Abstract
A group (n = 8) of healthy older (68 ± 6 yr) adults participated in a 36-session progressive resistance exercise training program targeting the thigh muscles to determine the relationship between muscle gene expression and gains in muscle size and strength. Biopsies were obtained from the vastus lateralis at baseline 72 h after an acute bout of exercise and 72 h after completion of the training program. Training increased thigh muscle size (7%) and strength for the three exercises performed: knee extension (30%) and curl (28%) and leg press (20%). We quantified 18 transcripts encoding factors that function in inflammation, growth, and muscle remodeling that were demonstrated previously to be regulated by aging and acute exercise. The gain in extension strength and muscle size showed a high number of significant correlations with gene expression. These gains were most strongly correlated (P ≤ 0.003, R ≥ 0.89) with the baseline mRNA levels for insulin-like growth factor-1, matrix metalloproteinase-2 and its inhibitor TIMP1, and ciliary neurotrophic factor. Moreover, strength gains were inversely correlated with the change in these mRNA levels after training (P ≤ 0.002 and R ≤ −0.90). Changes in gene expression after acute exercise were not associated with training outcomes. These results suggest that higher baseline expression for key genes in muscle conveys an adaptive advantage for certain older adults. Individuals with lower baseline expression of these genes show less adaptation to exercise despite increased gene expression in response to training. These genes hold promise as useful predictors of training outcomes that could be used to design more effective exercise regimens for maintaining muscle function in older adults.
Keywords: growth factors, metallopeptidase, ciliary neurotrophic factor, aging, resistance exercise, skeletal muscle
individuals can typically increase the size and strength of their skeletal muscles by participating in resistance exercise training. However, the magnitude of the response to training can vary significantly as has been shown for size (−3% to +59%) and strength (0% to +250%) changes of the biceps for a relatively large cohort of healthy young adults (14). Older adults also benefit from resistance exercise training, but the adaptive responses appear to be even more variable than for younger adults (10, 35). It seems likely that sarcopenia, the loss of muscle size and strength that occurs with aging due to a combination of external causes and age-associated physiological changes (28), will be more pronounced in individuals whose muscle displays less of an adaptive response to physical activity. Since sarcopenia has such detrimental effects on independence and health (32), understanding the molecular and cellular biology of skeletal muscle and using this information to predict an individual's response to training would be of great value.
Evidence is accumulating that genetic and molecular differences between individuals regulate muscle phenotype and response to exercise. A PubMed search in March 2009 for “skeletal muscle” and “genotype” or “gene expression” limited to humans returned 1,128 and 5,449 citations, respectively. For example, genotype of the angiotensin I-converting enzyme and alpha-actinin 3 genes influences the response to resistance training (5, 25), and myogenic regulatory factors such as MyoD and myogenin are common targets of gene expression studies (20, 27). Our efforts to define the molecular control of muscle in young and older adults have involved the study of a number of transcripts expressed in muscle (Table 1). We initially studied inflammation since acute inflammation is thought to play an important role in muscle repair and chronic inflammation has been associated with sarcopenia (29, 34). An initial study involving young males (21–46 yr) indicated that after a single bout of resistance exercise 72 h is better than 24 h as a postexercise time point for measuring the induction of cytokine transcripts (9). A follow-up study including older adults (65–80 yr) showed that compared with younger adults, older adults possessed higher levels of muscle cytokine transcripts at baseline and as a group did not display the 72 h postexercise induction of cytokines (26). The baseline and postacute exercise muscle was further compared between the young and older adults using quantitative gene expression profiling for 100 transcripts of interest (8). The profiling revealed that transcripts for a number of factors including insulin-like growth factor-1 and the vascular endothelial growth factors-A and -D were higher for the young than the older adults and that other transcripts including that of myostatin responded to acute exercise only for the young. Though significant differences were identified between younger and older adults at baseline and in the response to acute exercise for transcript expression, the amount of within-group variability was striking. We hypothesized that this molecular variability would have been associated with varied gains in muscle size and strength had these individuals undergone a resistance exercise training program. The current study tested this hypothesis by subjecting a group of healthy older adults to a progressive resistance exercise training program that targeted the thigh muscles to determine the relationship between the gains in muscle strength and size and muscle gene expression. Quantitative analysis of 18 transcripts (Table 1) potentially involved in muscle inflammation, growth, and remodeling was performed using vastus lateralis biopsy tissue collected from subjects at baseline (pre-exercise), 72 h postacute exercise, and 72 h after the resistance exercise training program.
Table 1.
