Abstract
Morbidity and mortality associated with acute lung injury (ALI) and acute respiratory distress syndrome remain substantial. Although many candidate genes have been tested, a clear understanding of the pathogenesis is lacking, as is our ability to predict individual outcome. Because ALI is a complex disease, single gene approaches cannot easily identify effectors that must be treated concurrently. We employed a strategy to help identify critical genes and gene combinations involved in ALI mortality. Using hyperoxia to induce ALI, a mouse model for genetic analyses of ALI survival time was identified: C57BL/6J (B) mice are sensitive (i.e., die early), whereas 129X1/SvJ (S) mice are significantly more resistant, but with low penetrance. Segregation analysis of reciprocal F2 mice generated from B and S strains revealed significant sex, cross, and parent of origin effects. Quantitative trait locus (QTL) analysis identified five chromosomal regions significantly linked to hyperoxic ALI survival time (named Shali1–Shali5). Further analyses demonstrated that both parental strains contribute resistance alleles to their offspring and that the phenotype demonstrated parent of origin effects. To validate earlier findings, we generated and tested mice from all eight possible B-S-derived backcrosses. Results from segregation and QTL analyses of 935 backcrosses, alone and combined with the previous 840 B-S-derived F2 population, further supported the highly significant QTLs on chromosomes 1 (Shali1) and 4 (Shali2) and confirmed that the sex, cross, and parent of origin all contribute to survival time with hyperoxic ALI.
Keywords: adult respiratory distress syndrome, mouse model, parent of origin effects, quantitative trait locus
despite significant advances in supportive care and improvements in ventilator management (1), acute lung injury (ALI) and adult respiratory distress syndrome (ARDS) are still associated with 30–45% mortality, which accounts for nearly 75,000 deaths/yr in the United States (36). Although more than four decades since its first description (2), the persistent high death rate related to ALI/ARDS continues to be a formidable challenge. Even results from recent meta-analyses have been contradictory as to whether ALI/ARDS mortality rates have changed over the last 15 yr (29, 50). Although anyone at any age can die after the development and progression of ALI/ARDS, older patients have an increased risk of dying (36, 39, 40, 51). Accordingly, as the population gets older, the future social and healthcare burdens surrounding ALI will continue to escalate. Consequently, alternative research strategies are needed to address this unremitting problem.
The pathophysiology of ALI has been described in detail (11, 18, 24, 25, 45, 46), yet little is known about the critical genes or gene products involved in the pathological mechanisms that lead to death. Because only ∼20% of ALI/ARDS patients die from refractory respiratory failure (15, 26, 43), the most important factors and mediators resulting in death are not obvious and may be far removed from lung-related phenotypes. This could, in part, explain the problems in identifying factors (and their associated genes) that lead to better prognoses. The difficulty is highlighted by the fact that no specific pharmacological therapies have reduced mortality in large phase III studies (7). Therefore, with the long-term goal of significantly decreasing ALI-associated mortality, we embarked on a very different strategy to traditional candidate gene approaches. Rather than predicting and testing genes directly or indirectly associated with ALI one at a time, quantitative trait locus (QTL) analysis initially identifies chromosomal regions of linkage without a priori prediction of the genes likely to harbor risk variants. Subsequent resolution of these QTLs will generate new hypotheses-driven studies aimed at decreasing ALI-associated mortality.
Using continuous hyperoxia, a well-established method to induce lung injury and death in animals (4, 5, 20, 44, 47), we (33) recently demonstrated that survival time with hyperoxic ALI (HALI) was a heritable phenotype in mice. Analysis of reciprocal F1 mice generated from sensitive C57BL/6J (B) mice and resistant 129X1/SvJ (S) mice [i.e., B × S (or B.S) and S × B (or S.B)] revealed a parent of origin effect. To further investigate this parent of origin difference, a large F2 population (n = 840 mice) was produced, consisting of 197 or more mice from each of the four possible intercrosses between B and S inbred mice. QTL analysis of the total F2 population identified five genomic regions (i.e., QTLs) significantly linked to HALI survival time, designated survival to hyperoxic acute lung injury 1–5 (Shali1–5) (32). Additional genetic analyses of F2 subpopulations identified significant sex, cross, and parent of origin effects on HALI survival time and demonstrated that a further increase in HALI survival time was due to a recombination of resistance alleles originating from both parental strains.
To validate these findings and further examine the mode of inheritance of HALI survival time, we generated and tested a large population (n = 935) of B-S-derived backcross mice. This population consisted of at least 94 mice from each of the eight possible backcross breeding schemes (Table 1). Separate segregation and QTL analyses were performed on males, females, and all mice for the group S backcrosses (i.e., the four backcrosses that yielded SS or SB genotypes), the group B backcrosses (i.e., the four backcrosses that yielded BB or SB genotypes), and the total backcross population. Segregation analyses verified significant cross and sex effects for HALI survival time among the backcross groups. QTL analyses results from the total backcross population verified Shali1 and Shali2 as highly significant linkages. Interestingly, when dividing the total population into the group B and S backcrosses, Shali1 was highly significant in group S backcrosses and Shali2 was highly significant in group B backcrosses. Overall, the backcross data validated that the S allele for Shali1 and the B allele for Shali2 are dominant or additive for HALI resistance. QTL analysis of the combined dataset, consisting of total B-S-derived backcrosses and F2 mice, confirmed Shali1–5 as significant loci.
Table 1.
The eight B-S-derived backcrosses
|
Group B Backcrosses (yielded B-B or B-S alleles) |
Group S Backcrosses (yielded S-S or B-S alleles)
|
||
|---|---|---|---|
| B dam | B sire | S dam | S sire |
| Cross 4: B × (B × S)F1 = B.BS | Cross 2: (B × S)F1 × B = BS.B | Cross 9: S × (B × S)F1 = S.BS | Cross 3: (B × S)F1 × S = BS.S |
| Cross 5: B × (S × B)F1 = B.SB | Cross 7: (S × B)F1 × B = SB.B | Cross 10: S × (S × B)F1 = S.SB | Cross 8: (S × B)F1 × S = SB.S |
B, C57BL/6J strain; S, 129X1/SvJ strain. Crosses 1 and 6 were assigned to different crosses being generated at the same time as these backcrosses and were not part of this study.
MATERIALS AND METHODS
Mice
From our initial screen of 18 inbred mouse strains, group B mice were identified as sensitive, whereas group S mice were significantly more resistant to HALI-induced mortality, although the penetrance of the resistance trait was low, ranging from 30% to 35% (33). Crosses of these strains (the B-S model) were used to generate mice for initial genetic and segregation analyses and the subsequent QTL analysis. First, reciprocal F1 lines were generated by breeding B females to S males (B.S) and S females to B males (S.B). The significant phenotype difference between these reciprocal F1 mice suggested a parent of origin effect, so reciprocal F1 offspring were systematically bred back to both parental lines, with the goal of generating ∼100 mice or more for each of the eight possible backcrosses (Table 1); a total of 935 backcross mice were used in these genetic experiments. Additional experiments were performed on a combined dataset consisting of this backcross population and the previously described (32, 33) F2 population (n = 840) in a single group of 1,775 recombinant mice.
Hyperoxia Exposure
Mice in standard shoebox cages with food and water ad libitum were placed inside a 0.13-m3 Plexiglas inhalation chamber (Stellar Plastics, Detroit, MI) and exposed continuously to >95% O2 until death. Each chamber housed up to nine cages with four mice per cage; individual mice were clearly visible from above and/or the sides. O2 level in the chamber was continuously controlled using a ProOx 110 portable O2 monitor (Biospherix, Redfield, NY), which was calibrated before each exposure using room air and 100% O2. The status of the exposures was closely monitored, such that the determined survival time for each mouse was within 5% error. Mice were exposed between 6 and 12 wk of age, and each exposure was continued until all mice within the chamber were dead. Mice included in these experiments were bred and exposed over a period of ∼5 yr; no significant seasonal differences were noted. Mice were handled in accordance with protocols approved by the Institutional Animal Care and Use Committee of Cincinnati Children's Hospital Medical Center.
DNA Preparation
Genomic DNA was isolated from 0.5-cm tail clips using a commercial DNA extraction kit (Wizard Genomic DNA, Promega, Madison, WI). A SpectraMAX 190 spectrophotometer (Molecular Devices, Sunnyvale, CA) and a 96-well quartz plate were used to analyze samples for purity (absorbance at 260/280 nm) and DNA content (absorbance at 260 nm). DNAs were diluted to 20 ng/μl for PCR requiring agarose gel separation or to 5 ng/μl for PCR using fluorescence genotyping.
