Abstract
New Zealand obese (NZO) mice present a metabolic syndrome of obesity, insulin resistance, and diabetes. To identify chromosomal segments associated with these traits, we intercrossed NZO mice with the lean and diabetes-resistant C57BL/6J (B6) strain. Obesity and hyperglycemia in the (NZO×B6)F2 intercross population were predominantly due to a broad quantitative trait locus (QTL) on chromosome 1 (Nob3; logarithm of the odds score 16.1, 16.0, 4.0 for body weight, body fat, and blood glucose, respectively), producing a difference between genotypes of 12.7 or 5.2 g of body weight and 12.0 or 4.0 g of body fat in females or males, respectively. In addition, significant QTL on chromosomes 3 and 13 and suggestive QTL on chromosomes 4, 6, 9, 12, 14, and 19 contributed to the obese phenotype. Distal chromosome 5 was significantly linked with plasma cholesterol (LOD score 10.7). Introgression of two segments of Nob3 into B6 confirmed the adipogenic effect of the QTL and suggested the presence of at least one causal gene. Haplotype mapping reduced the critical region of the distal part of the QTL to 31 Mbp containing the potential candidates Nr1i3, Apoa2, Atp1a2, Prox1, and Hsd11b1. We conclude that obesity and hyperglycemia of NZO is to a large part caused by variant genes located in Nob3 on chromosome 1. Since these exert robust effects on a B6 background, the QTL Nob3 is a prime target for identification of a novel diabesity gene.
Keywords: positional cloning, metabolic syndrome, diabetes, cholesterol, New Zealand obese mouse
the identification of obesity genes in monogenic mouse models has considerably helped to elucidate the mechanisms of energy balance and fat storage (16). In contrast, the genetic basis of polygenic obesity and diabetes in mice and humans is incompletely understood. In whole-genome scans of outcross populations of inbred mouse strains, numerous genomic segments [quantitative trait loci (QTL)] conferring susceptibility for adiposity and hyperglycemia have been identified (24, 31). However, most obesity QTL produce small effects that are markedly dependent on the background strain (27) and that frequently disappear after introgression into a lean strain. Thus, positional cloning of the responsible gene by conventional introgression of a QTL into a second strain has so far been successful in only a few cases such as Sorcs1 (6) and Tbc1d1 (4). Other strategies have therefore been used to identify adipogenic variants in the QTL by haplotype mapping, sequencing of candidates, mRNA profiling, and/or interspecies comparisons (3, 5, 8, 25, 28).
New Zealand obese (NZO) mice present a syndrome of morbid obesity, insulin resistance, hypertension, and hypercholesterolemia that resembles the human metabolic syndrome (1, 10, 17). As a consequence of this syndrome, male NZO mice develop Type 2-like diabetes characterized by marked hyperglycemia, low serum insulin levels, and β-cell destruction (7, 15). Thus, we and others have previously used outcross progeny of NZO with SJL, Small, or NON for identification of obesity and diabetes QTL (9, 12, 15, 19, 22, 29). In the present study, we generated a (NZO×B6)F2 population to study the effects of diabetogenic alleles from NZO on a diabetes-resistant background [C57BL/6J (13, 14)]. In a genome-wide scan of this population, we identified a major QTL on chromosome 1 (Nob3) that conferred obesity and hyperglycemia. We generated congenic lines carrying Nob3 on the B6 background, thereby successfully isolating the QTL and providing a strategy for positional cloning of the responsible gene.
MATERIALS AND METHODS
Animals and diets.
Female NZO mice from our own colony (NZO/HIBomDife: Dr. R. Kluge, German Institute of Human Nutrition, Nuthetal, Germany) and C57BL/6J (Charles River, Sulzfeld, Germany) were used throughout. Mice were singly housed at a temperature of 22°C with a 12:12 h light-dark cycle (lights on at 6:00 AM) in type II or type III macrolon cages with soft wood bedding. Standard chow (ssniff, Soest, Germany; maintenance diet for rats and mice, art. no. V153xR/M-H) contained 19% (wt/wt) protein, 3.3% fat, and 54.1% carbohydrates, with 26, 10, and 74% of total digestible energy (12.8 kJ/g) from protein, fat, and carbohydrates, respectively. The high-fat diet (purified, Altromin, Lage, Germany; art. no. C1057) contained (wt/wt) 18% protein, 15% fat, and 46% carbohydrates (4.8% monosaccharides, 11.1% disaccharides, 30.4% polysaccharides), with 18, 35, and 47% of total digestible energy (16.2 kJ/g) from protein, fat, and carbohydrates. The animals were kept in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals, and all experiments were approved by the Ethics Committee of the Ministry of Agriculture, Nutrition and Forestry (State of Brandenburg, Germany).
Breeding strategy.
NZO females were mated to C57BL/6J (B6) males to produce F1 hybrids, and F1 progeny was intercrossed to generate a total of 649 F2 mice (318 females, 331 males). Congenic strains B6.NZO-Nob3.91 [B6.NZO-Nob3(D1Mit468-D1Mit209)] and B6.NZO-Nob3.38 [B6.NZO-Nob3(D1Mit202-D1Mit209)] were generated from male (NZOxB6)F1 mice, which were backcrossed with B6 females. Single N2-N5 males selected for the presence of the NZO allele of QTL Nob3 and absence of other adipogenic QTL were mated with 6–12 B6 females. B6.NZO-Nob3.91 mice were characterized after three backcross generations. B6.NZO-Nob3.38 mice were backcrossed five times and intercrossed for homozygosity of Nob3 before characterization. Founders of N3 and N5 were genotyped for 114 polymorphic markers (see Supplementary Table S31 ) and carried <30% and <10% residual NZO genome, respectively. In N5, none of the other significant or suggestive QTL were present. To compare the effects of both lines B6.NZO-Nob3.91 and B6.NZO-Nob3.38, we analyzed female mice of a (NZO×B6)N4 generation obtained from an additional backcross of the N3 generation (Supplementary Fig. S4).