Transcripts related to inflammation, growth, and remodeling that are differentially expressed in muscle between young and older adults at baseline or in response to acute resistance exercise
| Transcript (Abbreviation) | Differences in Expression | Reference No. | ||
|---|---|---|---|---|
| Pro- and Anti-inflammatory Cytokines | ||||
| Chemokine ligand 18 (CCL18) | ARE Y>O | (26) | ||
| Interleukin-1β (IL1β) | BL O>Y, ARE Y>O | (9, 26) | ||
| Inteleukin-6 (IL6) | ND | (9, 26) | ||
| Interleukin-10 (IL10) | BL O>Y, ARE Y>O | (26) | ||
| Interleukin-1 receptor antagonist (IL1RA) | BL O>Y | (26) | ||
| Tumor necrosis factor-α (TNFα) | ND | (9) | ||
| Growth and Growth-Related Factors | ||||
| Ciliary neurotrophic factor (CNTF) | BL Y>O | (8) | ||
| Growth and differentiation factor 8 or myostatin (GDF8) | BL Y>O, ARE Y>O | (8) | ||
| Insulin-like growth factor-1 (IGF1) | BL Y>O | (8) | ||
| Insulin-like growth factor binding protein-5 (IGFBP5) | BL Y>O | (8) | ||
| Osteosarcoma viral oncogene homolog (c-fos) | BL Y>O | (8) | ||
| Vascular endothelial growth factor-A (VEGFA) | BL Y>O | (8) | ||
| Vascular endothelial growth factor-D (VEGFD) | BL Y>O | (8) | ||
| Remodeling Factors | ||||
| α-Cardiac actin (ACTC1) | ARE Y>O | (8) | ||
| Atrogin-1 or F-box protein 32 (MAFbx) | BL Y>O | (8) | ||
| Fibronectin (FN1) | BL Y>O | (8) | ||
| Matrix metallopeptidase-2 (MMP2) | BL Y>O | (8) | ||
| TIMP metallopeptidase inhibitor-1 (TIMP1) | ARE Y>O | (8) | ||
Differences in expression were previously identified in muscle gene expression between young and older adults: the 72 h response to acute resistance exercise was greater in the young than old (ARE Y>O), baseline values were greater in the old than young (BL O>Y), the baseline values were greater in the young than old (BL Y>O), or young was not different from the old (ND).
METHODS
Subjects.
Older male (n = 3) and female (n = 5) subjects were recruited from the community using newspaper advertisements [68.3 ± 5.6 yr, height 167 ± 8 cm, weight 69 ± 10 kg, body mass index (BMI) 25 ± 2 kg/m2 (mean ± SD)]. All subjects gave a written informed consent approved by the Institutional Review Board (IRB) of the University of Arkansas for Medical Sciences. Tissue and data analysis was certified as exempt from oversight by the IRB of the University of Kentucky. To determine eligibility, subjects underwent a thorough physical examination including blood chemistries and a resistance exercise cardiac stress test. Subjects were included if they were age 60 yr or older, had not participated in resistance exercise training in the previous 6 mo, were nonsmokers, had blood pressure <150/90 mmHg, had BMI ≤30 kg/m2, and were healthy as judged by the study physician. Subjects were excluded if they had any acute or chronic illness or disability that could place them at risk during exercise. Subjects were also excluded if they could not refrain from the use of anti-inflammatory medications for 3 days prior to and through the exercise phase of the protocol.
Resistance exercise protocol.
Subjects completed a single session of acute resistance exercise followed by a 36-session progressive resistance exercise training program. The training program was designed to increase mass and strength for the thigh muscles by exercising three times per week on nonconsecutive days for 12 wk. Due to scheduling convenience for the subjects, 15 ± 1 (mean ± SD) wk were required to complete the program. The acute resistance exercise session included 10 min of light cycling for warm-up and the determination of each subject's one-repetition maximum (1-RM) for bilateral knee extension, knee curl, and leg press (Keiser Pneumatic Strength Training Equipment; Keiser, Fresno, CA). Subjects rested for 5 min and then completed three sets of eight repetitions followed by a fourth set of repetitions to voluntary failure for each of the three exercises at 80% 1-RM. Subjects were given 2 min of rest between each set and 5 min between exercises. Subjects were supervised during each session to insure proper form and safety. 1-RM was determined every 2 wk, and the 80% 1-RM workload was readjusted.
CT scan.
The cross-sectional area of the nondominant thigh was measured by computed tomography (CT) prior to beginning the exercise training program and 5 days after training was completed. Subjects were supine for at least 30 min prior to the scan. The CT image was taken at the midpoint between the inguinal fold and proximal pole of the patella, along the femur. Muscle, fat, and bone areas were calculated with SliceOmatic software (Tomovision, Montreal, Canada).