Genotype Analysis
Primer pairs for polymorphic markers between the B and S strains were purchased from Research Genetics/Invitrogen (Frederick, MD) or IDT (Coralville, IA). PCR was performed in 15-μl volumes in 96-well plates (USA Scientific, Ocala, FL) using a four-block thermocycler (model PTC-225 or PTC-240, Bio-Rad) as previously described (33). Markers with PCR product allele sizes of 5% or more were separated by 2.5–4% agarose (ISC BioExpress, Kaysville, UT) gels and stained with ethidium bromide. Microsatellite markers <5% different in allele size were amplified from 20 ng of total DNA using fluorescent primers synthesized by ABI and the protocols provided. Fluorescent PCR products were separated using an ABI-3730xL sequencer located at the Cincinnati Children's Hospital DNA Sequencing Core Facility (http://dna.chmcc.org/), and genotypes were ascertained using GeneMapper software (version 3.5, ABI).
Segregation Analysis
Because survival times of the reciprocal F1 and F2 mice suggested a parent of origin effect in HALI survival (33), offspring from all eight possible backcross breeding schemes (Table 1) were generated to further investigate these cross and sex differences and to help predict trait heritability. Survival times were assessed in total mice and in males and females separately for each of the following cohorts: 1) the total backcross population; 2) the two groups of four combined backcrosses, based on whether the possible genotypes were heterozygous and homozygous S (group S backcrosses = SS or SB) or heterozygous and homozygous B (group B backcrosses = BB or BS); 3) the four backcross pairs generated with the same inbred strain as the dam or sire (i.e., S dam = crosses 9 and 10, S sire = crosses 3 and 8, B dam = crosses 4 and 5, and B sire = crosses 2 and 7); and 4) the combined backcross plus F2 dataset. Mean survival times (MSTs) were also assessed for total mice in the eight separate backcrosses.
Cross Comparisons and Assessment of Parental Effects
To confirm the cross differences seen in the F2 analysis and further assess these parent of origin effects, single and paired backcrosses within group B or S were compared. Within-group comparisons were valid to assess cross effects, since the genetic compositions of the lines are similar (i.e., group S backcrosses average 75% S alleles and 25% B alleles and group B backcrosses average ∼75% B alleles and 25% S alleles).
Single crosses.
Two comparisons were made for each of the eight backcrosses. Specifically, each backcross was compared with 1) the backcross in which the F1 strain of the dam or sire was switched with its reciprocal F1 strain (while maintaining the same inbred strain), and 2) its reciprocal backcross in which the male and female parental strains were reversed. This resulted in eight distinct comparisons: two comparisons of backcrosses with the same inbred dam but reciprocal F1 sires (i.e., crosses 4 vs. 5 and 9 vs. 10), two comparisons of backcrosses with the same inbred sire but reciprocal F1 dams (i.e., crosses 2 vs. 7 and 3 vs. 8), and four comparisons of reciprocal backcrosses (i.e., crosses 2 vs. 4, 3 vs. 9, 5 vs. 7, and 8 vs. 10).
Backcross pairs.
Of the 16 possible paired backcrosses (those with the same inbred or F1 strain as dam or sire), only the 4 pairs with the same inbred dam or same inbred sire were valid within-group comparisons to assess a parental effect. In particular, offspring of backcrosses from an S dam were compared with those from an S sire and progeny of backcrosses from a B dam were compared with those from a B sire.
Linkage Analysis and QTL Mapping
To assess linkage in the individual and combined backcross groups and to screen for global gene-gene interactions in the group B and S backcross populations, all 935 backcross mice were typed for 78 polymorphic microsatellite markers distributed at 20- to 25-cM intervals across the 19 autosomes; all markers were selected from the total set previously typed in the large F2 study (32). In addition, all mice were screened with four X chromosome (Chr) markers to assess whether the sex effect might be due to X Chr linkage. Chrs 1 and 4 were more densely typed to help further define the previously identified QTL interval(s). The phenotype used in all QTL analyses was log survival time, since log transformation better approximated a normal distribution. All phenotype and genotype data were analyzed for main effect QTLs using the freely available R/QTL (9) and GridQTL (http://www.gridqtl.org.uk/) computer packages where appropriate. Significance levels were determined using 10,000 permutations of the corresponding dataset for R/QTL analysis or 1,000 permutations for GridQTL and set at genome-wide significance levels of P < 0.001, 0.01, and 0.1 for highly significant, significant, or suggestive linkage, respectively.
To assess gene-gene interactions (i.e., additivity and epistasis), all pairwise combinations were evaluated separately in the group B and S backcross populations using the scantwo function of R/QTL. Empirical threshold significance values for pairwise effects were determined using 1,000 permutations of the respective log-transformed data; results did not differ from those for 500 permutations.
Estimating QTL Effects and Percent Penetrance
To estimate the contribution of the two major QTLs (Shali1 and Shali2) and this two-QTL combination to survival time, allelic effects were calculated and compared in the combined F2 plus backcross dataset. The QTL effects were assumed to correspond to the microsatellite marker nearest the peak LOD score. Mice with the same genotype (i.e., B-B, B-S, or S-S) for the representative peak marker of each QTL (i.e., D1Mit303 or D4Mit308) were combined, and the MST of the group was calculated and compared with the MSTs for groups of mice with the other possible genotypes at that marker. For example, the MST of all mice homozygous B for D1Mit303 was compared with the MST of mice that were homozygous S or heterozygous for D1Mit303. Importantly, beyond the similar genotypes at these desired loci, the remainder of the genome differs for the mice within these groups. A calculated difference between allelic groups supported a relative role for that QTL(s) in the overall phenotype, estimated the QTL effect, and suggested a likely mode of inheritance. These calculations were expanded to include combinations with the remaining QTLs (representative peak markers: D1Mit34, D9Mit355, and D15Mit184) and the major interacting QTL on Chr 18 (referred herein as Shali18i) identified in the original F2 analysis (i.e., D18Mit177). Percent penetrance was estimated as the number of mice within a group that had a resistant genotype and was resistant [i.e., a MST > 168 h, as previously defined (33)].
Statistical Analyses
Survival time distributions for all backcross cohorts were skewed towards sensitivity, in part due to decreased penetrance of long survival (i.e., resistance). We (33) have previously noted that the resistant S strain also had a skewed distribution, with resistance being 30–35% penetrant. Because data were skewed, MSTs were used for descriptive statistics (see Tables 2 and 4 and Suppl. Tables 1 and 2)1 and as the basis for group comparisons. Skewed data violated the normality requirement of most parametric analyses. In addition, because alternative nonparametric statistics are not suitable for modeling multiple factors and interactions at the same time, ANOVA was used with log transformation to satisfy assumptions. Survival analysis, such as the Cox proportional hazards model, was not needed because no censored data were included (i.e., all mice died). Statistical models were constructed with log survival time as the dependent variable and backcross group(s) as the independent variable. Specifically, backcross, dam-sire status, Group B and S backcrosses, sex, and their interactions were all used as predictors with the group of interest. Overall model P values from the F-test are reported as well as pairwise comparisons. Only prespecified comparisons were performed; in particular, parent of origin effects were assessed by comparing backcross groups in which 1) the inbred dams or sires were the same but the F1 dams or F1 sires were the reciprocal F1s or 2) the dam and sire were switched (i.e., reciprocal backcrosses). Multiple-comparison issues were addressed by applying a Sidak correction (38, 49) to ensure the overall family-wise type I error rate was <0.05. The Sidak correction is more accurate and less conservative than the Bonferroni adjustment. MST differences between groups are reported as the percentages of increase or decrease, since back transformation of the logarithm gave the geometric means.
Table 2.