Analysis of body composition.
Body fat and lean mass were determined with a nuclear magnetic resonance spectrometer (Bruker Minispec instrument; Echo Medical Systems, Houston, TX). Conscious mice were placed in an applied static field for 0.9 min (30). In addition, body weights were measured with an electronic scale.
Plasma parameters.
Blood glucose was determined with a Glucometer Elite (Bayer HealthCare, Leverkusen, Germany) after puncture of the tail vein of nonfasted mice at 8:30 AM. Blood samples for the determination of cholesterol and triglyceride levels were obtained from anesthetized nonfasted mice after cardiac puncture. Cholesterol and triglyceride levels were measured with the analyzing system VITROS DT60 II (Johnson & Johnson, Neckargemünd, Germany).
Rectal body temperature.
Rectal body temperature was measured with an electronic thermometer (physitemp model BAT-12; Physitemp Instruments, Clifton, NJ).
Feeding behavior.
Food intake was recorded with an automated drinking and feeding monitor system (TSE, Bad Homburg, Germany), consisting of macrolon type III cages equipped with baskets connected to weight sensors. The baskets contained standard chow pellets and were freely accessible to the mice. Mice were habituated to the test cages for 2 days before trials, and the measurement period lasted 5–6 days.
Genotyping.
DNA was prepared from mouse tails with a DNA isolation kit based on a salt precipitation method (InViTek, Berlin, Germany). Animals of the F2 generation were genotyped for 15 polymorphic microsatellite markers and 106 polymorphic single nucleotide polymorphisms (SNPs) (Supplementary Table S1). Residual donor DNA in congenic lines was determined by genotyping of 108 microsatellite markers and 6 SNPs (Supplementary Table S3). SNPs were genotyped by KBiosciences (KBiosciences, Hoddesdon, UK) with their competitive allele-specific PCR system (KASPar) that uses FRET quencher cassette oligonucleotides. Microsatellites were genotyped by PCR with oligonucleotide primers obtained from MWG (Ebersberg, Germany), and microsatellite length was determined by nondenaturing polyacrylamide gel electrophoresis.
Linkage analysis.
Normal distribution of all traits was checked by Kolmogorov-Smirnov test. Genetic map was constructed with MAPMAKER/EXP, version 3.0. Map order, genotyping errors, and linkage analysis for 600 animals (297 females, 303 males) were performed with the R/qtl 1.09-43 package of R (2) either with sex as additive and interacting covariate (Fig. 1, Table 2) or for males and females separately (Fig. 2). Linkage results obtained with all 600 animals were defined as sex-biased or sex-specific when the logarithm of the odds (LOD) scores differed (ΔLOD) by 0.9 or 3.1 units, respectively (26). Interactions were analyzed with two-QTL scans. Significance thresholds for linkage disequilibrium or interaction were estimated by 1,000 permutations.
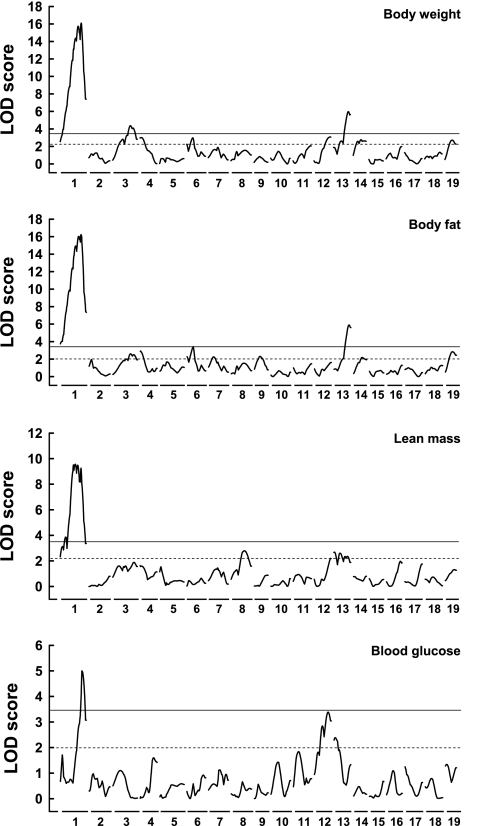
Fig. 1.
Genome-wide logarithm of the odds (LOD) score distribution in (NZO×B6)F2 for the traits body weight, body fat, lean mass, and blood glucose in week 22 with sex as additive covariate. Horizontal lines depict thresholds of significant (genome-wide testing P < 0.05) and suggestive LOD scores (P < 0.63), as determined by 1,000 permutations.