Muscle biopsy.
Muscle biopsies were taken from the vastus lateralis muscle just prior to acute exercise (baseline), 72 h postacute exercise, and 72 h after the resistance exercise training program. The 72 h time point was chosen based on our original interest in muscle inflammation after acute exercise (9, 26). The first biopsy was taken from the subject's nondominant leg, and the second and third biopsies were taken from the dominant leg. Tissue was obtained after local anesthetic (lidocaine HCl 1%, 3 ml), processed, and stored frozen until analysis as described previously (8).
Gene expression analysis.
Total RNA was purified from frozen muscle tissue as described previously to measure levels of 18 transcripts (Table 1) of interest and a control (18S ribosomal RNA) by quantitative reverse transcriptase real-time polymerase chain reaction (RT RT-PCR)(8). The RT RT-PCR reagents (iScript cDNA synthesis kit and 2× SYBR Green Master Mix) and the iQ5 Multicolor Real-Time PCR Detection System were used according to the manufacturer's instructions (Bio-Rad, Hercules, CA). A detailed description of the primers used to detect each transcript, data normalization to the control transcript, and calculation of relative expression values have been reported (8, 9, 26). In brief, parameters for each assay were carefully optimized for efficiency using standard curves that were generated from fourfold serial dilutions of a pooled-sample cDNA. Standards and individual samples were assayed at least in triplicate. Melting curves, gel electrophoresis of the PCR products, and controls lacking template were used to verify the single product specificity of the RT RT-PCR assays.
Statistics.
Exercise outcomes for muscle size and strength (pre- and posttraining as well as the percent change) were treated as continuous variables and summarized with descriptive statistics (mean and standard error of the mean, Table 2). Comparisons of exercise outcomes (pre- vs. posttraining) were made using the paired samples t-test. Gene expression data (pre-exercise baseline, and the fold-change from baseline after acute exercise or the fold-change after training) were also treated as continuous variables. The relationships between the percent change in the exercise outcomes and gene expression data were visualized using scatter plots. A curvilinear relationship was detected in these plots, which was corrected with a log transformation of the gene expression data. The associations between the percent change in the exercise outcomes and the transformed gene expression data were then evaluated using the Pearson test. The resulting correlation coefficients (R-values) are presented with the corresponding P values.
Table 2.
Thigh muscle size and strength pre- and postresistance exercise training
| Pretraining | Posttraining | % Change | P Value | |
|---|---|---|---|---|
| Muscle area, cm2* | 102.6±8.2 | 109.6±8.1 | 7.4±2.0 | 0.002 |
| Fat area, cm2* | 91.8±13.6 | 90.4±14.4 | −2.4±1.9 | 0.395 |
| Bone area, cm2* | 5.0±0.3 | 5.0±0.3 | 0.0±1.1 | 0.980 |
| Knee extension 1-RM, Nm† | 143.9±21.5 | 184.9±26.1 | 30.4±5.6 | <0.001 |
| Leg curl 1-RM, Nm† | 127.5±16.9 | 162.1±20.9 | 27.7±5.4 | 0.003 |
| Leg press 1-RM, N† | 1167.9±155.7 | 1388.1±177.4 | 19.8±2.8 | <0.001 |
Values are means ± SE.
Cross-sectional area for the midthigh was measured by CT scan.
Strength based on one-repetition maximum testing (1-RM). P values were calculated using paired samples t-test.
RESULTS
Training outcomes.
Eight older adults (age range 61–78 yr) completed an acute session of resistance exercise followed by 36 sessions of progressive resistance exercise training that targeted the thigh muscles. The pre- and posttraining outcomes for muscle size and strength are shown in Table 2. Training resulted in a significant increase of ∼7% in whole muscle size of the midthigh (P = 0.002), while fat (P = 0.395) and bone (P = 0.980) cross-sectional areas were not affected. Training also resulted in a significant 20–30% average improvement in strength (1-RM) for each of the three exercises: knee extension (P < 0.001), knee curl (P = 0.003), and leg press (P < 0.001), though for all measures there was considerable interindividual variability. The gain in muscle size was correlated with the gain in knee extension strength (P = 0.006, R = 0.87). However, other significant correlations between training outcomes did not occur.
Correlations between training outcomes and muscle gene expression.