Descriptive statistics of control strains and B-S-derived backcross populations
| Population | Sample Size |
Total Population |
Males
|
Females
|
|||
|---|---|---|---|---|---|---|---|
| MST, h | 25th and 75th percentiles | MST, h | 25th and 75th percentiles | MST, h | 25th and 75th percentiles | ||
| Control | |||||||
| B | 56 | 103 | (87, 126) | 114* | (96, 130)* | 98* | (78, 112)* |
| S | 47 | 106 | (97, 114) | 111 | (97, 128) | 105 | (89, 114) |
| Total backcrosses | 935 | 125 | (106, 146) | 126* | (111, 148)* | 123* | (105, 143)* |
| B.S × B | 97 | 118 | (106, 137) | 126 | (113, 138) | 113 | (106, 126) |
| B.S × S | 106 | 124 | (102, 153) | 114 | (101, 137) | 135 | (106, 153) |
| B × B.S | 154 | 120 | (112, 137) | 120 | (107, 137) | 121 | (112, 137) |
| B × S.B | 105 | 114 | (102, 129) | 119 | (105, 131) | 113 | (101, 128) |
| S.B × B | 100 | 137 | (119, 159) | 137 | (125, 183) | 129 | (113, 152) |
| S.B × S | 157 | 113 | (98, 137) | 120 | (99, 145) | 112 | (98, 135) |
| S × B.S | 122 | 137 | (103, 182) | 143 | (107, 198) | 137 | (100, 160) |
| S × S.B | 94 | 138 | (114, 185) | 144* | (127, 198)* | 130* | (106, 161)* |
| B dam | 259 | 119 | (106, 137) | 120 | (107, 135) | 114 | (106, 137) |
| S dam | 216 | 137 | (112, 184) | 144* | (113, 198)* | 135* | (105, 161)* |
| B sire | 197 | 129 | (113, 149) | 137 | (113, 153) | 125 | (112, 137) |
| S sire | 263 | 119 | (101, 143) | 119 | (101, 145) | 120 | (101, 138) |
| Group B | 456 | 123 | (112, 137) | 125* | (113, 138)* | 120* | (106, 137)* |
| Group S | 479 | 127 | (102, 154) | 129* | (102, 160)* | 126* | (102, 150)* |
Because F2 and backcross mice were frequently exposed in the same chambers, most of the B and S control mice reported herein were also included in the previous reports on F2 mice (32, 33). Sex effects were tested within each group using nested models based on log survival times, with Sidak adjustments. Medians are presented for descriptive purposes. MST, median survival time.
Statistically significant difference.
RESULTS
A total of 935 mice was generated from the 8 possible backcross breeding schemes (Table 1). All descriptive statistics for the total backcross population and the various population cohorts are shown in Table 2. MSTs of the parental B and S strains did not differ from each other in these backcross exposures, due to very low penetrance of resistance (11%) in the S strain. This differed from our previous findings, which reported ∼30–35% penetrance (33). However, because backcross and F2 mice were often exposed together, the fact that no difference was found between B and S strains in exposures that included backcross mice was likely due to chance. Nonetheless, it is noteworthy that the MST of the total backcross population (125 h) was significantly higher than that of either parental strain in these experiments, demonstrating an overall gain of resistance in the recombinant population. This validated our previous results showing that the sensitive B strain carries resistance alleles that recombined with resistant S alleles to add overall resistance in (recombinant) offspring (33). To further investigate cross and sex differences seen in the F2 analysis, the total population and males or females separately, in all but the individual crosses, were examined in detail in the different backcross cohorts.
Cross Comparisons
Single crosses.
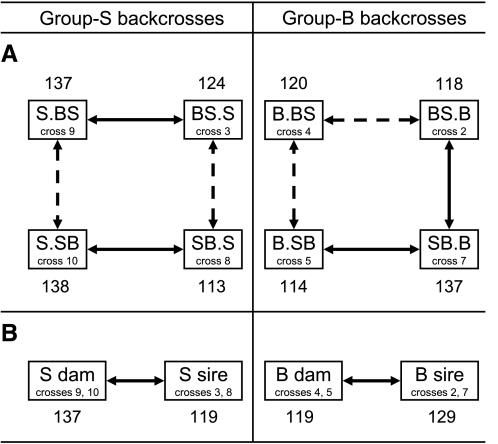
MSTs differed for the eight crosses (P < 0.0001 by F-test), demonstrating that HALI survival time depended on how the mice were bred (i.e., which strain or F1 was the dam or sire). MSTs for seven of the eight backcross populations were consistent with the phenotype of the maternal (or grandmaternal) inbred strain; only the MST of cross 8 did not agree. To confirm cross differences and to further assess possible parental effects, two within-group comparisons were made for each backcross. A visual representation of the results of these statistical comparisons is shown in Fig. 1A. Each line in Fig. 1A represents a prespecified comparison, with solid lines denoting significant differences and dashed lines representing no difference between compared groups. In total, four of the eight backcross comparisons were significant for a MST difference. One of four comparisons reached significance when crosses of reciprocal F1 dams or F1 sires were compared (Fig. 1A, vertical lines). Specifically, the comparison of crosses 2 and 7 revealed a 19-h MST difference (P < 0.0001) for mice generated with a B.S dam and B sire (n = 97) compared with those generated from a S.B dam and B sire (n = 100). The comparison of the four reciprocal backcrosses was especially interesting, because each reciprocal pair consisted of the same two strains (inbreds or F1 hybrids) except that the dam and sire strains were reversed. Three of the four reciprocal backcross comparisons (Fig. 1A, horizontal lines) had a significant difference in MSTs, supporting the previously identified parent of origin effect. In total, the MST of cross 7 was 19% longer than that of cross 2 and 25% longer than that of cross 5. The MST for cross 10 was 22% longer than that of cross 8, and the MST for cross 9 was 12% longer than that of cross 3.
Fig. 1.
Overview for statistical comparisons of 129X1/SvJ (group S) and C57BL/6J (group B) backcrosses. A: single backcross comparisons. Median survival times (MSTs; in h) were compared between backcrosses, which were split into group S (left; crosses 3, 8, 9, and 10, which yielded SB or SS genotypes) and group B (right; crosses 2, 4, 5, and 7, which yielded SB or BB genotypes) mating schemes. To assess parental effects, each backcross group was compared with two other backcrosses: 1) that with the same inbred dam or sire but a reciprocal F1 mate or 2) its reciprocal backcross. B: paired backcross comparisons. Within group B and S, the two backcrosses generated from the same inbred dam were combined, and the MST was compared with that for the two backcrosses generated from the same inbred sire. Solid lines between crosses represent statistical differences in MSTs (P < 0.05 by ANOVA with Sidak correction); dashed lines represent no differences between MSTs of offspring from the compared crosses.
Paired crosses.
To further assess parental effects, we combined pairs of backcrosses generated with the same inbred dam or inbred sire. Analysis results revealed significant differences between paired groups (P < 0.0001 by F-test). Comparisons for the four backcross pairs are shown in Fig. 1B. MSTs for both comparisons were significant. In particular, offspring from the backcross pair with an S dam survived longer than those from an S sire, and mice generated from crosses with a B sire survived significantly longer than those from a B dam (Table 2 and Fig. 1).
Group B versus S crosses.
Backcrosses were also combined based on similar genotypes (i.e., the four group B backcrosses vs. the four group S backcrosses). The importance of the group B and S pairings was suggested by previous findings demonstrating that resistance genes were present in both parental strains (32). MSTs of group B backcrosses differed from group S backcrosses (P = 0.009).
Assessing Backcross Groups for a Sex Difference
No interaction was found between backcross groups and sex (P = 0.74), demonstrating that the differences between males and females were similar in these backcrosses. Males survived longer than females when the total backcross population was assessed directly (P = 0.005) or when controlling for cross (P = 0.007), paired cross (P = 0.0002), or group B versus S crosses (P = 0.009). In all comparisons of the total backcross population, males survived 5% longer than females, thus verifying the sex difference found in the F2 population. However, when considering MSTs of individual crosses or cross pairs, a male-female difference (P < 0.1) was found only for cross 10 and the backcross pair generated with an S dam, although a trend toward a sex difference was apparent in several crosses (Table 2).
QTL Analysis
With phenotype data in hand, all 935 mice were genotyped for 82 microsatellite markers distributed throughout the 19 autosomes and the X Chr. QTL analysis was performed on the total backcross population and independently for total males and total females using GridQTL (http://www.gridqtl.org.uk/). Complete QTL results of the total backcross population are shown in Table 3; the two highly significant loci are plotted in Fig. 2. Importantly, QTL analysis identified Shali1 and Shali2 as highly significant linkages, confirming the two major QTLs previously identified with F2 analysis. Total backcross females were linked to Shali1, but total males were not. Shali3 was significant for linkage, but Shali4 and the male-specific Shali5 locus on distal Chr 1 were not linked in the total backcross population. Suggestive QTLs were also detected on Chrs 2 and 5 (LOD = 3.1 each), which were not found in the earlier F2 analysis (data not shown). Although a sex difference in HALI MSTs was determined, X Chr linkage was not significant (or even suggestive) in the total backcrosses (LOD score = 0.8) or separate male (LOD score = 0.2) and female (LOD score = 2.1) populations. The result showing no linkage to the X Chr agrees with results from the F2 population.