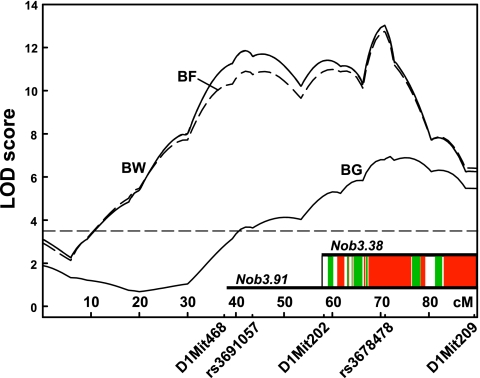
Fig. 2.
LOD score distribution of major metabolic phenotype QTL Nob3 on chromosome 1 for the traits body fat (BF), body weight (BW), and blood glucose (BG) in (NZO×B6)F2 females at week 22. Thresholds of genome-wide significance (P < 0.05) were 3.56, 3.52, and 3.56 for the traits BF, BW (horizontal line), and BG, respectively, as determined by 1,000 permutations. Horizontal bars depict the chromosomal segments Nob3.91 and Nob3.38 that were introgressed into the C57BL/6J strain (Fig. 3 and Supplementary Figs. S2–S4). Inset: polymorphic (red) and nonpolymorphic (green) haplotype blocks of a comparison of NZO with B6 (detailed information on the genotyped markers is given in Supplementary Fig. S5).
Statistical analysis.
Values are presented as means ± SE unless otherwise stated. Statistical significance was determined by ANOVA followed by either a Bonferroni or a Games-Howell posttest after testing for homogeneity of variances by Levene's test. Statistical analysis for Fig. 3 and Supplementary Figs. S2 and S3 was performed by two-tailed Student's t-test. Differences were considered significant when P < 0.05.
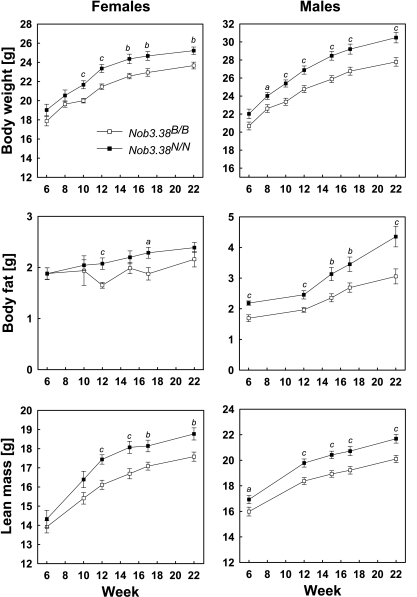
Fig. 3.
Development of body weight, body fat, and lean mass in female and male mice of congenic line B6.NZO-Nob3.38. Littermates of F2N5 were raised on a standard chow diet. Data represent means ± SE of 12–16 mice, and differences between homozygous carriers of Nob3.38 and controls (Nob3.38B/B) were tested for statistical significance by unpaired t-test (aP < 0.05; bP < 0.01; cP < 0.005).
RESULTS
Characterization of parental strains, F1, and F2 progeny.
Table 1 summarizes the characteristics of the parental strains, F1 hybrids, and F2 progeny. At 22 wk of age, body fat was >10 times higher in both male and female NZO than in C57BL/6J mice. In addition, blood glucose levels were higher in NZO than in B6. In the F1 and F2 progeny, however, mean blood glucose levels were not different from those in B6.
Table 1.
Characteristics of the parental strains B6 and NZO, F1 hybrids, and the F2 progeny at week 22
| B6 | NZO | F1 | F2 | |||||
|---|---|---|---|---|---|---|---|---|
| Females | ||||||||
| n | 10 | 9 | 12 | 318 | ||||
| BW, g | 24.1±1.4 | 62.7±7.6b | 42.4±6.9b | 44.2±10.6b | ||||
| BF, g | 3.6±0.7 | 39.7±8.0b | 18.0±7.1b | 22.8±10.1b | ||||
| LM, g | 18.4±0.9 | 27.2±2.3b | 25.0±2.6b | 23.8±3.2b | ||||
| BG, mM | 6.9±0.7 | 10.3±2.7a | 7.5±1.2 | 7.2±1.2 | ||||
| Males | ||||||||
| n | 6 | 7 | 7 | 331 | ||||
| BW, g | 30.5±2.9 | 71.6±7.6b | 62.1±4.8b | 58.3±7.9b | ||||
| BF, g | 3.1±1.2 | 42.0±11.6b | 26.6±4.2b | 26.4±7.2b | ||||
| LM, g | 23.6±2.0 | 29.4±4.9 | 34.8±1.5b | 32.3±3.0b | ||||
| BG, mM | 11.5±2.5 | 19.1±8.1 | 9.7±2.0 | 11.6±5.2 | ||||
Data are means ± SD. n, Number of mice; BW, body weight; BF, body fat; LM, lean mass; BG, blood glucose. Differences to B6 were analyzed by ANOVA followed by Bonferroni or Games-Howell as posthoc test
( P < 0.05;
P < 0.0001).
Genome-wide scan of the F2 progeny.