Muscle biopsies were taken from the vastus lateralis at baseline, and 72 h postacute exercise and 72 h posttraining to determine if the percent gains in muscle size and strength were related to the expression of specific genes. Transcripts for 18 genes that we previously characterized as being regulated by acute exercise and/or aging in human muscle were measured (Table 1). The results of a correlation analysis between the training outcomes and gene expression are shown in Table 3 as the number of “strong” (strong is arbitrarily defined as absolute value of R ≥ 0.85) and/or significant (P ≤ 0.05) relationships. The gain in extension strength after training was significantly related to 24 gene expression values (n = 13 at baseline and n = 11 posttraining) and strongly related to 12 of those values (n = 6 at baseline and n = 6 posttraining). Though fewer than for extension, the gain in muscle size was significantly related to 15 gene expression values with 4 of those being strong. By contrast, the gain in strength for press and curl did not have any strong relationships and relatively few total (n = 7 press, n = 1 curl) significant relationships with gene expression. None of the training outcomes was strongly related to gene expression postacute exercise, and only one significant relationship existed.
Table 3.
Number of significant correlations between resistance training outcomes and muscle gene expression
|
Training Outcome |
||||||||
|---|---|---|---|---|---|---|---|---|
|
Extension |
Size | Press | Curl | |||||
| Time of biopsy | R ≥ 0.85 | P ≤ 0.05 | R ≥ 0.85 | P ≤ 0.05 | R ≥ 0.85 | P ≤ 0.05 | R ≥ 0.85 | P ≤ 0.05 |
| Baseline | 6 | 13 | 1 | 6 | 0 | 3 | 0 | 0 |
| Postacute exercise | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Posttraining | 6 | 11 | 3 | 9 | 0 | 3 | 0 | 1 |
Correlations between training outcomes and gene expression values were determined by the Pearson test. An absolute value of R ≥ 0.85 was used arbitrarily to define a “strong” relationship. Training outcomes were the dependent variables. Outcomes were calculated as the percent gain posttraining for the change in thigh muscle size and strength for knee extension, knee curl, and leg press. 3Gene expression values were measured for muscle biopsies taken at baseline (pre-exercise), postacute exercise, or posttraining. Postacute and posttraining gene expression was calculated as the fold-change compared with baseline.
Strongest relationships between extension strength and transcript expression.
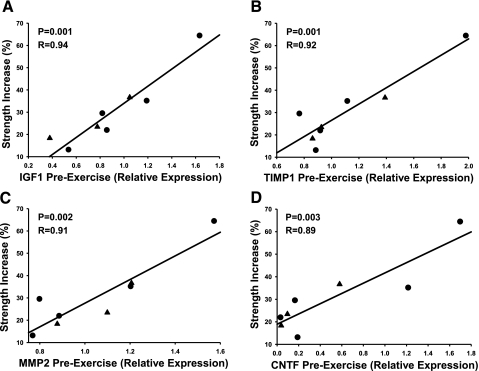
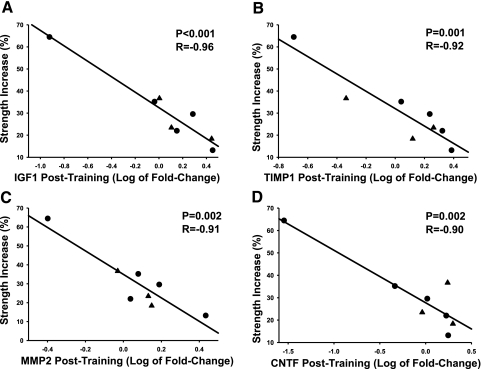
Since the majority of the strong and significant correlations between training outcomes and gene expression occurred between the gain in knee extension strength and gene expression at baseline and posttraining, these relationships are further described (Table 4). Extension strength was strongly and significantly related to the expression of IGF1, TIMP1, MMP2, and CNTF transcripts at both baseline and post-training (Table 4, group 1). Those individuals with the highest baseline levels of these transcripts and the least induction following training showed the greatest extension strength gain. The positive correlations to baseline expression and the negative correlations to post-training expression are shown graphically in Fig. 1 (baseline) and Fig. 2 (posttraining). Also in Figs. 1 and 2, one subject can be seen as a consistent outlier for strength and gene expression. Analysis after removal of this subject from the dataset produced similar overall results (data not shown).
Table 4.
