration in a Xenopus oocyte nucleus is 100,000-fold lower than in a somatic cell nucleus. Moreover, in contrast to somatic cells, the chromosomes of amphibian oocytes are not associated with the NE (20,50). In the method presented here, Xenopus oocytes were used to express the NE protein prelamin A. Lamin A, which is normally not expressed in Xenopus oocytes, forms filaments that tightly associate with the endogenous lamina, giving rise to a prominent lamina layer (Fig. 1). The stiffness of isolated nuclei of lamin A expressing oocytes was probed by AFM and compared with that of control nuclei. With this experimental system, the contribution of lamin A to NE mechanics can be studied directly since chromatin does not significantly contribute to nuclear mechanics.
Although there is some variation between different batches of oocytes, i.e., oocytes from different females analyzed on different days, the differences in stiffness between lamin A-expressing and control oocytes of the same batch are highly significant. We measured an average stiffness of the oocyte nuclei between 1.9 mN/m for noninjected oocytes and ∼7 mN/m in the case of oocytes injected with a high concentration of plasmid DNA (110 μg/mL) with values of up to 16 mN/m. Kramer et al. (31) reported values of 6 mN/m for wild-type NEs, which is somewhat higher than our values. However, in their experiment the NEs were attached to a support, which may increase the apparent stiffness somewhat. Using a pipette aspiration technique, Dahl et al. (32) obtained values of 28 mN/m in intact, unsupported oocyte nuclei. In this technique, very large deformations have to be applied, so their data were obtained under experimental conditions very different from ours. Possibly, a stiffening of the NE or lamin network occurred due to large deformations. Our data, which were obtained at rather small deformations (indentations up to 2–3 μm) of intact oocyte nuclei, yield somewhat lower values in the control specimens, but nevertheless of the same order of magnitude. This makes it conceivable that the differences discussed here stem from the different experimental conditions.
The mechanical properties (elastic modulus) of the lamin A layer can be inferred from the average stiffness by using Eq. 3. However, the assumptions behind this equation are that a spherical shell of radius R and thickness h of homogeneous and isotropic material is indented locally by a force. In a strict sense, none of these assumptions are justified in our case, mainly because 1), the mechanical response comes from the NE plus an additional lamin A layer; 2), the underlying nucleoskeleton may also contribute; and 3), the thickness of the lamin A layer is not homogeneous and varies strongly among different nuclei even with the same amount of DNA injected.
When we assume typical values for the radius of nuclei (0.25 mm) and thickness of the lamin A layer plus NE (200 nm), we obtain an elastic modulus of 200 MPa for the average stiffness measured in our experiments (7.3 mN/m). This value is comparable to data obtained from microtubules using AFM (∼100 Mpa) (51), and much smaller than values obtained from very stiff protein structures, such as spider silk (10 GPa) (52).
In future studies we will need to determine the thickness of the lamin A layer, which will probably require TEM measurements after mechanical experiments by AFM, and to develop a realistic mechanical model of the combined responses of the NE, lamin A layer, and actin nucleoskeleton, which will probably require an extensive finite element simulation by computer. Such a model could provide experimental values of the mechanical properties (elastic modulus) of the components of the nuclear membrane (e.g., the lamin layer) under near in vivo conditions, in contrast to the rough estimate we calculated above. The contribution of nuclear actin to the mechanical stability of Xenopus oocyte nuclei could be experimentally tested, e.g., by coexpressing lamin A and exportin 6, since it has been shown that introducing exportin 6 into oocyte nuclei results in depletion of nuclear actin (21).
Conclusions
The method presented here allows one to quantify the contribution of individual NE components to the mechanics of the NE. It therefore complements other approved methods, many of which measure nuclear properties in the context of intact cells (42). The method is straightforward and will be particularly useful for analyzing the large number of mutant lamins that have been identified by human geneticists as causes of laminopathies. Moreover, the experimental setup is not restricted to the analysis of lamins and will be applicable for any NE protein.
Acknowledgments
We thank Emmajane Newton, Christine Richardson, and Martin Goldberg (School of Biological and Biomedical Sciences, Durham University, Durham, UK) for help with the electron microscopy, and Holger Doschke (Institute of Biophysics, University of Bremen, Bremen, Germany) for assistance with data analysis.
