Abstract
Cdc25B is a dual-specificity phosphatase that catalyzes the dephosphorylation of the Cdk2/CycA protein complex. This enzyme is an important regulator of the human cell cycle, and has been identified as a potential anti-cancer target. In general, protein tyrosine phosphatases are thought to bind the dianionic form of the phosphate and employ general acid catalysis via the Asp residue in the highly conserved WPD-loop. However, the Cdc25 phosphatases form a special subfamily based on their distinct differences from other protein tyrosine phosphatases. Although Cdc25B contains the (H/V)CX5R catalytic motif present in all other protein tyrosine phosphatases, it lacks an analogous catalytic acid residue. No crystallographic data currently exists for the complex of Cdc25B with Cdk2/CycA, so in addition to its natural protein substrate, experimental and theoretical studies are often carried out with small molecule substrates. In an effort to gain understanding of the dephosphorylation mechanism of Cdc25B with a commonly used small molecule substrate, we have performed simulations of the rate-limiting step of the reaction catalyzed by Cdc25B with the substrate p-nitrophenyl phosphate using the recently developed QM/MM Minimum Free Energy Path method (Hu, et al., J. Chem. Phys. 2008, 034105). We have simulated the first step of the reaction with both the monoanionic and dianionic forms of the substrate, and our calculations favor a mechanism involving the monoanionic form. Thus, Cdc25 may employ a unique dephosphorylation mechanism among protein tyrosine phosphatases, at least in the case of the small molecule substrate p-nitrophenyl phosphate.
I. INTRODUCTION
One of the most common post–translational modifications encountered in proteins is reversible protein phosphorylation.[1] Enzymes that catalyze phosphate monoester hydrolysis (phosphatases) operate in an intricate balance with those that catalyze phosphorylation (kinases) and are therefore vitally important in cellular signaling and regulation. Two distinct families of protein phosphatases are known. These include the metal cation–dependent serine/ threonine phosphatases (PSTPases) and the protein tyrosine phosphatases (PTPases) (See Ref. 2 for a review of protein phosphatase reactions). The dual–specificity phosphatases (DSPs), of which Cdc25 phosphatases are examples, are a subclass of the PTPases. DSPs are uniquely able to catalyze the dephosphorylation of both phosphoserine/threonine and phosphotyrosine.
Three Cdc25 isoforms exist and each performs a distinct role in the regulation of the cell cycle. Cdc25A is involved in the checkpoint between the G1/S phase of the cell cycle and Cdc25B and Cdc25C mediate the G2/M checkpoint. Cdc25B is responsible for the activation of the Cdk/CycA complexes, which facilitate mitosis. Thus, inhibition of Cdc25 provides an avenue for halting cell division. It has also been noted that increased levels of expression of Cdc25 phosphatases are associated with several types of cancer.[3] Accordingly, Cdc25 phosphatases exhibit potential as targets for antitumor therapeutics.[4] For a recent review on the structure, specificity, and mechanism of the Cdc25 phosphatases, see Ref. 5.
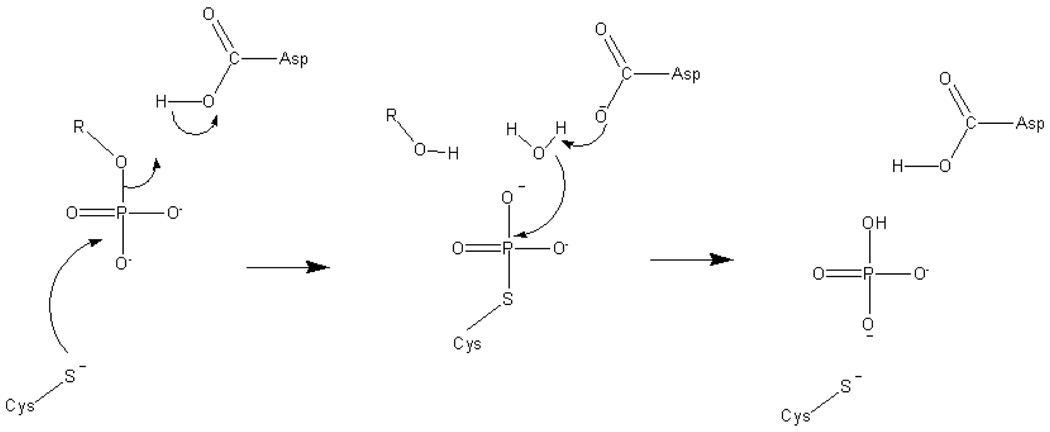
Cdc25B and other PTPases are responsible for the dephosphorylation of phosphate monoesters and proceed in two distinct chemical steps, as shown in Scheme 1. In the first step, a Cys residue performs an in-line nucleophilic attack on the P-O ester bond to form a covalent phosphocysteine intermediate. In the second step, the phosphocysteine is hydrolyzed to liberate phosphate and regenerate the active form of the enzyme. The natural substrate for Cdc25B is the Cdk2/CycA complex. In addition to the protein substrate, Cdc25B also dephosphorylates small molecule substrates such as O-methylfluorescein phosphate (mFP), para–nitrophenyl phosphate (pNPP), and phenyl phosphate, although at significantly reduced rates.
