Abstract
Alhydrogel® (aluminum hydroxide) is a widely used adjuvant in the US. Regulatory authorities require that vaccines be tested to determine the antigen content in the final vaccine product. The level of formulated antigen is currently determined in our laboratory by the o-Phthalaldehyde (OPA) fluorescent protein assay, and antigen identity and integrity are determined by Western blot and SDS-PAGE. However, OPA assay is non-specific and only limited to detection of total protein content, and it is often not sensitive enough to detect antigens in low dose formulations. Furthermore, antigens used in identity and integrity tests must be extracted from vaccines using an extraction procedure which is time-consuming and may not completely recover antigens for analysis or may alter the structures of antigens during extraction. The present study developed a Direct Alum Formulation Immunoassay (DAFIA) which was designed to directly (without antigen extraction), accurately, and sensitively determine the antigen content, identity and integrity on alum. The AMA1-C1/Alhydrogel formulation was used as a model vaccine in assay development and validation. The results showed that the DAFIA is highly antigen-specific, accurate (87–100%), sensitive (0.16 µg/ml), reproducible, and simple with a linear detection range of 0.16–10 µg/ml. These results demonstrate that DAFIA is an excellent assay to determine antigen content, identity and integrity of antigens bound to alum and may be used in routine vaccine quality control for testing antigens in Alhydrogel-based vaccines.
Keywords: DAFIA, Alhydrogel, vaccine, antigen content, Plasmodium falciparum apical membrane antigen 1
1. Introduction
Currently, the only adjuvants approved for human vaccine use in the US are aluminum containing compounds1–4, including aluminum hydroxide (Al(OH)3) or Alhydrogel®, aluminum phosphate (AlPO4), and potassium aluminum sulfate (KAl(SO4)2·12H2O) or alum3–5. To ensure vaccine quality, regulatory authorities require to quantitate vaccine antigen content in the final product; for example, at least 80% for tetanus vaccine by the World Health Organization6. In particular, it is essential to determine the amount as well as the identity and integrity of the antigens bound to aluminum containing adjuvants following formulation. Alhydrogel used in our formulations has a fibrous morphology with a primary particle size of 4.5 × 2.2 × 10 nm and exists as loose aggregates ranging from 1 to 10 µm5, 7–10. The presence of such aggregates in solution prevents direct quantitation of protein content in formulations using assays such as Lowry, BCA, or Bradford protein assay, not to mention that these assays are all non-specific and low in sensitivity. The o-Phthalaldehyde (OPA) fluorescent protein assay can directly and accurately determine protein content in vaccine formulations with a moderate sensitivity (20–500 µg/ml), but it only measures total proteins and lacks specificity to the protein antigens in the samples11–13 Furthermore, conventional ELISA is also unsuitable to directly measure protein content in formulations due to the presence of aggregates, albeit very high specificity. Presently, protein content in vaccine formulations is determined by OPA assay whose use has been limited to detection of levels of total proteins in formulations with concentrations of 20 µg/ml or higher. Antigen identity and integrity are determined by Western blot and SDS-PAGE, but vaccine antigens used in these assays that must be extracted from Alhydrogel formulations by a laborious and time-consuming extraction procedure whose extraction efficiency and possibility to alter the structures of antigens have been in question. Taken together, the vaccine research and development fields are in need of an assay that can quickly determine the content, identity, and integrity of formulated vaccine proteins with high accuracy, sensitivity, and specificity.
The present study developed a Direct Alhydrogel Formulation Immunoassay (DAFIA) which was designed to directly (without prior antigen extraction), accurately, sensitively, and specifically determine antigen content, identity and integrity on Alhydrogel. The Plasmodium falciparum apical membrane antigen 1 (AMA1)-C1 (AMA1-FVO and AMA1-3D7 were mixed at a ratio of 1:1) formulated on Alhydrogel was used as a model vaccine in the assay development and validation14. The present study used 3 AMA1-C1-specific monoclonal antibodies (mAb) including mAbs 1G4, 2E3, and 1E9, recognizing the domains I/II, domain III, and an unknown epitope of the AMA-1, respectively, and a penta-His mAb which recognizes the C-terminal His5-tag15. Our results indicated that this robust assay has greatly improved vaccine quality control process with high specificity, dramatically increased accuracy and sensitivity and decreased operational time needed. This assay may be widely used in vaccine research and development with a potential to monitor vaccine integrity in Alhydrogel-based formulations.