Table 3.
QTL analyses results for B-S-derived crosses
| B-S Population | Sample Size | Shali1 QTL | Shali5 QTL | Shali2 QTL | Shali4 QTL | Shali3 QTL |
|---|---|---|---|---|---|---|
| Total backcrosses | 935 | 6.0* | 5.5* | 3.7† | ||
| Males | 496 | |||||
| Females | 439 | 3.4 | ||||
| Group B | 456 | 6.0* | ||||
| Males | 242 | 2.6† | ||||
| Females | 214 | 3.4* | ||||
| Group S | 479 | 2.9† | 2.6† | |||
| Males | 254 | |||||
| Females | 225 | |||||
| B dam | 259 | 3.2* | ||||
| S dam | 216 | |||||
| B sire | 197 | 2.2 | 3.5* | |||
| S sire | 263 | 2.5 | 1.9 | |||
| Backcross + F2 | 1775 | 16.0* | 3.7† | 12.0* | 4.6 * | 8.0* |
| Males | 913 | 6.4* | 8.0* | 3.8† | ||
| Females | 862 | 10.4* | 5.3* | 4.4* |
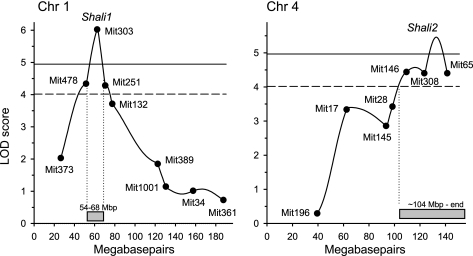
Quantitative trait locus (QTL) analysis (GridQTL) of log survival time was performed separately for males, females, and all mice in the total backcross population, all backcross pairs, and the backcross + F2 cohorts. R/QTL was used to analyze males, females, and total mice for separate group B and S backcrosses. Each significant or suggestive linkage is presented as a peak LOD score. Chromosomal positions were as follows: Shali1 [chromosome (Chr) 1], 54–68 Mbp; Shali5 (Chr 1), 140 Mbp to Chr end; Shali2 (Chr 4), 104 Mbp to Chr end; Shali4 (Chr 9), 88 Mbp to Chr end; and Shali3 (Chr 15), 0-50 Mbp. Significance threshold levels were determined for each population using 10,000 permutations (R/QTL) or 1,000 permutations (GridQTL):
highly significant linkage and
significant linkage. No symbol indicates a suggestive linkage. QTL peaks did not always correlate between groups; thus LOD scores are not always additive.
Fig. 2.
Results from quantitative trait locus (QTL) analysis of the total backcross population. QTL analysis (GridQTL) of 935 backcrosses generated from all 8 possible backcross mating schemes identified 2 highly significant QTLs on chromosome (Chr) 1 (Shali1; LOD score = 6.0) and Chr 4 (Shali2; LOD score = 5.5). The 1.5-LOD score confidence intervals are shown at the bottom of each plot and were determined at 54–68 Mbp for Shali1 and 104 Mbp to the terminus for Shali2. Symbols represent Build 37 locations of the tested MIT microsatellite markers polymorphic between B and S strains of mice. Solid and dashed lines denote highly significant (LOD score = 4.98) and significant (LOD score = 4.02) threshold values, respectively, based on 1,000 permutations of the total backcross dataset.
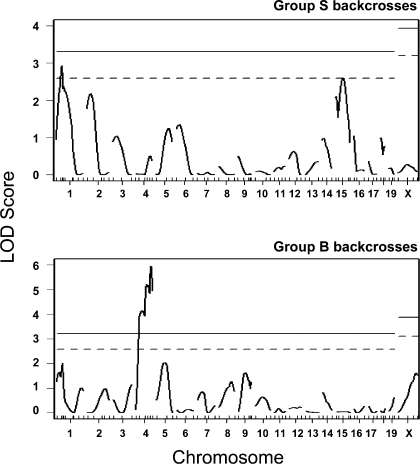
Using the R/QTL package (9), linkage analysis was also performed on total mice and separately for males and females for the group B and S backcrosses. Results were checked using GridQTL, with only slight differences in peak LOD scores noted between the two analysis programs. Genome-wide QTL analysis results for total mice in the separate group B and S backcrosses are shown in Fig. 3. LOD scores for these and the additional subpopulations are shown in Table 3. Shali1 on Chr 1 was significant in the total group S backcrosses but not in the separate group S males (LOD = 1.0) or females (LOD = 2.0) or any of the group B backcrosses (all LOD scores ≤ 2.2). Shali2 on Chr 4 was highly significant in the group B total backcrosses and group B females and significant for group B males but was not significant for any of the group S backcrosses (all LOD scores ≤ 0.7). These results clearly separate the Chr 1 and 4 QTL effects into the group B and S backcrosses (Fig. 3) and together closely correlate with results from the total F2 (32) and total backcrosses (Fig. 2). Linkage for Chr 15 Shali3 in the total backcrosses appears to be primarily due to significant linkage in the total group S backcrosses. Shali4 on Chr 9 and the male-specific QTL Shali5 on distal Chr 1 were not detected in these backcross cohorts. Again, no X Chr linkage was identified in these cohorts.
Fig. 3.
Results from QTL analyses of group B and S backcross populations. QTL analysis was performed separately on group B and S backcrosses using R/QTL (9). Shali1 (Chr 1, LOD score = 2.9) was highly significant and Shali3 (Chr 15, LOD score = 2.6) was significant in group S backcrosses, whereas only Shali2 (Chr 4, LOD score = 6.0) was highly significant in group B backcrosses. X Chr linkage was assessed only in the segregating backcrosses, i.e., crosses 2 and 7 (group B) and crosses 3 and 8 (group S). Significant (dashed lines) and highly significant (solid lines) linkages were based on 10,000 permutations of each dataset. Permutations for the X Chr were performed separately, as recommended (8).
The large backcross population also allowed us to run separate genome-wide QTL analyses on the eight individual backcrosses and for the four backcross pairs. The goal of these different analyses was to see if QTLs tracked with specific crosses, parental strains, or sexes. Although no significant linkage was detected in the individual backcrosses (likely due to the small sample sizes), Shali1–3 QTLs were at least suggestive for linkage in one or more of the paired backcross groups (Table 3). Interestingly, while having the highest MST, the backcross pair generated with an S dam was the only pair that did not show linkage to any Shali QTL.
QTL analysis was also performed on the combined backcross and F2 populations using GridQTL. Not unexpected, highly significant linkage was identified for the previously identified Shali1–4 QTLs in this combined population. Separate analysis of total males and total females detected highly significant linkage for Shali1 and Shali2 but not Shali4 or Shali5. Shali3 was highly significant in females but only suggestive in males. Total LOD scores for all Shali QTLs (except Shali2) approximated the sum of the separate analyses, suggesting a similar QTL peak in both the backcross and F2 datasets. The total LOD score for Shali2 was less than additive, suggesting different QTL peaks for males and females. Separate LOD plots for males and females supported this; males had LOD peaks near 130 and 114 Mbp, whereas females had a single LOD peak near 140 Mbp (data not shown). In agreement, total backcrosses (Fig. 2), group B backcrosses (Fig. 3), and total F2 mice (32) identified several peaks across a large distance of Chr 4 at different levels of significance.
Gene-Gene Interactions
We next looked for evidence of additive and/or epistatic interactions in the backcross population using R/QTL (i.e., twoscan). Since R/QTL is currently unable to analyze all backcrosses together, group B and S backcrosses were assessed separately. Based on 1,000 permutations of the respective datasets, neither group B nor S backcrosses reached suggestive significance for any pairwise interactions. Among the QTLs, the group S backcrosses showed additive effects for Shali1 and Shali5 (LOD = 5.1) and for Shali1 and Shali3 (LOD = 6.6). Group B backcrosses had additive effects for Shali1 and Shali2 (LOD = 8.1) and for Shali2 and Shali4 (LOD = 8.0).