Significant correlations between the training induced gain in knee extension strength and transcript expression at baseline and posttraining1
|
Time of Biopsy |
||||||||
|---|---|---|---|---|---|---|---|---|
|
Baseline |
Posttraining
|
|||||||
| R value | P value | R value | P value | |||||
| Group 1: absolute value of R ≥ 0.85 for both baseline and posttraining correlations | ||||||||
| IGF1 | 0.94 | 0.001 | −0.96 | <0.001 | ||||
| TIMP1 | 0.92 | 0.001 | −0.92 | 0.001 | ||||
| MMP2 | 0.91 | 0.002 | −0.91 | 0.002 | ||||
| CNTF | 0.89 | 0.003 | −0.90 | 0.002 | ||||
| Group 2: absolute value of R ≥ 0.85 for either baseline or posttraining correlations | ||||||||
| VEGFD | 0.93 | 0.001 | −0.81 | 0.014 | ||||
| IGFBP5 | 0.75 | 0.033 | −0.90 | 0.002 | ||||
| IL1RA | 0.86 | 0.007 | −0.74 | 0.035 | ||||
| GDF8 | 0.74 | 0.036 | −0.85 | 0.008 | ||||
| Group 3: absolute value of R < 0.85 for both baseline and posttraining correlations | ||||||||
| IL10 | 0.79 | 0.021 | −0.73 | 0.041 | ||||
| FN1 | 0.79 | 0.021 | −0.76 | 0.027 | ||||
| IL1β | 0.78 | 0.022 | −0.77 | 0.027 | ||||
Correlations between the gain in knee extension strength (dependent variable) after training and transcript expression (independent variable) was determined by the Pearson test. Transcripts were grouped according to the strength and consistency of their relationship with extension strength. An absolute value of R ≥ 0.85 was used arbitrarily as a cutoff value.
Fig. 1.
Baseline (pre-exercise) relationship between muscle gene expression and the percent gain in knee extension strength posttraining. Gene expression was measured by quantitative RT RT-PCR. Relative expression was calculated using standard curves and normalized using 18S. Data for each subject is shown (males ▴, females •). Note that gene expression data were log transformed.
Fig. 2.
Posttraining relationship between muscle gene expression and the percent gain in knee extension strength posttraining. Gene expression was measured by quantitative RT RT-PCR. Relative expression was calculated using standard curves and normalized using 18S. Data for each subject is shown (males ▴, females •). Note that for gene expression data the fold-change from baseline to posttraining was log transformed.
Other relationships between extension strength and transcript expression.
Transcript expression for VEGFD, IGFBP5, IL1RA, and GDF8 was significantly related to extension strength both at baseline and posttraining, but the relationship was strong at only one of the two time points (Table 4, group 2). Correlations between transcript expression for IL10, fibronectin, and IL1β and extension strength were significant but were not strong at either time point (Table 4, group 3). Correlations with IL6, MAFbx, and c-fos were significant at only one time point, and correlations with ACTC1, CCL18, TNF-α, VEGFA and the control 18S were not significant (data not shown).
DISCUSSION
The older adults in this study showed a range of responses to resistance exercise training from −1% to +18% for muscle size and +8% to +65% for muscle strength. Correlation analysis between the data for these training outcomes and expression data for 18 transcripts presumed to function in muscle response to inflammation, growth, or remodeling yielded three main findings: 1) strong correlations between gene expression in the vastus lateralis and strength gain were restricted to the exercise that maximally activated that muscle, e.g., the knee extension as opposed to the leg press or knee curl (1); 2) training outcomes were positively correlated with the level of transcripts from each functional class at baseline and negatively correlated with changes in transcript levels after training; and 3) training outcomes were not related to the response of these genes 72 h postacute exercise. These results suggest that higher baseline expression of key genes in muscle conveys an adaptive advantage for certain older adults. Individuals with lower baseline expression of these genes show less adaptation to exercise despite increased gene expression in response to training. To our knowledge this is the first report showing that baseline levels of muscle gene expression may have predictive value for training outcomes in older adults. Other studies suggesting that these transcripts play an important role in the response to resistance exercise training will be discussed.
Cytokines.
Altered regulation of muscle cytokine production during aging may contribute to diminished muscle adaptability. The production of pro- and anti-inflammatory cytokines in response to damaging exercise as part of the repair process is impaired by aging (12, 17), and aging results in a rise in basal inflammation that is thought to contribute to sarcopenia by stimulating muscle catabolism (29, 31). Our prior findings involving older adults and less damaging acute resistance exercise were also consistent with this idea that cytokine dysregulation could cause varied outcomes for resistance exercise training (26). The present training study investigated three hypotheses related to cytokine expression. Hypothesis 1, that the magnitude of induction of cytokine expression in response to acute exercise would be predictive of individual adaptability to training, was not supported. Changes in neither inflammatory nor anti-inflammatory cytokine expression after acute exercise were related to training outcomes. Hypothesis 2, that higher baseline levels of proinflammatory cytokines such as IL1β would diminish training outcomes, was also not supported. In fact, the opposite relationship was observed, that baseline IL1β was positively correlated with strength gain. Hypothesis 3, that the training-induced decrease in IL1β would be related to better training outcomes, was supported. The individuals who gained the most strength also had the largest decrease in IL1β after training. Similar results were found for the IL1β receptor antagonist and IL10, suggesting that the anti-inflammatory effects of resistance training that has been seen by others is also an important part of adaptation (11). Thus, regulated inflammation appears to be closely tied to training outcomes. However, more direct indicators of muscle inflammation need to be developed to understand the optimal inflammatory response to exercise.