Scheme 1.
Reaction catalyzed by PTPases.
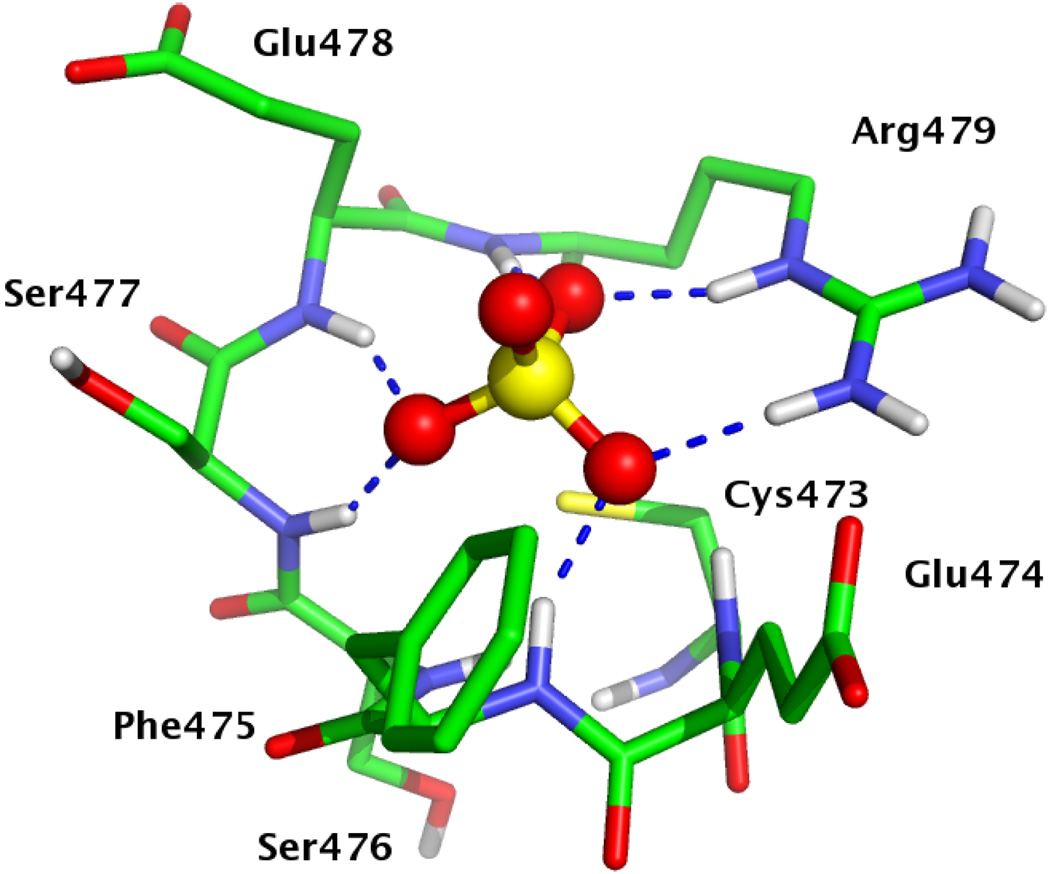
The active site of Cdc25B, shown in Figure 1, is characterized by the highly conserved HCX5R motif, also known as the phosphate binding loop, or P–loop. Binding and electrostatic stabilization of phosphate is facilitated by several inwardly– oriented backbone N–H bonds and the guanidinyl sidechain of Arg479. The nucleophile, Cys473 in Cdc25B, is deprotonated and has a pKa of 5.9.[6] Mutation studies have shown that Cys473 and Arg479 are required for enzyme activity.[7]
FIG. 1.