Figure 1 depicts the genome-wide distribution of LOD scores for the traits body weight, body fat, lean mass, and blood glucose in the combined male and female F2 progeny. The calculation of LOD scores with sex as additive covariate is summarized in Table 2. Significant linkage with traits of adiposity [body weight, body fat, or body mass index (BMI)] was detected for chromosomes 1, 3, and 13; suggestive linkage was found for chromosomes 4, 6, 9, 12, 14, and 19. The effects of some of the QTL were sex-dependent, and the adipogenic allele on chromosome 3 was contributed by the lean strain (Table 2). The predominant effect was contributed by a broad QTL with two LOD score maxima (40 and 70 cM) on chromosome 1 (Fig. 2), which accounted for a weight difference of 13 or 5 g in females or males, respectively (Table 3). We designated the QTL Nob3 in accordance with our previous designation of QTL identified in a cross of NZO with SJL (12). The QTL appeared to modify both lean and fat mass and produced a stronger effect in females than in males. In addition to the obesity-associated QTL, one significant (chromosome 5) and four suggestive QTL (chromosome 1, 9, 11, and 17) for plasma cholesterol were found (Table 2). One of these (chromosome 5) maps to the same locus as the cholesterol QTL previously identified in NZO (Chol/NZO1; 9) and NZB (18) outcross populations.
Table 2.
Summary of significant and suggestive QTL detected by genome-wide scan in (NZO×B6)F2 with sex as additive covariate
| Chr. | Peak, cM | Marker | Position, Mbp | Trait | LOD Score | Allele | ΔLOD Score | Sex Effect |
|---|---|---|---|---|---|---|---|---|
| 1 | 49.5 | rs3691057 | 125.3 | BW | 13.1‡ | NZO | 3.8 | F |
| BF | 13.8‡ | NZO | 4.8 | F | ||||
| LM | 9.1‡ | NZO | 3.2 | F | ||||
| BMI | 13.1‡ | NZO | 4.3 | F | ||||
| BL | 9.0‡ | NZO | 3.9 | F | ||||
| TG | 2.4* | B6 | 0.5 | |||||
| 1 | 75.3 | rs3678478 | 177.6 | BW | 16.0‡ | NZO | 3.0 | F |
| BF | 16.1‡ | NZO | 5.8 | F | ||||
| LM | 9.3‡ | NZO | 1.8 | F | ||||
| BMI | 18.2‡ | NZO | 4.0 | F | ||||
| BL | 8.5‡ | NZO | 4.0 | F | ||||
| BG | 4.0† | NZO | 0.1 | |||||
| Chol | 2.1* | NZO | 1.7 | F | ||||
| 2 | 73.2 | rs3693259 | 160.8 | TG | 2.9* | NZO | 1.3 | F |
| 3 | 54.4 | rs3712218 | 109.6 | BL | 4.2‡ | B6 | 0.8 | |
| TG | 4.1‡ | NZO | 0.8 | |||||
| 70.3 | rs4221957 | 129.5 | BW | 4.0† | B6 | 0.1 | ||
| BF | 2.3* | B6 | 0.2 | |||||
| BMI | 2.2* | B6 | 0.2 | |||||
| 4 | 1.0 | rs3698283 | 43.2 | BW | 3.0* | NZO | 0.1 | |
| BF | 2.9* | NZO | 0.7 | |||||
| BL | 3.6† | NZO | 0.6 | |||||
| 5 | 62.7 | D5Mit161 | 127.4 | Chol | 10.7‡ | NZO | 0.2 | |
| 6 | 22.7 | rs3717270 | 55.7 | BW | 3.0* | NZO | 0.1 | |
| BF | 3.4* | NZO | 0.2 | |||||
| BMI | 2.2* | NZO | 0.6 | |||||
| BL | 2.6* | NZO | 0.0 | |||||
| 7 | 54.9 | rs3692423 | 111.7 | BL | 3.3* | NZO | 0.1 | |
| 8 | 25.7 | rs3696502 | 78.2 | BL | 4.2‡ | NZO | 0.2 | |
| 8 | 40.4 | rs3710112 | 100.8 | LM | 2.7* | NZO | 0.9 | M |
| 9 | 25.3 | rs3666398 | 56.4 | BF | 2.1* | NZO | 0.1 | |
| Chol | 3.1* | NZO | 0.7 | |||||
| 11 | 63.8 | rs3705163 | 106.8 | Chol | 2.3* | NZO | 0.5 | |
| 12 | 58.4 | rs3023940 | 111.2 | BW | 3.1* | NZO | 0.0 | |
| LM | 2.2* | NZO | 0.2 | |||||
| BMI | 3.2* | NZO | 0.2 | |||||
| BG | 3.1* | NZO | 1.3 | F | ||||
| 13 | 58.2 | rs3686453 | 109.2 | BW | 5.6‡ | NZO | 0.4 | |
| BF | 5.7‡ | NZO | 1.0 | F | ||||
| BMI | 11.0‡ | NZO | 0.8 | |||||
| 14 | 32.5 | rs3676913 | 88.5 | BW | 2.6* | NZO | 0.6 | |
| BF | 2.2* | NZO | 0.7 | |||||
| BMI | 2.6* | NZO | 0.5 | |||||
| 16 | 54.7 | rs4216475 | 89.5 | BL | 2.1* | B6 | 0.2 | |
| 17 | 18.5 | rs3677240 | 44.3 | Chol | 2.1* | NZO | 0.8 | |
| 18 | 60.9 | rs3715760 | 87.6 | TG | 2.8* | NZO | 1.2 | M |
| 19 | 21.6 | rs3705022 | 36.5 | BW | 2.6* | NZO | 0.9 | F |
| BF | 2.8* | NZO | 2.2 | F | ||||
| BMI | 3.1* | NZO | 1.6 | F |
The analyzed traits are BW, BF, LM, body mass index (BMI), body length (BL), BG, plasma cholesterol (Chol), and plasma triglycerides (TG) at week 22. The logarithm of the odds (LOD) thresholds of genome-wide significance
(P < 0.63,
P < 0.05,
P < 0.01) were determined for each trait by 1,000 permutations and are presented in the supplementary data (Supplementary Table S2).