Allelic Analysis and Penetrance
Single QTL.
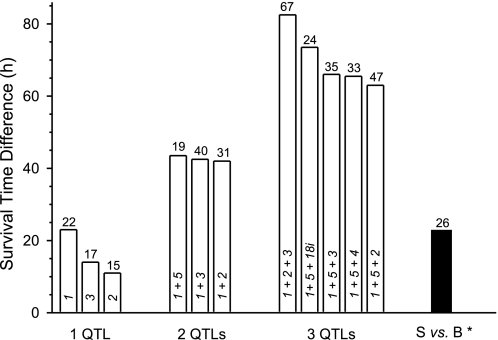
To estimate the phenotypic contribution of the Shali QTLs, the separate and combined allelic effects (i.e., the effect of genotype on the MST) of the peak markers for each Shali QTL were determined for total backcrosses, separate group B and S backcrosses, and the combined backcross plus F2 population. Results for the three genotypes of the Shali1 and Shali2 combination are shown for the different crosses in Table 4 and Suppl. Table 1. For total backcrosses, the marker with the largest difference in MSTs between mice that were homozygous SS versus BB was D1Mit303, as expected from QTL analysis results. Mice that were homozygous SS for D1Mit303 had a 16 h longer MST than mice that were homozygous BB for D1Mit303. Unexpected, mice that were SS, BS, or BB for D4Mit308 had no differences in MSTs, which differs from F2 results that clearly demonstrated the BB genotype imparted resistance. D4Mit308 (14 h) and D9Mit355 (11 h) had the largest differences in group B backcrosses, whereas D1Mit303 (16 h) and D15Mit184 (14.5 h) differed the most in group S backcrosses (data not shown). In the total F2 plus backcross population, mice that were homozygous SS for D1Mit303 (representing Shali1) had a MST of 23 h longer than mice that were homozygous BB for this marker (Fig. 4), with a penetrance of 22%. Shali2 and Shali3 each had a lesser effect on survival time.
Table 4.
Allelic effects of the two major QTLs on survival time in hyperoxia for total backcrosses and eight separate backcrosses
|
D1Mit303 (Shali1) |
D4Mit308 (Shali2)
|
|||||
|---|---|---|---|---|---|---|
| BB | BS | SS | BB | BS | SS | |
| Total backcross (n = 935*) | ||||||
| MST, h | 120 | 123 | 136 | 128 | 123 | 126 |
| 25th and 75th percentiles | (106, 137) | (105, 144) | (112, 161) | (113, 144) | (105, 146) | (100, 150) |
| n | 226 | 469 | 238 | 231 | 467 | 222 |
| Females (n = 439*) | ||||||
| MST, h | 114 | 114 | 136 | 127 | 119 | 124 |
| 25th and 75th percentiles | (103, 137) | (103, 137) | (112, 161) | (113, 145) | (104, 137) | (98, 147) |
| n | 104 | 227 | 108 | 104 | 219 | 111 |
| Males (n = 496*) | ||||||
| MST, h | 122 | 127 | 136 | 129 | 125 | 126 |
| 25th and 75th percentiles | (112, 137) | (111, 147) | (111, 172) | (114, 144) | (106, 151) | (102, 153) |
| n | 122 | 242 | 130 | 127 | 248 | 111 |
| Cross 2 (n = 97) | ||||||
| MST, h | 116 | 125 | 133.5 | 113 | ||
| 25th and 75th percentiles | (106, 137) | (112, 137) | (113, 141) | (105, 130) | ||
| n | 52 | 45 | 44 | 52 | ||
| Cross 4 (n = 154) | ||||||
| MST, h | 120 | 123 | 125 | 114 | ||
| 25th and 75th percentiles | (106, 137) | (113, 137) | (114, 137) | (106, 137) | ||
| n | 72 | 81 | 79 | 74 | ||
| Cross 5 (n = 105) | ||||||
| MST, h | 114 | 120 | 122 | 113 | ||
| 25th and 75th percentiles | (102, 130) | (103, 129) | (106, 135) | (95, 123) | ||
| n | 56 | 49 | 55 | 49 | ||
| Cross 7 (n = 100) | ||||||
| MST, h | 129 | 137 | 146 | 129 | ||
| 25th and 75th percentiles | (113, 151) | (127, 183) | (127, 174) | (113, 151) | ||
| n | 46 | 53 | 53 | 46 | ||
| Cross 3 (n = 106) | ||||||
| MST, h | 113 | 136 | 129 | 118 | ||
| 25th and 75th percentiles | (91.5, 140) | (113, 153) | (102, 161) | (101, 143) | ||
| n | 48 | 58 | 55 | 51 | ||
| Cross 8 (n = 157) | ||||||
| MST, h | 112 | 126 | 113 | 120 | ||
| 25th and 75th percentiles | (90, 130) | (103, 146) | (102, 136) | (98, 145) | ||
| n | 83 | 74 | 76 | 73 | ||
| Cross 9 (n = 122) | ||||||
| MST, h | 130 | 146 | 142 | 137 | ||
| 25th and 75th percentiles | (99, 160) | (113, 211) | (102, 196) | (111, 160) | ||
| n | 59 | 63 | 66 | 53 | ||
| Cross 10 (n = 94) | ||||||
| MST, h | 139 | 135 | 144 | 129 | ||
| 25th and 75th percentiles | (106, 184) | (115, 192) | (128, 198) | (105, 168) | ||
| n | 51 | 43 | 49 | 45 | ||
Within each backcross cohort, the MST of all mice with the same genotype at D1Mit303 or D4Mit303 (the peak markers representing the Shali1 and Shali2 effects, respectively) were calculated as described in materials and methods.
Total n will not add up to the population number due to missing genotypes. Only mice with all genotypes for the respective marker(s) were included in the calculations.
Fig. 4.
Additive effects of Shali QTLs. In the combined backcross plus F2 dataset, the contribution of each QTL or QTL combination to overall survival time was determined by calculating the MST difference between mice containing resistance alleles versus sensitivity alleles for the representative peak marker of each QTL (i.e., D1Mit303, D4Mit308, D9Mit355, D15Mit184, D1Mit34, and D18Mit177). For example, the calculated MST of all mice that were homozygous B for D1Mit303 was compared with the MSTs of mice that were homozygous S or heterozygous for D1Mit303. This was extended to include two- and three-QTL combinations. The largest difference between the most sensitive and resistant genotypes are presented for one QTL and for the best two- and three-QTL combinations (which always includes the Chr 1 QTL). Numbers within bars represent the Shali QTLs (i.e., 1 = Shali1, Chr 1; 2 = Shali2, Chr 4; 3 = Shali3, Chr 15; 4 = Shali4, Chr 9; 5 = Shali5, distal Chr 1; and 18i = Shali18i, interaction QTL on Chr 18). The number at the top of each bar corresponds to the percent penetrance of the resistance trait (i.e., the percentage of mice that carry the specific resistance allelic combination with a MST > 168 h), as previously defined (33). *The parental strain difference refers to all mice tested to date (n = 169 B mice and 180 S mice) and includes those previously reported (33).
Two QTLs.
In the total backcross population, several two-QTL combinations resulted in MST differences between sensitive and resistant alleles of ∼24 h, including Shali1 and Shali2, Shali1 and Shali3, Shali1 and Shali4, Shali3 and Shali4, and Shali4 and Shali18i. The calculated MSTs for the seven different Shali1–2 genotypes in the total backcrosses, males, females, and group B and S backcrosses are shown in Suppl. Table 2.
These differences were further highlighted in the combined backcross and F2 dataset, which includes all sensitive and resistant genotypes. When identifying the most extreme sensitive and resistant genotype combinations, Shali1 and Shali5 (MST difference = 45.5 h), Shali1 and Shali2 (MST difference = 42.5 h), and Shali1 and Shali3 (MST difference = 42 h) were most significant (Fig. 4). These results quantified findings from the twoscan analysis of R/QTL, which detected these QTL-QTL additive effects. The penetrance of the resistance trait in mice carrying these top three allelic combinations varied from 19% to 40% (Fig. 4).
Three or more QTLs.