Active site of Cdc25B with sulfate (PDB ID 1qb0). Cys473 is shown in the thiolate form; Glu474 and Glu478 are anionic. Nonpolar hydrogens have been omitted for clarity.
Among the various subfamilies of PTPases, there exists quite little sequence homology. In fact, often the only unifying feature is the (H/V)CX5R motif, and Cdc25B is no exception to this rule. All PTPases bear the (H/V)CX5R signature motif. However, the active sites of most other known PTPases also contain a Ser or Thr residue immediately following the Arg. Interestingly, Cdc25 lacks a Ser/Thr in this position, instead substituting a Gly residue.
Other PTPases also exhibit a conserved sequence motif known as the WPD-loop. The Asp residue in the WPD-loop is almost ubiquitously accepted as the catalytic acid in these PTPases. However, the WPD-loop is not present in Cdc25. Thus, there are no obvious candidates for catalytic acid residues in or near the active site of Cdc25.
Virtually all PTPase reactions are thought to proceed through binding of the dianionic form of the substrate followed by general acid catalysis.[2] In nearly all protein tyrosine phosphatases, acid–catalysis via the Asp residue in the WPD-loop has become the most commonly accepted mechanism. In addition to numerous experimental studies, several simulations [8–14] have been performed to determine the mechanisms of various PTPases. For example, Asthagiri et al. used density functional theory (DFT) calculations and a self-consistent reaction field approach to represent the protein and solvent in their simulation of the reaction of bovine heart phosphotyrosyl phosphatase (BPTP).[12] They predicted a mechanism in which the dianionic form of the phenyl phosphate substrate was favored, with protonation of the substrate leaving group by Asp129 in the WPD-loop.
Recently, Arantes used a specially calibrated semi-empirical QM parameterization[14] to compute the mechanism of the dual-specificity VHR (Vaccinia VH1-related) phosphatase with phenyl phosphate as the substrate.[15] The author concluded that VHR dephosphorylates phosphate monoester dianions, not monoanions, and that Asp92 is the catalytic acid.
One exception to the generally accepted PTPase mechanism was proposed by Kolmodin et al.[10] They used empirical valence bond calculations to compare the reaction mechanisms of low molecular weight (LMW) phosphatase with mono- and dianionic phenyl phosphate substrate. Although they predicted identical free energy barriers for both forms of the substrate, they found that the monoanion form had a much greater binding affinity. However, the validity of their results has been called into question recently.[15]
Because the WPD-loop is absent from the active site of all Cdc25 isoforms, Glu474 and Glu478 in the catalytic motif of Cdc25B have been proposed to perform the role of the catalytic acid. However, Chen et al. provided experimental evidence debunking this assertion.[16] They found that the overall activity of Cdc25B toward pNPP was 100-fold lower for the E474Q mutant, and was unchanged for the E478Q mutant, but the pH-rate profile for the reaction was unchanged. They also suggested that the reaction of Cdc25B with its natural protein substrate may proceed in an entirely different manner. In this case, the putative catalytic acid residue may instead be located on the protein substrate rather than on Cdc25B.[16] Such an arrangement would undoubtedly impart high specificity on the reaction. However, the catalytic acid would have to be oriented in such a way that it could protonate the leaving group of both phosphorylated residues, pThr14 and pTyr15, of Cdk2.[6] Recently, we proposed a docked model of Cdc25B with the Cdk2/CycA substrate based on a combination of rigid-body docking, molecular dynamics, and double mutant cycle experiments.[17] Although the predicted overall topology of the protein-protein complex is quite reasonable, it is unlikely that the detailed structure of the protein-protein interface in the vicinity of the active site is adequate for use in further simulation studies.
Even though pNPP is regarded as a poor substrate, it has been used extensively to study the mechanism of various PTPases, and a significant amount of experimental data exists for this substrate. It is often used as a mechanistic probe of the first step of the reaction of Cdc25 (formation of the phosphocysteine intermediate). The pKa of the leaving group oxygen (Olg) of pNPP is 7.0, which suggests that the P-Olg bond is able to dissociate without protonation of the leaving group. Kinetic isotope effects for pNPP hydrolysis by the closely related enzyme Cdc25A were found to resemble those obtained for other PTPases in which the catalytic acid had been removed by mutation.[18] In fact, Cdc25 is the only known phosphatase that does not utilize acid catalysis in the dephosphorylation of pNPP.