Growth factors.
Skeletal muscle contains numerous cell types (satellite and endothelial cells, fibroblasts, motoneurons, etc.) that support the muscle fiber and likely require a variety of growth factors to respond to exercise training. As candidates, we previously identified IGF1 and its binding protein IGFBP5, CNTF, VEGFD, and GDF8 as having reduced expression in muscle of older adults (26). Data presented here show lower baseline expression of these growth factor mRNAs is associated with limited adaptability to training. A body of knowledge exists for IGF1 and GDF8 in relation to human muscle and resistance exercise, but limited information exists for the other growth factors. Variations in the CNTF gene have a genetic influence on muscle mass and strength (2, 6, 30), but aside from our previous study (8), we are not aware that muscle CNTF expression has been measured. We are also not aware of other investigations of human muscle IGFBP5 and VEGFD and resistance exercise training.
IGF1 expression at baseline and posttraining was most consistently correlated with the gain in muscle size and strength (data only shown for strength). This is consistent with a role in skeletal muscle maintenance and adaptation through the constitutive expression of its IGF1Ea isoform (36). Our IGF1 primers did not distinguish between the IGF1Ea and IGF1Ec [mechano growth factor (MGF)] isoforms, but we believe the results are primarily due to IGF1Ea since its transcript levels are two to three orders of magnitude greater than MGF in human muscle (13). The work of Bamman et al. (3) also reached the conclusion that IGF1 expression is strongly associated with the variability in resistance training outcomes. Their study was designed to compare “responders” and “nonresponders” with respect to myofiber hypertrophy. The change in IGF1 expression 24 h after acute exercise was significantly related to myofiber hypertrophy, but IGF1 expression 24 h posttraining was not related to hypertrophy. This is an interesting contrast to our results, which found that strength gain was not related to IGF1 72 h after acute exercise but was strongly related 72 h posttraining.
GDF8 (myostatin) has been extensively studied as a negative regulator of muscle growth. The hypothesis that levels of GDF8 are elevated in muscle of older compared with younger adults has been recently supported (22), but our previous results showed the opposite relationship (8). Our current results show that higher GDF8 transcript levels at baseline were predictive of positive training outcomes, which supports the “novel paradox” proposed by Kim et al. (18), whereby individuals with higher baseline levels of the “antigrowth factor” GDF8 have greater potential for muscle growth. Our results are also consistent with many previous studies showing a decrease in GDF8 expression with exercise (7, 8, 18, 23, 27). GDF8 downregulation after acute exercise was strongly associated with muscle mass gains after training (15), and we show that the magnitude of GDF8 suppression by training was strongly related to strength gains. In contrast, Kim et al. (19) found that the GDF8 response was not related to myofiber hypertrophy after training. These contrasting results may be due to differences in study design or methods of analysis. We speculate that the conflicting results might also suggest that, of the numerous complementary molecular changes required for adaptation to exercise training, certain ones may be more closely tied to specific outcomes (i.e., myofiber or whole muscle hypertrophy vs. strength gain).
Remodeling-associated factors.
The matrix metalloproteinase MMP2, the MMP inhibitor TIMP1, and the extracellular matrix (ECM) glycoprotein fibronectin were identified by our previous study as factors whose expression was altered by aging and acute resistance exercise (8). The relationships identified in the current study between expression of these factors and strength gain suggests that cleavage of muscle ECM or altered production thereof is an important process during training adaptation. MMP2 and TIMP1 have been mostly studied in human muscle during conditions of accelerated remodeling such as ischemia or paralysis (4, 21), and fibronectin is known to be a structural component of the sarcolemma (33). We are not aware of other studies that have examined these factors after resistance training, and thus, their function in this context remains to be determined. Collagen IV of the basement membrane is a typical substrate for MMP2, though fibronectin is also cleaved by MMP2 and other MMPs that are inhibited by TIMP1 (24). Cleavage of the ECM may be a structural change that allows for adaptive processes such as satellite cell migration and fusion to the myofiber, or it may play a bioactive role in regulating cell proliferation and differentiation (16, 24).
Limitations and conclusion.