Table 3.
Effects of the QTL on chromosome 1 (Nob3) on adiposity-related and metabolic traits in week 22
|
chr.1 (rs3691057) |
chr.1 (rs3678478)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B/B | N/B | N/N | B/B | N/B | N/N | |||||||
| Females | ||||||||||||
| n | 79 | 152 | 61 | 77 | 142 | 73 | ||||||
| BW, g | 37.7±0.9 | 44.7±0.8c | 50.4±1.5c | 36.9±0.9 | 44.8±0.8c | 49.6±1.4c | ||||||
| BF, g | 16.9±0.9 | 23.2±0.8c | 28.5±1.4c | 15.9±0.8 | 23.4±0.8c | 27.9±1.3c | ||||||
| LM, g | 21.9±0.3 | 24.0±0.2c | 25.4±0.4c | 21.7±0.3 | 24.1±0.3c | 24.9±0.4c | ||||||
| BG, mM | 6.8±0.1 | 7.3±0.1a | 7.5±0.1b | 6.7±0.1 | 7.3±0.1c | 7.6±0.2c | ||||||
| BL, cm | 10.8±0.05 | 11.1±0.03c | 11.2±0.06c | 10.7±0.05 | 11.1±0.04c | 11.2±0.05c | ||||||
| BMI | 0.32±0.01 | 0.36±0.01c | 0.39±0.01c | 0.32±0.01 | 0.36±0.01c | 0.39±0.01c | ||||||
| Chol, mM | 3.2±0.1 | 3.4±0.1 | 3.4±0.1 | 3.0±0.1 | 3.3±0.1a | 3.5±0.1c | ||||||
| TG, mM | 1.1±0.1 | 1.1±0.1 | 1.0±0.1 | 1.0±0.1 | 1.0±0.1 | 1.2±0.1a | ||||||
| Males | ||||||||||||
| n | 73 | 134 | 86 | 71 | 138 | 89 | ||||||
| BW, g | 55.5±1.0 | 58.9±0.6a | 59.9±0.8a | 55.0±0.9 | 58.8±0.6b | 60.2±0.8c | ||||||
| BF, g | 24.2±0.9 | 26.6±0.6a | 28.0±0.8a | 24.0±0.8 | 26.5±0.6a | 28.0±0.8b | ||||||
| LM, g | 31.4±0.4 | 32.7±0.2a | 32.4±0.3 | 31.3±0.3 | 32.6±0.3a | 32.5±0.3a | ||||||
| BG, mM | 11.1±0.6 | 11.8±0.5 | 11.8±0.5 | 10.4±0.5 | 12.1±0.5a | 12.0±0.6 | ||||||
| BL, cm | 11.6±0.04 | 11.7±0.03a | 11.7±0.03a | 11.6±0.04 | 11.7±0.03 | 11.7±0.03 | ||||||
| BMI | 0.41±0.01 | 0.43±0.01a | 0.44±0.01a | 0.41±0.01 | 0.43±0.01b | 0.44±0.01c | ||||||
| Chol, mM | 4.1±0.1 | 4.1±0.1 | 4.3±0.1 | 4.1±0.1 | 4.3±0.1 | 4.1±0.1 | ||||||
| TG, mM | 1.3±0.1 | 1.2±0.1 | 1.1±0.1a | 1.3±0.1 | 1.2±0.1 | 1.1±0.1a | ||||||
Data represent means ± SE. n, Number of animals; B/B, homozygotes for B6 allele; B/N, heterozygotes; N/N, homozygotes for NZO allele. Differences to B/B genotype were tested for statistical significance by ANOVA followed by either a Bonferroni or a Games-Howell posttest
(P < 0.05;
P < 0.001;
P < 0.0001).
In parental NZO, severe hyperglycemia resulting from loss of β-cells occurs in >90% of males on a high-fat diet. In contrast, the prevalence of severe hyperglycemia was much lower in the F2 progeny; blood glucose levels exceeded 300 mg/dl (16.6 mM) in only 17% of male F2 mice (Supplementary Fig. S1). Thus, NZO-derived diabetes appeared markedly suppressed by the C57BL/6J background in the F2 progeny. A significant LOD-score value for the trait blood glucose at week 22 was obtained only for distal chromosome 1 (Fig. 1, Table 2). The effect of this QTL on blood glucose was not sex-biased (Table 2) and produced a difference between genotypes of 0.9 or 1.6 mM in females or males, respectively. As anticipated, hyperglycemia in F2 mice appeared dependent on the body weight (Supplementary Fig. S1). However, exclusion of mice with lower body weight or introduction of body weight as covariate failed to yield additional QTL for the trait blood glucose. Thus, the intercross identified only a single diabetogenic QTL that does not fully explain the severe hyperglycemia of the parental NZO males.
In addition to the single-QTL genome scan, we performed a two-QTL scan for the traits body weight, body fat, lean mass, BMI, and blood glucose to test the F2 population for interactions. Only suggestive interactions were found for chromosomes 3 and 5 (body weight, LODint = 5.3), chromosomes 7 and 17 (body fat, LODint = 6.1), and chromosomes 9 and 16 (body fat, LODint = 5.1). There was no interaction of the two LOD peaks on chromosome 1 with each other or with any other chromosome.