The pH-rate profile of kcat/Km for Cdck2-pTpY/CycA protein substrate is bell-shaped, with a slope of +2 for the acidic limb and -1 for the basic limb.[16] The ionizations in the acidic limb correspond to the pKas of the phosphothreonine substrate (6.1) and Cys473 (5.9), and the ionization in the basic limb corresponds to the pKa of the catalytic acid (6.4). Recently, a pH-rate profile has been determined for Cdc25B at low pH and for thiophosphorylated protein substrate.[19] In that study, it was observed that the pH-rate profile for thiophosphorylated substrate was still bell-shaped and the basic limb was unchanged, but the slope of the acidic limb was +1. At physiological pH, there exists an equilibrium between the monoanionic and dianionic forms of phosphate monoesters, and this ionization is manifested in the acidic side of the pH-rate profile for Cdc25B with protein substrate. Thiophosphate monoesters, however, have pKa values that are approximately 1.5 pK units lower than phosphate monoesters, and are therefore present only as dianions at physiological pH. Thus, the difference in the acidic side of the pH-rate profile between phosphorylated substrate (slope = +2) and thiophosphorylated substrate (slope = +1) clearly demonstrates that a phosphoryl proton is not required for catalysis, and Cdc25B dephosphorylates dianionic phosphates on the protein substrate. Unfortunately, analogous experiments for pNPP were not possible based on its poor reactivity and limitations of the colorimetric assay.
Most likely, the mechanism employed by Cdc25B in the dephosphorylation of pNPP is different than that of protein substrate. In fact, McCain et al. showed that the mechanism for Cdc25A with various small molecule substrates is different based on whether the pKa of the leaving group is above or below about 8[20] When the pKa is higher than 8, the mechanism is acid catalyzed. However, when the leaving group pKa is lower than 8 (as is the case for pNPP), no catalytic acid is required.
In this work, we seek to determine the dephosphorylation mechanism of Cdc25B with the small molecule substrate pNPP. We have chosen pNPP because its dephosphorylation reaction is well characterized experimentally and its reaction with Cdc25B has been shown to proceed without acid catalysis.[20] For the pNPP substrate, hydrolysis of the phosphocysteine is known to be rapid (i.e., non-rate-limiting) from experimental studies.[16] Because the formation of the phosphocysteine intermediate is rate-limiting, we only simulate the reaction up to that step. The QM/MM-MFEP method[21] is used to compute the free energy profiles for two possible reaction mechanisms involving the monoanionic or dianionic form of pNPP and compare them to the experimental rate constant[16] (kcat = 0.17 s−1, ΔG‡ = 18.5 kcal mol−1 at 298 K) using classical transition state theory. To our knowledge, this work represents the first quantum chemical and free energy based simulation of a Cdc25 phosphatase enzyme.
II. METHODS
The structure of the Δ25B1 catalytic domain (residues 374–551) of Cdc25B phosphatase co-crystallized with sulfate[22] was obtained from the Protein Data Bank (PDB ID 1QB0). The MolProbity server[23] was used to add hydrogens to the protein and assess the assignment of sidechain rotamers. Protonation states were further investigated with the H++ server.[24–26]
The H++ server correctly predicted that Cys473 is in the thiolate form, consistent with the experimentally measured pKa of 5.9.[6] Glu474 and Glu478 were also predicted to be in the anionic form, providing further support that these residues do not serve as catalytic acid residues. Cys473 was modeled in the deprotonated form, and all other residues were simulated in their standard pH 7 protonation states. For the six histidine residues, hydrogens were placed at the ε-position for His375, His395, His519, and His533, and at the δ-position for His436, His472. This configuration was found to yield the lowest RMSD relative to the crystal structure in 2 ns classical MD simulations. The total charge of the protein was −1, and no counterions were included in the simulation.
For the pNPP monoanion system, the QM subsystem consisted of the sidechains of Cys473 and Arg479, the monoanionic pNPP substrate and one water molecule. For the pNPP dianion system, the QM subsystem contained the sidechains of Cys473 and Arg479 and the dianionic pNPP substrate. All other atoms were represented by the Charmm22 force field[27] and the TIP3P water model.[28] The pseudobond method[29] was used to describe the interface between the QM and MM subsystems and all QM/MM calculations were performed with the program Sigma[30–32] interfaced with a modified version of Gaussian 03.[33] The B3LYP exchange–correlation functional[34, 35] and the 6–31G(d) basis set were used for all QM calculations, and the Merz-Singh-Kollman[36] scheme was used for ESP charge fitting.