This work was supported by grants from the Deutschen Forschungsgemeinschaft (Sti98/7-1 to R.S.) and the University of Bremen (FNK 01/115/04).
Contributor Information
Manfred Radmacher, Email: radmacher@uni-bremen.de.
Reimer Stick, Email: stick@uni-bremen.de.
References
- 1.Mattout A., Dechat T., Adam S.A., Goldman R.D., Gruenbaum Y. Nuclear lamins, diseases and aging. Curr. Opin. Cell Biol. 2006;18:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison C.J., Worman H.J. A-type lamins: guardians of the soma? Nat. Cell Biol. 2004;6:1062–1067. doi: 10.1038/ncb1104-1062. [DOI] [PubMed] [Google Scholar]
- 3.Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 4.Rankin J., Ellard S. The laminopathies: a clinical review. Clin. Genet. 2006;70:261–274. doi: 10.1111/j.1399-0004.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann H., Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 6.Broers J.L., Machiels B.M., Kuijpers H.J., Smedts F., van den Kieboom R. A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 1997;107:505–517. doi: 10.1007/s004180050138. [DOI] [PubMed] [Google Scholar]
- 7.Lehner C.F., Stick R., Eppenberger H.M., Nigg E.A. Differential expression of nuclear lamin proteins during chicken development. J. Cell Biol. 1987;105:577–587. doi: 10.1083/jcb.105.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 9.Vergnes L., Peterfy M., Bergo M.O., Young S.G., Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fawcett D.W. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- 12.Ghadially F.N. Butterworths; London: 1988. Ultrastructural Pathology of the Cell and Matrix. [Google Scholar]
- 13.Höger T.H., Grund C., Franke W.W., Krohne G. Immunolocalization of lamins in the thick nuclear lamina of human synovial cells. Eur. J. Cell Biol. 1991;54:150–156. [PubMed] [Google Scholar]
- 14.Goldberg M.W., Huttenlauch I., Hutchison C.J., Stick R. Filaments made from A- and B-type lamins differ in structure and organization. J. Cell Sci. 2008;121:215–225. doi: 10.1242/jcs.022020. [DOI] [PubMed] [Google Scholar]
- 15.Delbarre E., Tramier M., Coppey-Moisan M., Gaillard C., Courvalin J.C. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum. Mol. Genet. 2006;15:1113–1122. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- 16.Nigg E.A., Kitten G.T., Vorburger K. Targeting lamin proteins to the nuclear envelope: the role of CaaX box modifications. Biochem. Soc. Trans. 1992;20:500–504. doi: 10.1042/bst0200500. [DOI] [PubMed] [Google Scholar]
- 17.Beck L.A., Hosick T.J., Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J. Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber K., Plessmann U., Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg M.W., Fiserova J., Huttenlauch I., Stick R. A new model for nuclear lamina organization. Biochem. Soc. Trans. 2008;36:1339–1343. doi: 10.1042/BST0361339. [DOI] [PubMed] [Google Scholar]
- 20.Gall J.G., Wu Z., Murphy C., Gao H. Structure in the amphibian germinal vesicle. Exp. Cell Res. 2004;296:28–34. doi: 10.1016/j.yexcr.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Bohnsack M.T., Stuven T., Kuhn C., Cordes V.C., Gorlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat. Cell Biol. 2006;8:257–263. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 22.Lourim D., Kempf A., Krohne G. Characterization and quantitation of three B-type lamins in Xenopus oocytes and eggs: increase of lamin LI protein synthesis during meiotic maturation. J. Cell Sci. 1996;109:1775–1785. doi: 10.1242/jcs.109.7.1775. [DOI] [PubMed] [Google Scholar]
- 23.Stick R. cDNA cloning of the developmentally regulated lamin LIII of Xenopus laevis. EMBO J. 1988;7:3189–3197. doi: 10.1002/j.1460-2075.1988.tb03186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radmacher M. Studying the mechanics of cellular processes by atomic force microscopy. Methods Cell Biol. 2007;83:347–372. doi: 10.1016/S0091-679X(07)83015-9. [DOI] [PubMed] [Google Scholar]