Introgression of distal chromosome 1 (Nob3) into the C57BL/6J strain and characterization of the congenic lines.
As an attempt to define a critical segment of Nob3 with a conventional strategy of positional cloning, we introgressed a 91 Mbp fragment defined by the markers D1Mit468 and D1Mit209 (see Fig. 2) from NZO into the C57BL/6J strain and reduced the remaining NZO genome from other chromosomes by three backcrosses to <15%. Supplementary Fig. S2 illustrates the characterization of female B6.NZO-Nob3.91 mice (N3 generation, heterozygous Nob3 carriers, with littermates homozygous for the B6 allele as controls). Weight gain in NZO mice depends on the fat content and the caloric density of the diet. Thus, we tested both standard chow and high-fat diet to optimize conditions for discrimination of the congenic lines. As anticipated, body weight, body fat, and lean mass were significantly increased in B6.NZO-Nob3.91 mice on both standard chow and high-fat diet. Significant although smaller differences between Nob3 carriers and controls were also observed in the male N3 progeny (Supplementary Fig. S3). Although the N3 progeny carried only <15% unlinked residual NZO genome, it cannot fully be excluded that other NZO QTL contributed to the weight difference. Thus, we generated additional backcross generations of B6.NZO-Nob3.91.
By backcross of B6.NZO-Nob3.91 to B6, we generated a second congenic line carrying a smaller fragment of chromosome 1 that corresponded with the distal peak of Nob3 (B6.NZO-Nob3.38, see Fig. 2). Homozygous carriers of Nob3.38 (generated after five backcrosses, with <5% residual NZO genome) exhibited significantly higher body weight, body fat, and lean mass than their littermates carrying B6 alleles of Nob3 (Fig. 3). These differences (∼2 g of body weight and 0.5–1 g of body fat) appeared smaller than those observed in the line B6.NZO-Nob3.91 (Supplementary Fig. S2; 5 g of body weight and 2–3 g of body fat).
To assess the presence of more than one adipogenic gene variant we compared the effects of both fragments Nob3.91 and Nob3.38. Since the residual NZO genome differed in both congenic lines (N3 vs. N5 generation), we performed an additional, direct comparison of the N4 generations of the two lines (Supplementary Fig. S4). Indeed, B6.NZO-Nob3.91 gained weight faster than B6.NZO-Nob3.38.
Food intake in the congenic B6.NZO-Nob3.38 (homozygous carriers of Nob3 allele) was not significantly different from controls (homozygous carriers of B6 allele): female Nob3.38, 3.92 ± 0.15 g/day; female B6, 3.55 ± 0.17; male Nob3.38, 3.85 ± 0.1; male B6, 3.61 ± 0.14. In contrast, female, but not male, B6.NZO-Nob3.38 mice exhibited a significantly lower body temperature: female Nob3.38, 37.2 ± 0.1°C; B6, 37.6 ± 0.1°C; P < 0.005; male Nob3.38, 36.6 ± 0.1°C; B6, 36.6 ± 0.1°C; not significant.
Haplotype mapping of Nob3.
Based on the assumption that an ancestral gene variant is responsible for the effect of Nob3, we generated a haplotype map of the segment introgressed in B6.NZO-Nob3.38 to narrow down its critical region (inset in Fig. 2). Nob3.38 contains large polymorphic blocks (NZO vs. B6) that account for more than half of the interval; one of them corresponds with the peak region of the LOD-score curve. A search for candidate genes in the polymorphic blocks that have been studied previously in a context of the observed traits (21, 24, 31) identified the following genes: constitutive androstane receptor CAR (Nr1i3), apolipoprotein a2 (Apoa2), Na/K-ATPase α-2 subunit (Atp1a2), the homeobox transcription factor Prox1, and 11-β-hydroxy-steroid dehydrogenase 1 (Hsd11b1). Since linkage data from other crosses of NZO with NON or SM are available (22, 29), we generated a second map including these strains (full information in Supplementary Fig. S5). This map could guide future breeding of subcongenic lines and help to prioritize candidates to be studied.
DISCUSSION
The present data describe the successful introgression of a QTL for body weight on chromosome 1 of NZO, Nob3, into the lean C57BL/6J strain. This finding is of interest because many previous attempts to isolate obesity QTL by introgression have been unsuccessful. On the C57BL/6J background, Nob3 produced a moderate but robust difference in weight gain of the congenic strain; this effect will allow positional cloning of the responsible gene variant by a conventional breeding strategy that narrows down the critical region. QTL for body weight on distal chromosome 1 have also been identified in previous genome-wide scans of outcross populations of NZO with NON (15) and with SM mice (29). Thus, the analysis of haplotype blocks in Nob3 can guide selection of substrains carrying smaller fragments and, together with a search for variants that are unique for NZO, can ultimately lead to the responsible gene.