A multiple time step algorithm was used in the MD simulations. Step sizes of 1 fs and 4 fs were used for short and medium range forces, respectively. The nonbonded pair list was updated every 20 fs, and a nonbonded cutoff of 8.0 Å was used for short range interactions. No cutoff was used for the long range interactions. The temperature was maintained at 300 K using a Berendsen thermostat.[37] For each MD simulation in a given QM/MM-MFEP optimization cycle, an initial period of 16 ps of equilibration was followed by 64 ps of sampling.
QM/MM-MFEP method
Several studies demonstrating the utility of QM/MM simulations of enzyme reactions have been performed in our lab[21, 29, 38–43] and by others. A comprehensive review of QM/MM methods and applications can be found in Ref. 44.
In an effort to compute reaction free energies directly and circumvent the difficulties associated with optimization on potential energy surfaces and bias stemming from single starting conformations, we have recently developed the QM/MM Minimum Free Energy Path (MFEP) method for simulating reactions in enzymes and in solution.[21, 45] In this approach, the QM atoms are optimized in the environment of the fluctuating MM subsystem. A finite ensemble is generated via molecular dynamics simulations and the QM conformations are optimized on the free energy, or potential of mean force (PMF), surface. Once the QM subsystem is optimized, its geometry and charges are then used to obtain more accurate sampling for the MM subsystem. This process is then iterated until convergence is achieved.
In this work we use the Quadratic String Method (QSM)[46] to optimize each reaction path, or chain-of-replicas. Once the reactant and product geometries have been obtained, a discretized, linear interpolation is generated. The path is then optimized by integrating in the descent direction perpendicular to the path. The individual replicas are then redistributed using a cubic spline interpolation to enforce equal geometric spacing along the path.
For further information regarding the simulation setup and an overview of the QM/MM-MFEP method, see the Supporting Information.
III. RESULTS AND DISCUSSION
In this section, we report the optimized structures and convergence behavior of important QM/MM-MFEP optimized stationary points for the complexes of Cdc25B with the pNPP monoanion and dianion substrates. Note that we refer to the phosphocysteine intermediate as the product state because its formation is known to be rate-limiting and we did not simulate the subsequent phosphocysteine hydrolysis step.
Geometry optimizations for the reactant, product and intermediate states for the monoanion system proceeded efficiently with the QM/MM-MFEP method. The reactant state optimization required a total of 12 cycles, the intermediate state required 10 cycles, and the product state needed only 6 cycles to achieve convergence. Transition state 2 was optimized to a true transition state (i.e., a single imaginary eigenvalue) in only 3 cycles. The initial guess coordinates for transition state 2 were taken from the highest energy point on a QSM path connecting the intermediate to the product state. Because of the complexity of the energy surface, several atoms were constrained to enable faster convergence. The constrained atoms included Cα and Cβ of Cys473, all of Arg479, and the phenoxide leaving group of pNPP. These constraints were reasonable because the quality of the initial guess geometry was quite high.