2. Material and Methods
2.1 Monoclonal antibodies
Anti-AMA1 monoclonal antibodies were generated by A&G Pharmaceutical, Inc. (Columbia, MD) with the specificity to recognize AMA1 domains I/II (1G4), domain III (2E3), or an unknown epitope of the AMA1 (1E9). Penta-His™ antibody was purchased from Qiagen (Hilden, Germany). mAb 1G4, 2E3 and 1E9 were used at 1:200 dilutions and penta-His mAb used at 1:100 dilutions in 3% skim milk (Difco™, Becton, Dickinson and Company, Franklin Lakes, NJ) in 1xTBS.
2.2 Preparation of AMA1-C1/Alhydrogel formulations
The clinical grade P. falciparum apical membrane antigen 1 (AMA1)-FVO lot WRAIR0932 and AMA1-3D7 lot WRAIR0941 were purified and characterized by Malaria Vaccine Development Branch (MVDB), National Institute of Allergy and Infectious Disease, National Institutes of Health, with the methods developed in MVDB16. Protein concentration was measured by UV spectrum at 280nm and calculated using extinction coefficient of 1.206 or 1.205 for AMA1-FVO or AMA1-3D7, respectively. Purified AMA1-FVO and AMA1-3D7 were mixed at 1:1 ratio, each concentration (0.02, 0.04, 0.08, 0.16, 0.31, 0.63, 1.25, 2.5, 5, 10, 20, or 40 µg/ml) of the mixture (assay standards) was prepared individually in Alhydrogel (1,600 µg/ml in saline, Brenntag Biosector, Denmark), aliquoted and stored at 4°C until used. The mixture was then rotated in a rotary spinner (Appropriate Technical Resources, Laurel, MD) at 16–24 rpm for 1 h at room temperature. Test samples (AMA1-C1/Alhydrogel or BSA/Alhydrogel) at 10, 40 or 160 µg/ml were prepared similarly. The standards or test samples were aliquoted and kept at 4°C until use. Test samples were further diluted to a final concentration of 1 µg/ml with 1,600 µg/ml Alhydrogel (placebo) prior to analysis by DAFIA. Samples for OPA assay were used without dilutions.
2.3 Western blot
AMA1-FVO or AMA1-3D7 (1 µg per lane) were run on pre-cast 4–20% gradient Tris-glycine SDS-PAGE gels under reducing and non-reducing conditions at a constant current of 30 mA per gel for approximately 40 min using an XCell SureLock elecrophoresis Mini-Cell apparatus (Invitrogen Corp., Carlsbad, CA), and then transferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA) at 30 V constant for 60 min. After transfer, the membranes were stained with Ponceau S solution (Sigma-Aldrich Co., St. Louis, MO) to visualize the protein bands. The lanes of each membrane were then cut into individual strips, rehydrated in 1x Tris buffered saline (TBS), and blocked with blocking buffer (3% skim milk in TBS) for 30 min, followed by incubation for 1 h at room temperature with individual mAbs. After extensive washing in TBS/0.05% Tween 20 (TBS/T, Sigma-Aldrich Co.), the strips were incubated with goat anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate (KPL Labs, Gaithersburg, MD) for 1 h at room temperature. Following further washes, the blots were developed by incubation with 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT substrate, Kirkegaard and Perry Labs, Gaithersburg, MD) according to the manufacturer’s instructions. The reaction was stopped when additional bands ceased to develop.
2.4 OPA assay
OPA reagent was purchased from Pierce (Rockford, IL) and the assay was performed in a 96-well black flat-bottom plate (Thermo Fisher Scientific, Waltham, MA) following manufacturer’s instructions.