It should be noted that Nob3 is a very broad QTL spanning a total of >40 cM with two apparent peaks. Its proximal peak appears to correspond with a QTL that was previously identified in an NZO×NON outcross (22). A QTL corresponding to its distal peak has been identified in an NZO×SM outcross (29). We successfully introgressed the complete QTL (91 Mbp) as well as a smaller segment comprising only the distal peak (38 Mbp) into the B6 background. The segment corresponding with the complete QTL appeared to exert a greater effect on body weight than the distal segment. However, it should be noted that in the initial characterization the B6.NZO-Nob3.91 line (N3 progeny) carried considerably more residual unlinked NZO genome (∼15%) than the B6.NZO-Nob3.38 line (N5, <5%). Therefore, direct comparison of the data in Supplementary Figs. S2 and S3 cannot unequivocally prove the presence of more than one causal gene. However, a comparison of the N4 generations (Supplementary Fig. S4, residual NZO genome ∼7.5%) replicated the difference. Thus, we conclude that the distal segment of the QTL (Nob3.38) contains a gene variant responsible for part of the adiposity and hyperglycemia in NZO. In addition, since the Nob3.38 segment appeared to produce a smaller effect on body weight and adiposity than the complete QTL (Nob3.91), it cannot be excluded that the proximal part contains additional adipogenic variants.
So far, only the approximate chromosomal positions of the adipogenic and diabetogenic genes have been determined, whereas their cellular and molecular functions are still incompletely known. From a previous characterization of the parental strain it follows that obesity in NZO is the result of both hyperphagia and reduced energy expenditure (11). Consistent with this finding, the present data show a reduced body temperature in females of the B6.NZO-Nob3.38 line but failed to demonstrate a statistically significant difference in food intake between the two genotypes. It should be noted, however, that the calculated difference in food intake that explains the higher weight gain of B6.NZO-Nob3.38 (5 g/30 wk corresponding with 24 mg/day) is only 0.74 kJ/d, which is only 1.5% of the total energy consumed per day. Such a difference is much smaller than the standard deviation of the daily food intake and therefore hard to detect.
For the present outcross experiment, we have chosen the diabetes-resistant strain C57BL/6J, assuming that this strain might be particularly suitable to identify diabetogenic alleles from NZO. Indeed, the prevalence of severe hyperglycemia was markedly lower in (NZO×B6)F2 than in outcross populations with other strains such as NON and SJL that contributed diabetogenic alleles (15, 19, 20). However, the distal part of Nob3 was the only locus that was in significant linkage disequilibrium for the trait hyperglycemia. Furthermore, none of the previously reported QTL for diabetes, contributed by NZO chromosome 11 in (NZO×NON)F2 (15) and 15 in (NZO×SJL)N2 (19), were detected here. This finding could indicate that the previously reported QTL represent diabetes-suppressing alleles contributed by the lean strains. Similarly, the hyperglycemic effect of distal Nob3 seems so far unique for our outcross experiment and could therefore represent a diabetes-suppressing variant from B6.
According to the haplotype map of NZO compared with B6, five candidate genes with functional association to the investigated traits are located in polymorphic blocks. However, this analysis is based on the assumption that the effects of the QTL are caused by ancestral gene variants that are in linkage disequilibrium with adjacent markers. Recently, we identified an obesity-suppressing mutation in Tbc1d1 that was unique for the SJL strain and therefore arose much later than the single nucleotide and microsatellite polymorphisms. Thus, the haplotype map may guide future restriction of the QTL, but direct evidence linking the phenotype to a critical region will still be required.
GRANTS
The study was supported by grants from the European Union (EUGENE2) and the German Bundesministerium für Bildung und Forschung [NGFN2 (01GS0487) and NGFN-Plus (01GS0821)].
Supplementary Material
Acknowledgments
The expert technical assistance of Elvira Steinmeyer, Aljona Borschewski, Sabine Frenzel, and Monika Niehaus is gratefully acknowledged.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Bielschowsky M, Bielschowsky F. A new strain of mice with hereditary obesity. Proc Univ Otago Med School 31: 29–31, 1953. [Google Scholar]
- 2.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890, 2003. [DOI] [PubMed] [Google Scholar]
- 3.Buchmann J, Meyer C, Neschen S, Augustin R, Schmolz K, Kluge R, Al-Hasani H, Jürgens H, Eulenberg K, Wehr R, Dohrmann C, Joost HG, Schürmann A. Ablation of the cholesterol transporter adenosine triphosphate-binding cassette transporter G1 reduces adipose cell size and protects against diet-induced obesity. Endocrinology 148: 1561–1573, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Chadt A, Leicht K, Deshmukh A, Jiang LQ, Scherneck S, Bernhardt U, Dreja T, Vogel H, Schmolz K, Kluge R, Zierath JR, Hultschig C, Hoeben RC, Schürmann A, Joost HG, Al-Hasani H. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet 40: 1354–1359, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Chiu S, Kim K, Haus KA, Espinal GM, Millon LV, Warden CH. Identification of positional candidate genes for body weight and adiposity in subcongenic mice. Physiol Genomics 31: 75–85, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET, Stapleton DS, Gray-Keller MP, Young MB, Stoehr JP, Lan H, Boronenkov I, Raess PW, Flowers MT, Attie AD. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet 38: 688–693, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Crofford OB, Davis CK Jr. Growth characteristics, glucose tolerance and insulin sensitivity of New Zealand obese mice. Metabolism 14: 271–280, 1965. [DOI] [PubMed] [Google Scholar]