- 25.Rotsch C., Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys. J. 2000;78:520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prass M., Jacobson K., Mogilner A., Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzke R., Jacobson K., Radmacher M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat. Cell Biol. 2001;3:607–610. doi: 10.1038/35078583. [DOI] [PubMed] [Google Scholar]
- 28.Dufrêne Y. Atomic force microscopy of microbial cells. Microscopy and Analysis. 2001;15:27–29. [Google Scholar]
- 29.Arnoldi M., Kacher C., Bäuerlein E., Radmacher M., Fritz M. Elastic properties of the cell wall of Magnetospirillum gryphiswaldense investigated by atomic force microscopy. Appl. Phys. 1997;66:S613–S617. [Google Scholar]
- 30.Kramer A., Ludwig Y., Shahin V., Oberleithner H. A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small macromolecules. J. Biol. Chem. 2007;282:31437–31443. doi: 10.1074/jbc.M703720200. [DOI] [PubMed] [Google Scholar]
- 31.Kramer A., Liashkovich I., Oberleithner H., Ludwig S., Mazur I. Apoptosis leads to a degradation of vital components of active nuclear transport and a dissociation of the nuclear lamina. Proc. Natl. Acad. Sci. USA. 2008;105:11236–11241. doi: 10.1073/pnas.0801967105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahl K.N., Kahn S.M., Wilson K.L., Discher D.E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 33.Sive H.L., Grainger R.M., Harland R.M. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. Early Development of Xenopus laevis: A Laboratory Manual. [Google Scholar]
- 34.Ralle T., Grund C., Franke W.W., Stick R. Intranuclear membrane structure formations by CaaX-containing nuclear proteins. J. Cell Sci. 2004;117:6095–6104. doi: 10.1242/jcs.01528. [DOI] [PubMed] [Google Scholar]
- 35.Gibbons M.M., Klug W.S. Nonlinear finite-element analysis of nanoindentation of viral capsids. Phys. Rev. E. 2007;75:031901. doi: 10.1103/PhysRevE.75.031901. [DOI] [PubMed] [Google Scholar]
- 36.Hertz H. Über die Berührung fester elastischer Körper. J. Reine Angew. Mathematik. 1882;92:156–171. [Google Scholar]
- 37.Krohne G., Waizenegger I., Höger T.H. The conserved carboxy-terminal cysteine of nuclear lamins is essential for lamin association with the nuclear envelope. J. Cell Biol. 1989;109:2003–2011. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 39.Schäfer A., Radmacher M. Influence of myosin II activity on stiffness of fibroblast cells. Acta Biomater. 2005;1:273–280. doi: 10.1016/j.actbio.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Domke J., Radmacher M. Measuring the elastic properties of thin polymer films with the AFM. Langmuir. 1998;14:3320–3325. [Google Scholar]
- 41.Lammerding J., Dahl K.N., Discher D.E., Kamm R.D. Nuclear mechanics and methods. Methods Cell Biol. 2007;83:269–294. doi: 10.1016/S0091-679X(07)83011-1. [DOI] [PubMed] [Google Scholar]
- 42.Rowat A.C., Lammerding J., Herrmann H., Aebi U. Towards an integrated understanding of the structure and mechanics of the cell nucleus. Bioessays. 2008;30:226–236. doi: 10.1002/bies.20720. [DOI] [PubMed] [Google Scholar]
- 43.Dahl K.N., Engler A.J., Pajerowski J.D., Discher D.E. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Röber R.A., Weber K., Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 45.Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 46.Broers J.L., Peeters E.A., Kuijpers H.J., Endert J., Bouten C.V. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 47.Lee J.S., Hale C.M., Panorchan P., Khatau S.B., George J.P. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys. J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., Discher D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 51.Kis A., Kasas S., Babic B., Kulik A.J., Benoit W. Nanomechanics of microtubules. Phys. Rev. Lett. 2002;89:248101. doi: 10.1103/PhysRevLett.89.248101. [DOI] [PubMed] [Google Scholar]
- 52.Osaki S., Ishikawa R. Determination of elastic modulus of spider's silks. Polym. J. 2002;34:25–29. [Google Scholar]