2.5 DAFIA Procedure
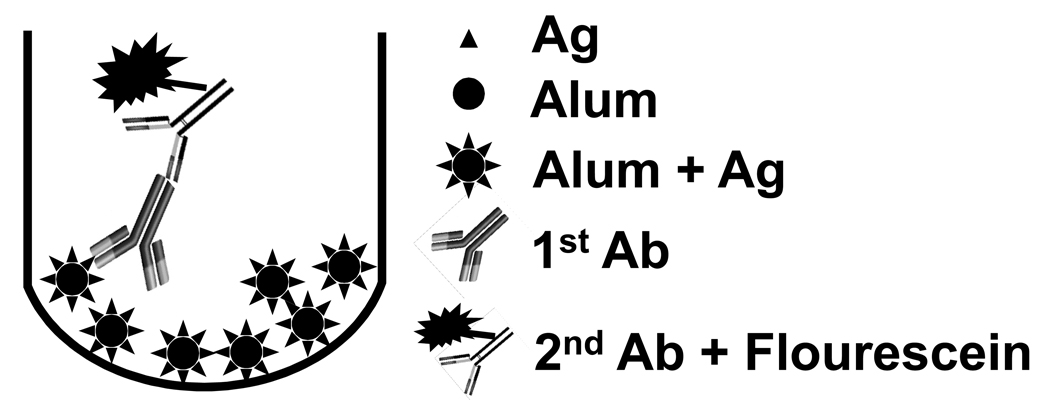
The DAFIA procedure is depicted in Fig. 1. Briefly, standards or test samples (200 µl/well) were added to black, opaque, 96-well U-bottom plates (Coring Inc. NY) and washed 3 times with phosphate-buffered saline (PBS, pH 7.4) by centrifugation at 1,000 g for 4 min at room temperature. The plates were then blocked with 200µl 3% BSA/PBS at room temperature for 1.5 h, followed by 3 washes with PBS. Mouse anti-AMA1-C1 monoclonal antibodies as primary reagent in 100 µl were added and incubated for 1 h at room temperature, followed by 3 washes with PBS. Goat anti-mouse IgG (H+L) conjugated to fluorescein (1:100, Pierce, Rockford, IL) as secondary reagent in 100µl was added to plates and incubated for 1 h at room temperature and washed 3 times with PBS. Following the final wash, pellets were resuspended in 100 µl PBS and read by a fluorometer (PerkinElmer, Waltham, MA) at 485nm/535 nm. Controls included Alhydrogel alone (1,600 µg/ml, placebo) with first and secondary Abs, Alhydrogel with secondary Ab alone, and AMA1-C1/Alhydrogel formulation at 1.25 µg/ml with secondary antibody alone.
Fig. 1.
A schematic representation of the DAFIA procedure. The assay consists of 4 steps: Step 1, addition of Alhydrogel formulation and wash (3x), followed by blocking with 3% BSA/PBS and wash (3x); Step 2, addition of and incubation with primary antibody and wash (3x); Step 3, addition of and incubation with secondary antibody-flourescein and wash (3x); and Step 4, determination of fluorescent intensity by a fluorometer at 485nm/535 nm.
2.6 Data analysis
The standard curves were generated with four-parameter nonlinear regression and the amount of standards or test samples were calculated from the equation of Y = ((b/(a-log(X)))^(d)-1)/c, where a, b, c or d are value of the parameters, Y is the amount determined and X is fluorescence readings. The back calculation was performed by converting the observed fluorescence readings at 485/535 nm of the standards to concentrations of the antigens using the four-parameter nonlinear regression equations. The percent accuracy was calculated by the following formula: percent accuracy = (|calculated concentration – nominal concentration| / nominal concentration) × 100. Inter-assay variation was calculated for the same samples (from different aliquot) that were tested in different assays and reported as coefficient of variation (CV) which is the standard deviation divided by the mean.