The application of these results is limited in several ways: 1) The sample size was small and lacked the power to identify possible confounding variables such as sex, which can affect muscle phenotype and gene expression (37); 2) The study involved only older adults, and thus, whether expression of these genes has predictive power in healthy young adults or other populations is not yet known; and 3) At this point, the genes of interest to this study are simply promising candidates as markers of training outcomes. The list included the extensively studied IGF1 and GDF8 genes, which have been hypothesized to regulate satellite cell function in response to resistance training (18, 19), whereas the novel relationships identified between expression of the other genes and muscle adaptation provide fuel for future mechanistic hypotheses. Despite the limitations, the results of this study are particularly convincing in that the transcripts (IGF1, TIMP1, MMP2, and CNTF) that displayed the strongest relationships with training outcomes for older adults here also showed the greatest disparity between young and older adults in our previous study. Baseline gene expression was not related to muscle size and strength prior to training (data not shown). Thus, these transcripts appear to reflect the adaptive potential of the complex muscle environment and hold excellent potential as predictive markers of resistance training outcomes at least in healthy older adults and possibly other populations in need of personalized exercise and rehabilitation routines.
GRANTS
This research was supported in part by funds provided to the University of Arkansas for Medical Sciences Microarray Facility through Act 1 of The Arkansas Tobacco Settlement Proceeds Act of 2000; by National Center for Research Resources grants through the BRIN/INBRE Program (P20 RR-16460) and the General Clinical Research Center of the University of Arkansas for Medical Sciences (M01-RR14288) located in the John L. McClellan Memorial Veterans Hospital of the Central Arkansas Veterans Healthcare System; and by National Institute on Aging Grant AG-012411 to C. A. Peterson.
Acknowledgments
We thank William Evans for assistance with the clinical protocol and Melinda M. Bopp for proofreading the manuscript.
REFERENCES
- 1.Andersen LL, Magnusson SP, Nielsen M, Haleem J, Poulsen K, Aagaard P. Neuromuscular activation in conventional therapeutic exercises and heavy resistance exercises: implications for rehabilitation. Phys Ther 86: 683–697, 2006. [PubMed] [Google Scholar]
- 2.Arking DE, Fallin DM, Fried LP, Li T, Beamer BA, Xue QL, Chakravarti A, Walston J. Variation in the ciliary neurotrophic factor gene and muscle strength in older Caucasian women. J Am Geriatr Soc 54: 823–826, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Bamman MM, Petrella JK, Kim JS, Mayhew DL, Cross JM. Cluster analysis tests the importance of myogenic gene expression during myofiber hypertrophy in humans. J Appl Physiol 102: 2232–2239, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Baum O, Ganster M, Baumgartner I, Nieselt K, Djonov V. Basement membrane remodeling in skeletal muscles of patients with limb ischemia involves regulation of matrix metalloproteinases and tissue inhibitor of matrix metalloproteinases. J Vasc Res 44: 202–213, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Hoffman EP. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol 99: 154–163, 2005. [DOI] [PubMed] [Google Scholar]
- 6.De Mars G, Windelinckx A, Beunen G, Delecluse C, Lefevre J, Thomis MA. Polymorphisms in the CNTF and CNTF receptor genes are associated with muscle strength in men and women. J Appl Physiol 102: 1824–1831, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Deldicque L, Atherton P, Patel R, Theisen D, Nielens H, Rennie MJ, Francaux M. Effects of resistance exercise with and without creatine supplementation on gene expression and cell signaling in human skeletal muscle. J Appl Physiol 104: 371–378, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Dennis RA, Przybyla B, Gurley C, Kortebein PM, Simpson P, Sullivan DH, Peterson CA. Aging alters gene expression of growth and remodeling factors in human skeletal muscle both at rest and in response to acute resistance exercise. Physiol Genomics 32: 393–400, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis RA, Trappe TA, Simpson P, Carroll C, Huang BE, Nagarajan R, Bearden E, Gurley C, Duff GW, Evans WJ, Kornman K, Peterson CA. Interleukin-1 polymorphisms are associated with the inflammatory response in human muscle to acute resistance exercise. J Physiol 560: 617–626, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frontera WR, Meredith CN, O'Reilly KP, Knuttgen HG, Evans WJ. Strength conditioning in older men: skeletal muscle hypertrophy and improved function. J Appl Physiol 64: 1038–1044, 1988. [DOI] [PubMed] [Google Scholar]
- 11.Greiwe JS, Cheng B, Rubin DC, Yarasheski KE, Semenkovich CF. Resistance exercise decreases skeletal muscle tumor necrosis factor alpha in frail elderly humans. FASEB J 15: 475–482, 2001. [DOI] [PubMed] [Google Scholar]