Looking at three or more QTLs simultaneously further highlighted their additive effects and the different allelic combinations that could produce a significant survival difference. For instance, in the combined backcross and F2 dataset, the MSTs of mice with the best resistant and sensitive allelic combinations differed by 82.5 h for markers representing Shali1, Shali2, and Shali3 (BB-SS-SS = 183.5 h vs. SS-BB-H = 101 h). Other 3-QTL combinations had MST differences of 63–73.5 h between the sensitive and resistant allelic combinations (Fig. 4). Interestingly, the best 2-QTL combination was included in all but the best 3-QTL combination, suggesting an additive role of Shali2, Shali3, Shali4 or Shali18i to the Shali1-Shali5 combination. Penetrance of the survival trait in mice with resistance alleles for 3-QTL combinations was again improved, to as high as 67% in those with resistance alleles for Shali1–3 (8 of 12 mice). Further supporting this allelic combination, only 1 of 24 mice with sensitive alleles at Shali1–3 demonstrated resistance (data not shown). Though additional QTLs in combination suggested similar additive trends, sample sizes for individual sensitive and resistant allelic combinations were smaller and less reliable.
Mode of Inheritance
In addition to identifying allelic combinations for sensitivity and resistance, this large backcross dataset, with and without the previous F2 dataset, allowed us to predict the likely mode of inheritance of each QTL. Three inheritance patterns were identified, including the S allele dominant or additive for resistance, the B allele dominant or additive for resistance, and the B allele dominant or additive for sensitivity (Table 5). While almost always intermediate in the different F2 crosses (32), survival times of mice heterozygous at peak markers depended on the QTL; MSTs of heterozygous mice were intermediate to mice with homozygous genotypes for Shali1, similar to the BB genotype for Shali4 and Shali18i, or showed no difference between homozygous genotypes for Shali2 and Shali5. From this, we predicted the best sensitive and resistant allelic genotypes for recombinant mice (Table 5).
Table 5.
Predicted heritability patterns for each Shali QTL
| Shali1 | Shali2 | Shali3 | Shali4 | Shali5 | Shali18i | |
|---|---|---|---|---|---|---|
| Sensitive genotypes | BB | SS | BB | SS | BB SB | BB |
| Resistant genotypes | SS SB | BB SB | SS SB | BB SB | SS | SS SB |
| Heritability | S dominant or additive for resistance | B dominant or additive for resistance | S dominant or additive for resistance | B dominant or additive for resistance | B dominant or additive for sensitivity | S dominant or additive for resistance |
| Most sensitive genotype | BB | SS | BB | SS | BB | BB |
| Most resistant genotype | SS | BB | SS | BB | SS | SS |
Chromosomal locations are as follows: Shali1, Chr 1; Shali2, Chr 4; Shali3, Chr 15; Shali4, Chr 9; Shali5, Chr 1; and Shali18i, Chr 18.
DISCUSSION
Mortality associated with ALI remains higher than acceptable, and treatments beyond supportive care to reduce this level have not proven to be reliably effective. Because the death of ALI patients more often results from causes beyond refractory respiratory failure (15, 26, 43), we established a mouse model to identify genes linked to mortality with ALI. Using a mouse model of HALI, we previously embarked on a forward genetics approach using QTL analysis to identify critical regions controlling outcome differences in mice. Not surprisingly, this trait is complex, with at least six QTLs affecting the survival time difference between B and S strains of inbred mice. Unexpectedly, we revealed that the sensitive parental strain carried resistance QTLs, which contributed to prolonged survival in recombinant offspring. Overall survival times were also affected by sex and cross, with clear parent of origin effects in reciprocal F2 populations (32, 33). To validate these F2 results and to further characterize the inheritance pattern and relative importance of the significant QTLs, we generated and analyzed a large backcross population consisting of mice from all eight possible mating schemes.
Significant cross and/or sex differences were determined between many of the backcross cohorts. For example, MSTs of backcrosses generated with a maternal or grandmaternal S strain were resistant except when an inbred S mouse was the sire. Similarly, backcrosses generated from a maternal or grandmaternal B strain were more sensitive, although less so when an inbred S mouse was the sire. On average, males survived 5% longer than females. Phenotype differences between the sexes are not uncommon, having been reported in many diseases, including several lung-related phenotypes (14, 17, 31, 41, 42). Whether this sex difference is biologically relevant is still to be determined. Backcross data were also consistent with a maternal inheritance pattern for HALI survival time, a finding that has now been supported by all four F2 crosses and seven of the eight backcrosses. Further evidence in F2 and backcross mice suggested that paternal inheritance of one or more genes may also be important (32). Accordingly, progeny from the backcross pair generated with a B sire were more resistant than those from an S sire. Interestingly, F2 analysis was consistent with HALI survival time tracking with the Y Chr. However, the MST for six of eight backcrosses disagreed with the paternal grandfather strain, effectively ruling out Y Chr inheritance. These results confirmed the sex, cross, and parent of origin effects previously identified in the F2 population.
Parent of origin effects can be caused by several genetic and epigenetic phenomena, including imprinting and other sources of differential allelic expression and maternal (including mitochondrial inheritance) and paternal inheritance as well as nongenetic maternal effects (i.e., intrauterine and rearing effects). Because homozygous mothers can only give rise to one of the reciprocal heterozygotes, maternal effects can mimic patterns of phenotypic variation expected for various forms of imprinting (19). Our results were consistent with maternal inheritance but could also be explained by an imprint that turns on maternal gene expression or turns off paternal gene expression. Because mitochondrial DNA is almost entirely inherited through the maternal germline, mitochondrial inheritance could have a role in HALI survival time. As with maternal inheritance, seven of eight backcrosses were consistent with mitochondrial inheritance; only the SB.S backcross disagreed. Mitochondrial dysfunction has been implicated in many diseases (30, 37), including numerous lung diseases (6, 10, 35). In addition, HALI is known to damage the pulmonary endothelium and vasculature (12), and hyperoxia induced apoptosis of epithelial cells involved mitochondrial pathways (28). Mitochondrial electron transport chain activity was compromised in lung epithelial cells isolated from rats after HALI (3). These data suggest that mitochondrial damage is an important factor in lung injury. However, mitochondrial diseases are multisystemic and virtually every organ system can be involved. Since ALI mortality often involves other organs, mitochondrial DNA mutations in nonpulmonary organs and tissues could be also relevant. Nonetheless, although not ruled out specifically, mitochondrial inheritance seems less likely to contribute to HALI survival time than maternal inheritance of nuclear genes.
QTL analysis of total backcrosses verified the significant linkages of Shali1–3. Combining this backcross population with the previous F2 group identified Shali1–5 as highly significant, with Shali1–2 being highly significant in separate cohorts of males and females. Because the backcross population was generated from the same two inbred strains used for the large F2 population, we anticipated the confirmation of these QTLs; however, we also expected to further resolve the QTL intervals. The Shali1 interval determined for the backcross population was 54–68 Mbp on Chr 1. This interval fits within the Shali1 interval identified for the F2 population (i.e., 50–71 Mbp), thereby refining Shali1 to this 14-Mbp interval. The QTL interval for Shali2 was also refined to some extent, although suggestive evidence remains for at least three separate QTLs on Chr 4. QTL results of backcrosses suggested that the highest linkage is for the distal third of Chr 4, with the 1.5-LOD confidence interval spanning from 104 Mbp to the terminus. This region appears to harbor two of the three aforementioned QTLs identified in the F2 analysis. The more proximal QTL, which was highly significantly linked in the F2 population, did not reach significance in the backcrosses. Nonetheless, QTL analysis results from the combined F2 and backcross populations identified three peaks for Shali2 with LOD scores over 10, spanning from 58 Mbp to the terminus. The peak for Shali3 was again mapped to the central region of Chr 15 (near 40 Mbp), but, as seen in the F2 population, the peak was broad and shallow and still contained much of Chr 15. Shali4 at the distal end of Chr 9 was confirmed in combined analysis. The broad (Shali3) and multiple-peaked (Shali2) QTLs can be ascribed to many causes, including several loci within the QTL intervals, decreased penetrance of the trait, gene-gene interactions, and other genetic and epigenetic phenomena. For these collective QTL intervals, conservative congenic lines or full Chr substitution strains will be required for Shali2 and Shali3, but refined congenic lines should suffice for Shali1 and Shali4.