3. Results
3.1 Western blot analysis of AMA1-FVO and AMA1-3D7 by mAbs
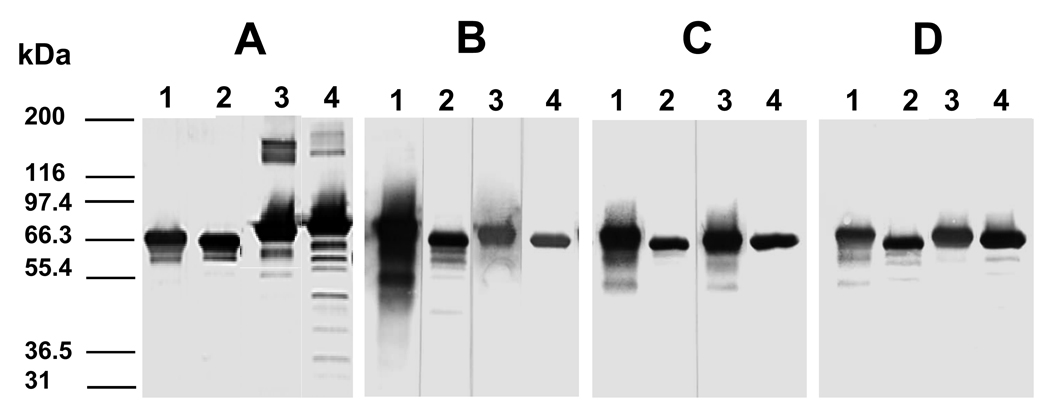
The specificity of the monoclonal antibodies to AMA1-FVO and AMA1-3D7 was determined by Western blot (Fig. 2). All mAb used in this study recognized both AMA1-FVO and AMA1-3D7 under reducing and non-reducing conditions (Fig. 2).
Fig. 2.
The specificities of monoclonal antibodies to AMA1-FVO and AMA1-3D7 were determined by Western blot. A. 1G4, B. 1E9, C. 2E3, and D. anti-his. Lane 1, AMA1-3D7 under non-reducing condition; Lane 2, AMA1-3D7 under reducing condition; Lane 3, AMA1-FVO under non-reducing condition; Lane 4, AMA1-FVO under reducing condition. Antigen was loaded at 1 µg per lane and mAbs were used at 1:500 dilution.
3.2 Establishment and validation of DAFIA
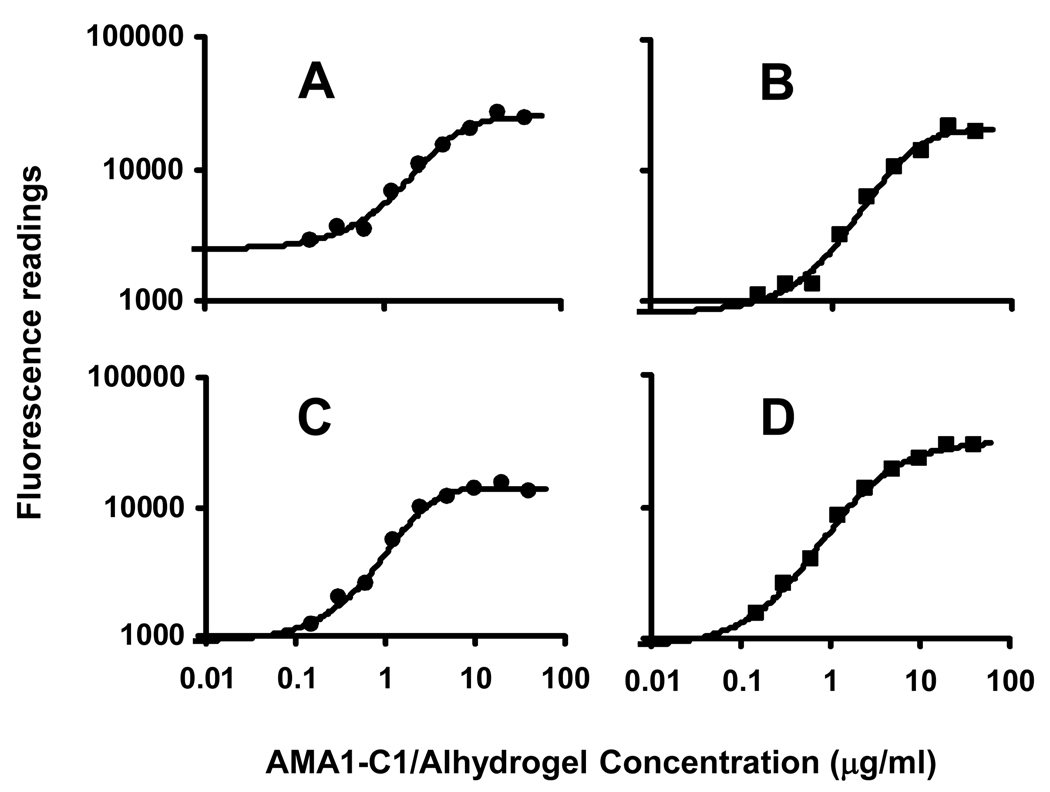
Standard samples were tested between the range of 0.02 and 40 µg/ml by DAFIA using 4 mAbs and standard curves were generated between the concentrations of 0.16 and 40 µg/ml with four-parameter nonlinear regression analysis, with a correlation coefficient (R2) greater than 0.99 (Fig. 3). The lower doses (0.02 – 0.08 µg/ml) were tested, but the data points were eliminated due to poor correlations to the nominal concentrations (data not shown).