- 12.Hamada K, Vannier E, Sacheck JM, Witsell AL, Roubenoff R. Senescence of human skeletal muscle impairs the local inflammatory cytokine response to acute eccentric exercise. FASEB J 19: 264–266, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol 547: 247–254, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubal MJ, Gordish-Dressman H, Thompson PD, Price TB, Hoffman EP, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Clarkson PM. Variability in muscle size and strength gain after unilateral resistance training. Med Sci Sports Exerc 37: 964–972, 2005. [PubMed] [Google Scholar]
- 15.Hulmi JJ, Ahtiainen JP, Kaasalainen T, Pollanen E, Hakkinen K, Alen M, Selanne H, Kovanen V, Mero AA. Postexercise myostatin and activin IIb mRNA levels: effects of strength training. Med Sci Sports Exerc 39: 289–297, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Ingber DE Fibronectin controls capillary endothelial cell growth by modulating cell shape. Proc Natl Acad Sci USA 87: 3579–3583, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Molecular characteristics of aged muscle reflect an altered ability to respond to exercise. Int J Sport Nutr Exerc Metab 11: S7–S13, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab 288: E1110–E1119, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Kim JS, Petrella JK, Cross JM, Bamman MM. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: a cluster analysis. J Appl Physiol 103: 1488–1495, 2007. [DOI] [PubMed] [Google Scholar]
- 20.Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol 101: 531–544, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Koskinen SO, Kjaer M, Mohr T, Sorensen FB, Suuronen T, Takala TE. Type IV collagen and its degradation in paralyzed human muscle: effect of functional electrical stimulation. Muscle Nerve 23: 580–589, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Leger B, Derave W, De Bock K, Hespel P, Russell AP. Human sarcopenia reveals an increase in SOCS-3 and myostatin and a reduced efficiency of Akt phosphorylation. Rejuvenation Res 11: 163–175B, 2008. [DOI] [PubMed] [Google Scholar]
- 23.Mascher H, Tannerstedt J, Brink-Elfegoun T, Ekblom B, Gustafsson T, Blomstrand E. Repeated resistance exercise training induces different changes in mRNA expression of MAFbx and MuRF-1 in human skeletal muscle. Am J Physiol Endocrinol Metab 294: E43–E51, 2008. [DOI] [PubMed] [Google Scholar]
- 24.Ohtake Y, Tojo H, Seiki M. Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J Cell Sci 119: 3822–3832, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Pescatello LS, Kostek MA, Gordish-Dressman H, Thompson PD, Seip RL, Price TB, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, Zoeller RF, Devaney JM, Hoffman EP. ACE ID genotype and the muscle strength and size response to unilateral resistance training. Med Sci Sports Exerc 38: 1074–1081, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Przybyla B, Gurley C, Harvey JF, Bearden E, Kortebein P, Evans WJ, Sullivan DH, Peterson CA, Dennis RA. Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp Gerontol 41: 320–327, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol 101: 53–59, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Rolland Y, Czerwinski S, Abellan Van Kan G, Morley JE, Cesari M, Onder G, Woo J, Baumgartner R, Pillard F, Boirie Y, Chumlea WM, Vellas B. Sarcopenia: its assessment, etiology, pathogenesis, consequences and future perspectives. J Nutr Health Aging 12: 433–450, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth SM, Metter EJ, Ling S, Ferrucci L. Inflammatory factors in age-related muscle wasting. Curr Opin Rheumatol 18: 625–630, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Roth SM, Schrager MA, Ferrell RE, Riechman SE, Metter EJ, Lynch NA, Lindle RS, Hurley BF. CNTF genotype is associated with muscular strength and quality in humans across the adult age span. J Appl Physiol 90: 1205–1210, 2001. [DOI] [PubMed] [Google Scholar]
- 31.Roubenoff R Physical activity, inflammation, and muscle loss. Nutr Rev 65: S208–S212, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Roubenoff R Sarcopenia and its implications for the elderly. Eur J Clin Nutr 54, Suppl 3: S40–S47, 2000. [DOI] [PubMed] [Google Scholar]
- 33.Stenman S, Vaheri A. Distribution of a major connective tissue protein, fibronectin, in normal human tissues. J Exp Med 147: 1054–1064, 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tidball JG Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol 288: R345–R353, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Villareal DT, Holloszy JO. DHEA enhances effects of weight training on muscle mass and strength in elderly women and men. Am J Physiol Endocrinol Metab 291: E1003–E1008, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Wallis M New insulin-like growth factor (IGF)-precursor sequences from mammalian genomes: the molecular evolution of IGFs and associated peptides in primates. Growth Horm IGF Res 19: 12–23, 2009. [DOI] [PubMed] [Google Scholar]
- 37.Welle S, Tawil R, Thornton CA. Sex-related differences in gene expression in human skeletal muscle. PLoS ONE 3: e1385, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]