Dividing the total backcross population into group S and B backcrosses provided an interesting validation of the total backcross and F2 QTL results. Whereas the total backcross and total F2 populations identified all five Shali QTLs, group S backcrosses identified Shali1 and Shali3 and group B backcrosses detected only Shali2. These results further supported that Shali1 and Shali3 are dominant or additive for resistance in the S strain (group S backcrosses were significantly linked to Shali1 and Shali3) and Shali2 is dominant or additive resistance QTLs in the B strain (group B backcrosses were highly significant linked to Shali2). Because the total backcross population contains all three genotypes (SS, SB, and BB), all significant loci could be detected. However, since the separate group B and S backcross sets only contain heterozygous and one of the homozygous genotypes, and heterozygosity yielded a reduced survival time, the major Shali loci were sorted in the backcrosses primarily due to their homozygous dominant effects. These findings agree with previous QTL and segregation analyses results that have suggested the existence of dominant HALI resistance alleles in both parental strains (32, 33).
To estimate the specific contributions that the linked QTLs had on survival time, we calculated the separate and combined allelic effects in mice carrying the different genotypes for the peak markers representing the Shali loci in the total backcrosses, group B and S backcrosses, and combined populations of F2 and total backcrosses. Compared with the previous F2 results, backcrosses were less likely to detect significant MST differences between genotypes for the established Shali QTLs, and, when detected, the differences were smaller. This was expected because mice heterozygous for the different peak markers demonstrated variable sensitivity; heterozygotes were intermediate in phenotype for Shali1, similar to the sensitive homozygotes for Shali4 and Shali18i, or not significantly different than either homozygote for Shali2, Shali3, and Shali5. Still, the combination of the F2 and backcross data confirmed the significant differences for all Shali QTLs. In fact, significance of the major Shali loci was further supported by including both homozygous genotypes in a single file (as in the F2 dataset) and by increasing the number of mice with specific sensitive and resistant genotypes for the markers of interest.
Combining alleles of peak markers for multiple QTLs was useful to identify and quantify the QTL-QTL additive effects between the major loci. The estimated contributing effects for the different Shali combinations revealed that many different combinations are potentially effective and suggested possible functional overlap among the loci. The data were the strongest and most consistent for three-QTL combinations, reaching MST differences between sensitive and resistant genotypes of 3 days or more, with improved penetrance for several combinations of three loci. Although additional QTLs in combination suggested similar additive trends, sample sizes for the individual sensitive and resistant allelic combinations were smaller and less reliable. Nonetheless, the trend highlighted the potential importance of all identified Shali QTLs. For example, backcross mice with resistance alleles for Shali1–5 had a MST of 184 h (n = 9), whereas mice with sensitivity alleles for all five QTL peak markers had a MST of 89 h (n = 9), an average survival difference of 4 days, representing a doubling of the MSTs between the groups. Moreover, six of nine mice with resistance alleles had a resistant phenotype (67% penetrance), whereas zero of nine mice with sensitivity alleles for these five loci had a resistant phenotype. This correlation was stronger than expected, given that the remainder of the genome of each mouse varied outside these QTL markers and gives hope that, with the right combination of genes and alleles, survival time and penetrance can be improved considerably over those for the S strain.
From these results, we predicted the most sensitive and resistant allelic genotypes for recombinant mice (Table 5). These data suggest that S mice can be made more resistant by introducing resistant B alleles at Shali2 and/or Shali4 and B mice can potentially be made more sensitive by introducing sensitive S alleles at Shali2 and/or Shali4. As proof for this concept, we are currently generating reciprocal consomic or congenic lines for all six Shali loci (Shali1–5 and Shali18i). When constructed, we will have the ability to generate mice containing any combination of sensitive and resistant QTLs in either background strain, which are deemed important in individual congenic strain analyses.
Many previously identified QTLs relate to oxidant lung injury, inflammation, or function and overlap the Shali intervals. Two QTLs, including the modifier of LPS response 2 (Mol2) and a suggestive locus for nickel-induced acute lung injury, map to the Shali1 interval (23, 34). The Shali2 interval contains at least three QTLs of interest, including Lhyp, a locus controlling ozone-induced lung hyperpermeability (21), a significant but unnamed QTL for silica-induced pulmonary fibrosis (27), and a suggestive locus for pulmonary tolerance to zinc oxide exposure (48). The Shali3 interval includes Bhr2, a locus for bronchial hyperresponsiveness (13), and several QTLs controlling pulmonary function (16). A QTL for macrophages in bronchoalveolar lavage after butylated hydroxytoluene administration (22) and a suggestive locus for nickel-induced acute lung injury (34) map within Shali4 on distal Chr 9. Specific candidate genes for these QTLs, found in the original reports, can be considered as candidates for the Shali QTLs and, together with those previously described for the Shali loci (33), make up a preliminary list of positional candidate genes for HALI survival time. As noted earlier, however, mortality often results from nonpulmonary causes, so candidate genes associated with other organs and tissues must also be considered.
In summary, the two major QTLs (Shali1 and Shali2) previously identified with highly significant linkage to HALI survival time in a large F2 population were confirmed in a large backcross population generated from the same B and S inbred strains. Most evidence supports maternal inheritance for survival time, with significant cross and sex differences. Results further support that the two major loci, together in different combinations with several other modifying loci, can enhance survival time and increase penetrance. These findings have set the stage for generating congenics to further resolve these major QTLs and to begin to predict and test viable candidate genes. In addition, multicongenics that carry combinations of beneficial QTLs should lend insight into the potential additive gene effects for improved HALI outcome.
GRANTS
This work was initially supported by the Division and Program in Human Genetics at Cincinnati Children's Hospital Research Foundation and subsequently by National Heart, Lung, and Blood Institute Grant HL-75562 (to D. R. Prows).
Supplementary Material
Acknowledgments
The authors thank Erin Full, Amanda Hafertepen, Andrea Hogan, Michelle Horner, Matt Monfils, Dusti Folger-Snider, and Shannon Speelman for excellent technical assistance. R/QTL support was received from Dr. Karl Broman (Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, WI).
Footnotes
Supplemental material for this article is available online at the Physiological Genomics website.