Michaelis complex - pNPP monoanion
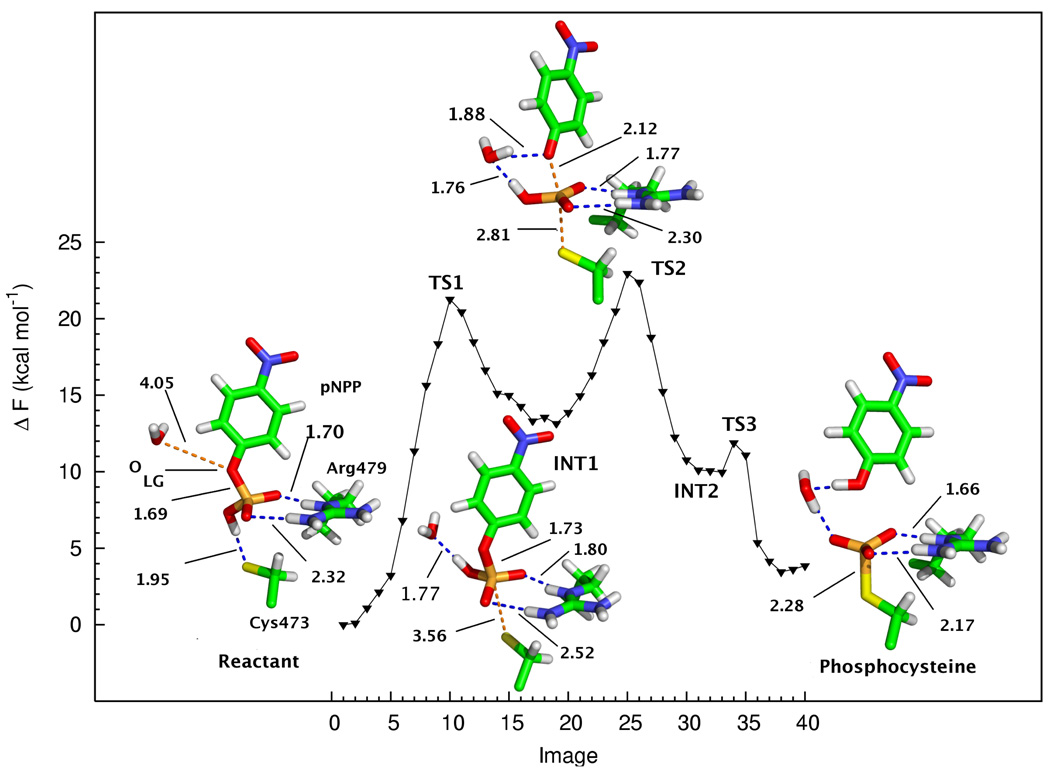
The QM/MM-MFEP optimized reactant state for Cdc25B with the pNPP monoanion substrate centered in the phosphate binding loop is shown in Figure 2. From the Figure, it is evident that the hydroxyl proton of the phosphate is involved in a hydrogen bond with the thiolate sulfur of Cys473, characterized by an Sγ-H distance of 1.95 Å. The other two nonbridging oxygens of the phosphate are involved in a bidentate hydrogen bonding interaction with the guanidinyl sidechain of Arg479. The hydrogen bond distances between the N-H groups of Arg479 and the phosphate oxygens are 1.70 and 2.32 Å, respectively. The P-Olg bond distance, at 1.69 Å, is longer than what might be expected based on available crystal structures of pNPP monoanions (See Supporting Information). The nucleophilic thiolate Sγ is 3.78 Å from the P atom of pNPP, and the Sγ-P-Olg angle is 149.4 degrees. This conformation precludes an in-line nucleophilic attack on phosphate by Cys473, particularly considering that the thiolate group is hydrogen bonded to the phosphate hydroxyl group.
FIG. 2.
Free energy profile and stationary points for the reaction of Cdc25 with the pNPP monoanion. Only atoms in the QM subsystem are shown. Important distances (in Å) are labeled.
Additionally, a water molecule is hydrogen bonded to Glu474 and one of the nonbridging oxygens of the phosphate throughout the molecular dynamics trajectories (not shown). In the crystal structure, an analogous water forms a hydrogen bond bridge between the apical oxygen of the bound sulfate and Glu474. This water molecule does not appear to have a role other than helping to stabilize the substrate in the correct conformation. The next-nearest water molecule to Olg is > 4 Å away. Thus, although the active site is located at the protein–solvent interface, it is relatively hydrophobic upon substrate binding of the pNPP monoanion.
Intermediate state - pNPP monoanion
A stable intermediate structure was found for the pNPP monoanion reaction in which the hydrogen bond between the hydroxyl proton of the phosphate and the thiolate of Cys473 has been broken and the Olg-P-O-H dihedral has rotated from−179.9 degrees to −12.7 degrees. In this structure, shown in Figure 2, the hydroxyl group has been reoriented so that it forms a hydrogen bond with a water molecule that has approached the substrate from the bulk solvent. The O-H distance between the phosphoryl oxygen and the water molecule is 1.77 Å. Although there is no significant hydrogen bonding interaction between the water molecule and Olg, the distance between the two oxygen atoms is only 3.09 Å. The interactions between the N-H groups of Arg479 and the nonbridging oxygens of pNPP remain largely intact, although the H-O distances have increased to 1.80 and 2.52 Å, respectively.