Fig. 3.
DAFIA standard curves for AMA1-C1/Alhydrogel formulations using 4 monoclonal antibodies specific to AMA1-C1. A. Penta-his5 mAb; B. mAb 1G4; C. mAb 2E3; and D. mAb 1E9. Results showed the detection range of 0.16 – 10 µg/ml using all 4 monoclonal antibodies. All standard curves were fitted by 4-parameter nonlinear regression with R2 greater than 0.99.
Back calculation was used to confirm the reliability of the standard curves and determine the detection range of the assay. For each data point of standard curve, the protein concentration of AMA1-C1/Alhydrogel was back calculated using the fluorescent readings and the four parameter nonlinear regression equation (Table 1). The protein concentrations determined by back calculation agreed well with the nominal amounts of AMA1-C1(with a percent accuracy of 84% or higher) between a detection range of 0.16 and 10 µg/ml, indicating the standard curves within the detection range generated by four parameter nonlinear regression were useful to extrapolate AMA1-C1/Alhydrogel concentrations in vaccines. All anti-AMA1-C1 mAbs had a similar detection limit of 0.16 and 10 µg/ml with some variations in the lower detection range, with an exception for penta-His mAb which had a detection limit of 0.31 and 10 µg/ml (Fig. 2; Table 1).
Table 1.
Back calculation and percent accuracy of standard curvesa
| Nominal concentration | Concentrations detected by DAFIA (µg/ml)c |
||||
|---|---|---|---|---|---|
| (µg/ml)b | anti-his | 1G4 | 2E3 | 1E9 | Mean ± STDEV |
| 10 | 9.8 ± 0.6 | 9.8 ± 0.5 | 9.4 ± 0.1 | 9.7 ± 0.8 | 9.5 ± 0.4 |
| % accuracy | 95.6 ± 3.5 | 96.5 ± 2.7 | 93.7 ± 0.7 | 94.3 ± 4.4 | 94.5 ± 2.5 |
| 5 | 5.0 ± 0.6 | 5.3 ± 0.6 | 5.4 ± 0.4 | 4.5 ± 0.5 | 5.2 ± 0.5 |
| % accuracy | 92.0 ± 6.1 | 92.2 ± 8.6 | 90.2 ± 4.9 | 90.2 ± 9.4 | 91.8 ± 5.2 |
| 2.5 | 2.5 ± 0.1 | 2.4 ± 0.2 | 2.6 ± 0.3 | 2.7 ± 0.1 | 2.53 ± 0.2 |
| % accuracy | 96.3 ± 2.5 | 94.8 ± 7.2 | 91.4 ± 5.5 | 93.5 ± 5.3 | 93.5 ± 5.0 |
| 1.25 | 1.34 ± 0.04 | 1.29 ± 0.08 | 1.28 ± 0.10 | 1.34 ± 0.05 | 1.31 ± 0.08 |
| % accuracy | 92.7 ± 2.8 | 95.3 ± 4.3 | 93.3 ± 4.3 | 92.5 ± 3.9 | 93.3 ± 3.6 |
| 0.63 | 0.67 ± 0.08 | 0.60 ± 0.07 | 0.58 ± 0.05 | 0.57 ± 0.03 | 0.59 ± 0.06 |
| % accuracy | 91.5 ± 11.1 | 92.3 ± 5.6 | 91.6 ± 7.2 | 90.8 ± 4.6 | 91.5 ± 6.1 |
| 0.31 | 0.32 ± 0.09 | 0.34 ± 0.03 | 0.33 ± 0.03 | 0.33 ± 0.02 | 0.33 ± 0.04 |
| % accuracy | 84.0 ± 2.2 | 92.2 ± 9.5 | 91.5 ± 8.8 | 94.7 ± 3.5 | 91.2 ± 7.2 |
| 0.16 | NDd | 0.14 ± 0.03 | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.15 ± 0.02 |