REFERENCES
- 1.ARDS Network. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. The Acute Respiratory Distress Syndrome Network. N Engl J Med 342: 1301–1308, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet 2: 319–323, 1967.4143721 [Google Scholar]
- 3.Bassett DJ, Elbon CL, Reichenbaugh SS. Respiratory activity of lung mitochondria isolated from oxygen-exposed rats. Am J Physiol Lung Cell Mol Physiol 263: L439–L445, 1992. [DOI] [PubMed] [Google Scholar]
- 4.Bhandari V, Choo-Wing R, Homer RJ, Elias JA. Increased hyperoxia-induced mortality and acute lung injury in IL-13 null mice. J Immunol 178: 4993–5000, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Bhandari V, Choo-Wing R, Lee CG, Zhu Z, Nedrelow JH, Chupp GL, Zhang X, Matthay MA, Ware LB, Homer RJ, Lee PJ, Geick A, de Fougerolles AR, Elias JA. Hyperoxia causes angiopoietin 2-mediated acute lung injury and necrotic cell death. Nat Med 12: 1286–1293, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet S, Michelakis ED, Porter CJ, Andrade-Navarro MA, Thebaud B, Bonnet S, Haromy A, Harry G, Moudgil R, McMurtry MS, Weir EK, Archer SL. An abnormal mitochondrial-hypoxia inducible factor-1alpha-Kv channel pathway disrupts oxygen sensing and triggers pulmonary arterial hypertension in fawn hooded rats: similarities to human pulmonary arterial hypertension. Circulation 113: 2630–2641, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Bosma KJ, Lewis JF. Emerging therapies for treatment of acute lung injury and acute respiratory distress syndrome. Expert Opin Emerg Drugs 12: 461–477, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM, Churchill GA. The X chromosome in quantitative trait locus mapping. Genetics 174: 2151–2158, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Carraway MS, Suliman HB, Kliment C, Welty-Wolf KE, Oury TD, Piantadosi CA. Mitochondrial biogenesis in the pulmonary vasculature during inhalational lung injury and fibrosis. Antioxid Redox Signal 10: 269–275, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow CW, Herrera Abreu MT, Suzuki T, Downey GP. Oxidative stress and acute lung injury. Am J Respir Cell Mol Biol 29: 427–431, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Crapo JD, Barry BE, Foscue HA, Shelburne J. Structural and biochemical changes in rat lungs occurring during exposures to lethal and adaptive doses of oxygen. Am Rev Respir Dis 122: 123–143, 1980. [DOI] [PubMed] [Google Scholar]
- 13.De Sanctis GT, Merchant M, Beier DR, Dredge RD, Grobholz JK, Martin TR, Lander ES, Drazen JM. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nat Genet 11: 150–154, 1995. [DOI] [PubMed] [Google Scholar]
- 14.de Torres JP, Cote CG, Lopez MV, Casanova C, Diaz O, Marin JM, Pinto-Plata V, de Oca MM, Nekach H, Dordelly LJ, Aguirre-Jaime A, Celli BR. Gender differences in mortality in patients with COPD. Eur Respir J 33: 528–535, 2009. [DOI] [PubMed] [Google Scholar]
- 15.Ferring M, Vincent JL. Is outcome from ARDS related to the severity of respiratory failure? Eur Respir J 10: 1297–1300, 1997. [DOI] [PubMed] [Google Scholar]
- 16.Ganguly K, Stoeger T, Wesselkamper SC, Reinhard C, Sartor MA, Medvedovic M, Tomlinson CR, Bolle I, Mason JM, Leikauf GD, Schulz H. Candidate genes controlling pulmonary function in mice: transcript profiling and predicted protein structure. Physiol Genomics 31: 410–421, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Gharaee-Kermani M, Hatano K, Nozaki Y, Phan SH. Gender-based differences in bleomycin-induced pulmonary fibrosis. Am J Pathol 166: 1593–1606, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunther A, Walmrath D, Grimminger F, Seeger W. Pathophysiology of acute lung injury. Semin Respir Crit Care Med 22: 247–258, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Hager R, Cheverud JM, Wolf JB. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics 178: 1755–1762, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, Kim HP, Chi M, Ifedigbo E, Ryter SW, Choi AM. Deletion of caveolin-1 protects against oxidative lung injury via up-regulation of heme oxygenase-1. Am J Respir Cell Mol Biol 39: 171–179, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleeberger SR, Reddy S, Zhang LY, Jedlicka AE. Genetic susceptibility to ozone-induced lung hyperpermeability: role of Toll-like receptor 4. Am J Respir Cell Mol Biol 22: 620–627, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Malkinson AM, Radcliffe RA, Bauer AK. Quantitative trait locus mapping of susceptibilities to butylated hydroxytoluene-induced lung tumor promotion and pulmonary inflammation in CXB mice. Carcinogenesis 23: 411–417, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Matesic LE, De Maio A, Reeves RH. Mapping lipopolysaccharide response loci in mice using recombinant inbred and congenic strains. Genomics 62: 34–41, 1999. [DOI] [PubMed] [Google Scholar]
- 24.Matthay MA, Bhattacharya S, Gaver D, Ware LB, Lim LH, Syrkina O, Eyal F, Hubmayr R. Ventilator-induced lung injury: in vivo and in vitro mechanisms. Am J Physiol Lung Cell Mol Physiol 283: L678–L682, 2002. [DOI] [PubMed] [Google Scholar]
- 25.Matthay MA, Zimmerman GA. Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am J Respir Cell Mol Biol 33: 319–327, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery AB, Stager MA, Carrico CJ, Hudson LD. Causes of mortality in patients with the adult respiratory distress syndrome. Am Rev Respir Dis 132: 485–489, 1985. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuka Y, Wang XT, Saito J, Ishida T, Munakata M. Genetic linkage analysis of pulmonary fibrotic response to silica in mice. Eur Respir J 28: 1013–1019, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Pagano A, Donati Y, Metrailler I, Barazzone Argiroffo C. Mitochondrial cytochrome c release is a key event in hyperoxia-induced lung injury: protection by cyclosporin A. Am J Physiol Lung Cell Mol Physiol 286: L275–L283, 2004. [DOI] [PubMed] [Google Scholar]
- 29.Phua J, Badia JR, Adhikari NK, Friedrich JO, Fowler RA, Singh JM, Scales DC, Stather DR, Li A, Jones A, Gattas DJ, Hallett D, Tomlinson G, Stewart TE, Ferguson ND. Has mortality from acute respiratory distress syndrome decreased over time?: a systematic review. Am J Respir Crit Care Med 179: 220–227, 2009. [DOI] [PubMed] [Google Scholar]
- 30.Pieczenik SR, Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp Mol Pathol 83: 84–92, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Postma DS Gender differences in asthma development and progression. Gend Med 4, Suppl B: S133–S146, 2007. [DOI] [PubMed]
- 32.Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ Jr, Liu CY, Nick TG. Genetic analysis of hyperoxic acute lung injury survival in reciprocal intercross mice. Physiol Genomics 30: 271–281, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ Jr, Nick TG. A genetic mouse model to investigate hyperoxic acute lung injury survival. Physiol Genomics 30: 262–270, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Prows DR, Leikauf GD. Quantitative trait analysis of nickel-induced acute lung injury in mice. Am J Respir Cell Mol Biol 24: 740–746, 2001. [DOI] [PubMed] [Google Scholar]
- 35.Ratner V, Starkov A, Matsiukevich D, Polin RA, Ten VS. Mitochondrial dysfunction contributes to alveolar developmental arrest in hyperoxia-exposed mice. Am J Respir Cell Mol Biol 40: 511–518, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 353: 1685–1693, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Schapira AH Mitochondrial disease. Lancet 368: 70–82, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Sidak Z Rectangular confidence region for the means of multivariate normal distributions. J Am Stat Assoc 62: 626–633, 1967. [Google Scholar]
- 39.Sloane PJ, Gee MH, Gottlieb JE, Albertine KH, Peters SP, Burns JR, Machiedo G, Fish JE. A multicenter registry of patients with acute respiratory distress syndrome. Physiology and outcome. Am Rev Respir Dis 146: 419–426, 1992. [DOI] [PubMed] [Google Scholar]
- 40.Suchyta MR, Clemmer TP, Elliott CG, Orme JF Jr, Weaver LK. The adult respiratory distress syndrome. A report of survival and modifying factors. Chest 101: 1074–1079, 1992. [DOI] [PubMed] [Google Scholar]
- 41.Van Winkle LS, Gunderson AD, Shimizu JA, Baker GL, Brown CD. Gender differences in naphthalene metabolism and naphthalene-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol 282: L1122–L1134, 2002. [DOI] [PubMed] [Google Scholar]
- 42.Vancza EM, Galdanes K, Gunnison A, Hatch G, Gordon T. Age, strain, and gender as factors for increased sensitivity of the mouse lung to inhaled ozone. Toxicol Sci 107: 535–543, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vincent JL, Zambon M. Why do patients who have acute lung injury/acute respiratory distress syndrome die from multiple organ dysfunction syndrome? Implications for management. Clin Chest Med 27: 725–731, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Ward NS, Waxman AB, Homer RJ, Mantell LL, Einarsson O, Du Y, Elias JA. Interleukin-6-induced protection in hyperoxic acute lung injury. Am J Respir Cell Mol Biol 22: 535–542, 2000. [DOI] [PubMed] [Google Scholar]
- 45.Ward PA Acute lung injury: how the lung inflammatory response works. Eur Respir J Suppl 44: 22s–23s, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Ward PA, Mulligan MS. Molecular mechanisms in acute lung injury. Adv Pharmacol 24: 275–292, 1993. [DOI] [PubMed] [Google Scholar]
- 47.Waxman AB, Einarsson O, Seres T, Knickelbein RG, Warshaw JB, Johnston R, Homer RJ, Elias JA. Targeted lung expression of interleukin-11 enhances murine tolerance of 100% oxygen and diminishes hyperoxia-induced DNA fragmentation. J Clin Invest 101: 1970–1982, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wesselkamper SC, Chen LC, Gordon T. Quantitative trait analysis of the development of pulmonary tolerance to inhaled zinc oxide in mice. Respir Res 6: 73, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westfall P, Tobias R, Rom D, Wolfinger R, Hochberg Y. Multiple Comparisons and Multiple Tests Using SAS. Cary, NC: SAS Institute, 1999.
- 50.Zambon M, Vincent JL. Mortality rates for patients with acute lung injury/ARDS have decreased over time. Chest 133: 1120–1127, 2008. [DOI] [PubMed] [Google Scholar]
- 51.Zilberberg MD, Epstein SK. Acute lung injury in the medical ICU: comorbid conditions, age, etiology, and hospital outcome. Am J Respir Crit Care Med 157: 1159–1164, 1998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.