- 8.Farber CR, Medrano JF. Dissection of a genetically complex cluster of growth and obesity QTLs on mouse Chromosome 2 using subcongenic intercrosses. Mamm Genome 18: 635–645, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Giesen K, Plum L, Kluge R, Ortlepp J, Joost HG. Diet-dependent obesity and hypercholesterolemia in the New Zealand obese mouse: identification of a quantitative trait locus for elevated serum cholesterol on the distal mouse Chromosome 5. Biochem Biophys Res Commun 304: 812–817, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Herberg L, Coleman DL. Laboratory animals exhibiting obesity and diabetes syndromes. Metabolism 26: 59–99, 1977. [DOI] [PubMed] [Google Scholar]
- 11.Jürgens HS, Schürmann A, Kluge R, Ortmann S, Klaus S, Joost HG, Tschöp MH. Hyperphagia, lower body temperature, and reduced running wheel activity precede development of morbid obesity in New Zealand obese mice. Physiol Genomics 25: 234–241, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Kluge R, Giesen K, Bahrenberg G, Plum L, Ortlepp JR, Joost HG. Quantitative trait loci for obesity and insulin resistance (Nob1, Nob2) and their interaction with the leptin receptor allele (LeprA720T/T1044I) in New Zealand obese mice. Diabetologia 43: 1565–1572, 2000. [DOI] [PubMed] [Google Scholar]
- 13.Leiter EH The genetics of diabetes susceptibility in mice. FASEB J 3: 2231–2241, 1989. [DOI] [PubMed] [Google Scholar]
- 14.Leiter EH, Herberg L. The polygenetics of diabesity in mice. Diabetes Rev 5: 131–148, 1997. [Google Scholar]
- 15.Leiter EH, Reifsnyder PC, Flurkey K, Partke HJ, Junger E, Herberg L. NIDDM genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes 47: 1287–1295, 1998. [DOI] [PubMed] [Google Scholar]
- 16.Naggert J, Harris T, North M. The genetics of obesity. Curr Opin Genet Dev 7: 398–404, 1997. [DOI] [PubMed] [Google Scholar]
- 17.Ortlepp JR, Kluge R, Giesen K, Plum L, Radke P, Hanrath P, Joost HG. A metabolic syndrome of hypertension, hyperinsulinaemia and hypercholesterolaemia in the New Zealand obese mouse. Eur J Clin Invest 30: 195–202, 2000. [DOI] [PubMed] [Google Scholar]
- 18.Pitman WA, Korstanje R, Churchill GA, Nicodeme E, Albers JJ, Cheung MC, Staton MA, Sampson SS, Harris S, Paigen B. Quantitative trait locus mapping of genes that regulate HDL cholesterol in SM/J and NZB/B1NJ inbred mice. Physiol Genomics 9: 93–102, 2002. [DOI] [PubMed] [Google Scholar]
- 19.Plum L, Kluge R, Giesen K, Altmüller J, Ortlepp JR, Joost HG. Type 2 diabetes-like hyperglycemia in a backcross model of NZO and SJL mice: characterization of a susceptibility locus on Chromosome 4 and its relation with obesity. Diabetes 49: 1590–1596, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Plum L, Giesen K, Kluge R, Junger E, Linnartz K, Schürmann A, Becker W, Joost HG. Characterisation of the mouse diabetes susceptibility locus Nidd/SJL: islet cell destruction, interaction with the obesity QTL Nob1, and effect of dietary fat. Diabetologia 45: 823–830, 2002. [DOI] [PubMed] [Google Scholar]
- 21.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 14: 529–644, 2006. [DOI] [PubMed] [Google Scholar]
- 22.Reifsnyder PC, Churchill G, Leiter EH. Maternal environment and genotype interact to establish diabesity in mice. Genome Res 10: 1568–1578, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reifsnyder PC, Leiter EH. Deconstructing and reconstructing obesity-induced diabetes (diabesity) in mice. Diabetes 51: 825–832, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt C, Gonzaludo NP, Strunk S, Dahm S, Schuchhardt J, Kleinjung F, Wuschke S, Joost HG, Al-Hasani H. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol Genomics 34: 42–53, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Schmolz K, Pyrski M, Bufe B, Vogel H, Nogueras R, Scherneck S, Nestler M, Zahn C, Rüschendorf F, Tschop MH, Meyerhof W, Joost HG, Schürmann A. Regulation of feeding behavior in normal and obese mice by neuromedin-U: A variant of the neuromedin-U receptor 2 contributes to hyperphagia in the New-Zealand Obese mouse. Obesity Metab 3: 28–37, 2007. [Google Scholar]
- 26.Shao H, Reed DR, Tordoff MG. Genetic loci affecting body weight and fatness in a C57BL/6J × PWK/PhJ mouse intercross. Mamm Genome 18: 839–851, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stylianou IM, Korstanje R, Li R, Sheehan S, Paigen B, Churchill GA. Quantitative trait locus analysis for obesity reveals multiple networks of interacting loci. Mamm Genome 17: 22–36, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Stylianou IM, Affourtit JP, Shockley KR, Wilpan RY, Abdi FA, Bhardwaj S, Rollins J, Churchill GA, Paigen B. Applying gene expression, proteomics and single-nucleotide polymorphism analysis for complex trait gene identification. Genetics 178: 1795–1805, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor BA, Wnek C, Schroeder D, Phillips SJ. Multiple obesity QTLs identified in an intercross between the NZO (New Zealand obese) and the SM (small) mouse strains. Mamm Genome 12: 95–103, 2001. [DOI] [PubMed] [Google Scholar]
- 30.Tinsley FC, Taicher GZ, Heiman ML. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obes Res 12: 150–160, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Wuschke S, Dahm S, Schmidt C, Joost HG, Al-Hasani H. A meta-analysis of quantitative trait loci associated with body weight and adiposity in mice. Int J Obes (Lond) 31: 829–841, 2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.