| % accuracy | - | 86.5 ± 15.7 | 90.9 ± 4.9 | 90.7 ± 5.4 | 90.0 ± 6.9 |
Back calculation was performed by converting the observed readings at 485nm/535 nm of the standards to concentrations of antigens using the four parameter nonlinear regression equations; percent accuracy is percent similarity between the amount of AMA1-C1 calculated by back calculation (detected by DAFIA) and the known amount of AMA1-C1. Percent accuracy = (|calculated concentration – nominal concentration| / nominal concentration) × 100. Results are means ± standard deviation, representing 2–4 independent experiments.
Known amount of AMA1-C1 (AMA1-FVO and AMA1-3D7 at 1:1 ratio) was fresh formulated on Alhydrogel.
Concentrations detected by DAFIA were obtained by back calculation using the four parameter nonlinear regression equations.
ND, not detectable.
Inter-assay variation analysis showed that coefficient of variation (CV) of the test samples were from 0 % to 16.0 % at 1.0 µg/ml (Table 2). In general, the acceptable CV between the assays should be less than 15%. For all 12 experiments performed, only one experiment (1E9 at 40 µg/ml dose) gave a higher than expected CV (16%), which indicate that the DADIA was repeatable within the defined detection range.
Table 2.
Analysis of test samples using DAFIA and OPAa
| Nominal | Concentration detected (µg/ml) |
|||||
|---|---|---|---|---|---|---|
| concentration | DAFIA |
OPA | ||||
| (µg/ml)b | anti-his | 2E3 | 1E9 | 1G4 | Mean ± STDEV | |
| 160 | 164.4 ± 0.3 | 152.9 ± 1.8 | 168.7 ± 1.8 | 160.2 ± 9.6 | 159.3 ± 9.5 | 167.7 ± 2.7 |
| % accuracy | 97.3 ± 0.2 | 95.6 ± 1.1 | 94.6 ± 1.1 | 95.8 ± 0.1 | 95.0 ± 2.7 | 95.2 ± 1.3 |
| CVc | 0.2 | 1.2 | 1.1 | 6.0 | - | 1.6 |
| 40 | 40.4 ± 6.0 | 45.1 ± 5.2 | 40.1 ± 6.4 | 42.0 ± 1.9 | 41.9 ± 5.0 | 39.7 ± 1.5 |
| % accuracy | 88.9 ± 1.3 | 87.3 ± 12.9 | 89.3 ± 1.3 | 94.8 ± 4.3 | 89.7 ± 6.0 | 97.5 ± 3.5 |
| CV | 14.9 | 11.5 | 16.0 | 4.5 | - | 3.8 |
| 10 | 10.6 ± 0.0 | 12.3 ± 1.0 | 10.8 ± 0.8 | 11.1 ± 1.0 | 11.3 ± 0.9 | NDd |
| % accuracy | 94.2 ± 0.4 | 76.6 ± 9.8 | 91.5 ± 8.3 | 89.0 ± 9.0 | 87.0 ± 9.1 | - |
| CV | 0 | 8.1 | 9.7 | 9.0 | - | - |
DAFIA was performed with four AMA1-C1-specific mAbs and OPA assay was conducted following the manufacturer’s instructions.
Known amount of AMA1-C1 (AMA1-FVO and AMA1-3D7 at 1:1 ratio) was fresh formulated on Alhydrogel.
CV, coefficient of variation between assays. CV is calculated as standard deviation divided by mean.
Test sample AMA1-C1-Alhydrogel formulations were prepared fresh and diluted to a final concentration of 1.0 µg/ml with placebo (1,600 µg/ml Alhydrogel).
ND, not detectable.
Alhydrogel alone (1,600 µg/ml, placebo) with first and secondary Ab, placebo with secondary Ab alone or AMA1-C1/Alhydrogel formulation at 1.25 µg/ml with secondary antibody alone were used as controls. BSA/Alhydrogel was formulated at 10, 40 or 160 µg/ml and used as irrelevant antigen control for the specificity of Penta-His™ antibodies. These controls had readings similar to those of the background (data not shown).
3.3 Comparative analysis of formulation samples by DAFIA and OPA
Freshly prepared Alhydrogel formulations at 10, 40 or 160 µg/ml were analyzed by DAFIA using 4 monoclonal antibodies. All samples were pre-diluted to a final concentration of 1 µg/ml by placebo prior to analysis. The results showed that a % accuracy of 87% or higher for all samples tested was achieved, with an exception that the % accuracy (76.6 ± 9.8) for sample formulation at 10 µg/ml analyzed with 2E3 was slightly lower than those of other mAbs (Table 2).
AMA1-C1/Alhydrogel formulation at 40 or 160 µg/ml doses was also tested by OPA assay to confirm the protein content. The 10 µg/ml dose was excluded due to insufficient assay sensitivity with OPA assay. The results of OPA assay were comparable to the results of DAFIA for 40 and 160 µg/ml doses (Table 2).
4. Discussion
There is no generic method available for the determination of protein content for the alum-based vaccines. Katz reported the use of ELISA to quantitate antigens formulated on Alhydrogel17. This report utilized a 2-step procedure for analysis, involving elution of antigens from Alhydrogel and followed by ELISA. However, the elution procedure was very tedious using 1.2 M potassium phosphate, and the efficacy of elution and the chance to alter the structure of antigens are in question. Our laboratory adapted OPA assay for the direct protein content determination of intact vaccines, but the sensitivity of detection of this assay is limited and unable to determine individual antigens in the multivalent vaccines due to lack of specificity.
The results from current study clearly showed that the DAFIA was highly accurate (87–100%), sensitive (0.16 µg/ml with 3 out of 4 mAbs), and simple to perform with a detection range of 0.16–10 µg/ml. In theory, DAFIA is able to detect protein content of vaccine formulations with concentrations ranging from 0.16 µg/ml to the highest concentrations formulated, because diluting seem not to compromise the accuracy of this test as shown with formulations between 10 and 160 µg/ml. At the least, this assay should be able to overcome the disadvantage of OPA assay and accurate detection of protein content in low dose formulations (10 µg/ml or lower) in our quality control process.
In addition to its sensitivity and accuracy for protein content determination, DAFIA has several important potential applications while protein content is simultaneously measured using single or multiple specific mAbs. First of all, since this is an antibody-based assay, DAFIA can specifically determine the protein identity on Alhydrogel without protein extraction. Secondly, it is highly feasible that DAFIA may be used to determine protein integrity while the intact formulation state is maintained. Our results clearly demonstrated that the epitopes recognized by all antibodies were present on AMA1-C1/Alhydrogel, in part depicting the intact state of the vaccine. Additional work is warranted using molecularly engineered truncated vaccines and multiple specific antibodies recognizing structures critical to the immunogenicity of the vaccine of interest. Thirdly, the use of DAFIA eliminates the laborious protein extraction process, and therefore prevents protein loss due to experimental and human errors and structure alterations including loss of critical epitopes by inevitable exposure to the extraction reagents and temperature variations. Finally, DAFIA can shorten the operational time and simplify the operation procedures. Moreover, it may be reasonably to speculate that DAFIA is particularly useful in determining protein content, identity, integrity and/or structural alterations of multivalent vaccines provided that all the specific antibodies to each of the vaccine components are available.
In summary, DAFIA represents a novel assay which may have broad applications in quality control of vaccine research and development or other similar research areas.
Acknowledgements
We thank David Narum and Richard Shimp, Jr. for making AMA1-FVO and AMA1-3D7 available, and this work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- DAFIA
Direct Alhydrogel Formulation Immunoassay
- OPA
o-Phthalaldehyde
- AMA1
Plasmodium falciparum apical membrane antigen 1
- AMA1-C1
AMA1-FVO and AMA1-3D7 were mixed at a ratio of 1:1
- STDEV
standard deviation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Singh M, O'Hagan DT. Recent advances in vaccine adjuvants. Pharm Res. 2002;19(6):715–728. doi: 10.1023/a:1016104910582. [DOI] [PubMed] [Google Scholar]
- 2.Peek LJ, Martin TT, Elk Nation C, Pegram SA, Middaugh CR. Effects of stabilizers on the destabilization of proteins upon adsorption to aluminum salt adjuvants. J Pharm Sci. 2007;96(3):547–557. doi: 10.1002/jps.20762. [DOI] [PubMed] [Google Scholar]
- 3.Baylor NW, Egan W, Richman P. Aluminum salts in vaccines--US perspective. Vaccine. 2002;31(20) Suppl 3:S18–S23. doi: 10.1016/s0264-410x(02)00166-4. [DOI] [PubMed] [Google Scholar]
- 4.Gupta RK. Aluminum compounds as vaccine adjuvants. Adv Drug Deliv Rev. 1998;6(32 3):155–172. doi: 10.1016/s0169-409x(98)00008-8. [DOI] [PubMed] [Google Scholar]
- 5.Hem SL, Hogenesch H. Relationship between physical and chemical properties of aluminum-containing adjuvants and immunopotentiation. Expert Rev Vaccines. 2007;6(5):685–698. doi: 10.1586/14760584.6.5.685. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. Technical Report Series No. 595. 1976:6–8.
- 7.Johnston CT, Wang SL, Hem SL. Measuring the surface area of aluminum hydroxide adjuvant. J Pharm Sci. 2002;91(7):1702–1706. doi: 10.1002/jps.10166. [DOI] [PubMed] [Google Scholar]
- 8.Morefield GL, HogenEsch H, Robinson JP, Hem SL. Distribution of adsorbed antigen in mono-valent and combination vaccines. Vaccine. 2004;7(2215–16):1973–1984. doi: 10.1016/j.vaccine.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 9.Lindblad EB. Aluminium compounds for use in vaccines. Immunol Cell Biol. 2004;82(5):497–505. doi: 10.1111/j.0818-9641.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindblad EB. Aluminium adjuvants--in retrospect and prospect. Vaccine. 2004;9(2227–28):3658–3668. doi: 10.1016/j.vaccine.2004.03.032. Review. [DOI] [PubMed] [Google Scholar]
- 11.Roth M. Fluorescence reaction for amino acids. Anal Chem. 1971;43(7):880–882. doi: 10.1021/ac60302a020. [DOI] [PubMed] [Google Scholar]
- 12.Porter DH, Swaisgood HE, Catignani GL. A rapid fluorometric assay for measurement of peptidase activity. Anal Biochem. 1982;123(1):41–48. doi: 10.1016/0003-2697(82)90620-0. [DOI] [PubMed] [Google Scholar]
- 13.Church FC, Porter DH, Catignani GL, Swaisgood HE. An o-phthalaldehyde spectrophotometric assay for proteinases. Anal Biochem. 1985;1(1462):343–348. doi: 10.1016/0003-2697(85)90549-4. [DOI] [PubMed] [Google Scholar]
- 14.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24(2):74–84. doi: 10.1016/j.pt.2007.12.002. Review. [DOI] [PubMed] [Google Scholar]
- 15.Kato K, Mayer DC, Singh S, Reid M, Miller LH. Domain III of Plasmodium falciparum apical membrane antigen 1 binds to the erythrocyte membrane protein Kx. Proc Natl Acad Sci U S A. 2005;102(15):5552–5557. doi: 10.1073/pnas.0501594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy MC, et al. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70(12):6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz J. Desorption of porcine parvovirus from aluminum hydroxide adjuvant with subsequent viral immunoassay or hemagglutination assay. Vet Res Commun. 1987;11(1):83–92. doi: 10.1007/BF00361329. [DOI] [PubMed] [Google Scholar]