The hydrogen bonding pattern between the phosphate and the QM water molecule suggests a mechanism in which the phosphate proton may be transferred via the QM water to the leaving group oxygen of the substrate. The geometry induced by this conformational rearrangement now permits nucleophilic attack by Cys473. The Sγ-P-O angle is nearly linear at 174.5 degrees, whereas it is only ~145 degrees in the reactant structure. The P-Olg bond has elongated slightly to 1.73 Å and the P-Sγ distance has decreased slightly from 3.78 Å in the reactant structure to 3.56 Å in the intermediate, thus facilitating an inline attack by the thiolate nucleophile.
Transition state 2 - pNPP monoanion
Transition state 2 (Figure 2) is characterized by a dissociative metaphosphate structure with a P-Olg distance of 2.12 Å and a P-Sγ distance of 2.82 Å. This predicted structure is in agreement with kinetic isotope effect experiments which suggest a dissociative, metaphosphate-like transition state with no protonation of the leaving group.[20] The heavy atoms of the protonated metaphosphate are coplanar and the Olg-P-Sγ angle is 169.7 degrees. The bidentate hydrogen bonding interaction between Arg479 and metaphosphate is preserved in the transition state, with O-H distances of 1.77 and 2.30 Å, respectively. Note that the metaphosphate remains protonated in this step and proton transfer to the leaving group does not occur until after formation of the phosphocysteine.
Product state - pNPP monoanion
In the product (i.e., phosphocysteinyl) state for the pNPP monoanion shown in Figure 2, the phosphoryl group has undergone a concerted, dual proton transfer to the leaving group via the nearby water molecule. The P-Sγ bond in the phosphocysteine structure is quite long at 2.28 Å, suggesting that this bond is relatively weak and easily hydrolyzed. This provides evidence that the final hydrolytic step of the reaction is likely to occur quite rapidly, consistent with experimental observations. The bidentate hydrogen bonds between the nonbridge oxygen atoms and Arg479 are slightly shorter than in the other states at 1.66 and 2.17 Å, respectively.
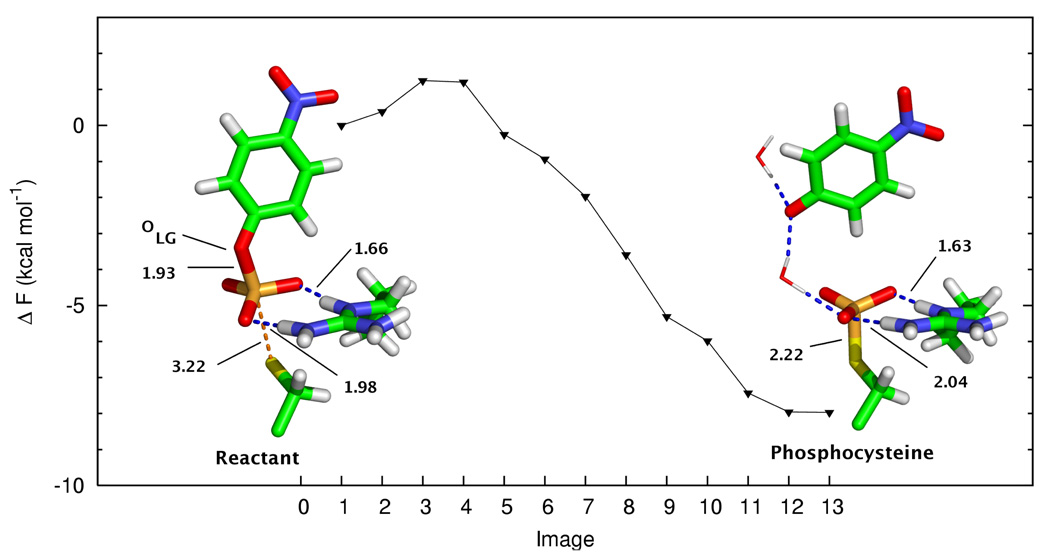
Reactant state - pNPP dianion
The reactant state for Cdc25B with the pNPP dianion is shown in Figure 3. Due to the greater negative charge on the phosphate, electrostatic interactions between the nonbridging oxygens and Arg479 are stronger than with the monoanionic substrate. The distances between the phosphate oxygens and the polar hydrogens of Arg479 are 1.66 and 1.98 Å, respectively. One quite interesting obseration for the dianionic substrate is that the P-Olg bond in the reactant state is almost completely dissociated at 1.93 Å. The P-Sγ distance is 3.22 Å, and the Olg-P-Sγ angle is nearly linear at 175.1 degrees.
FIG. 3.
Free energy profile and stationary points for the reaction of Cdc25b with the pNPP dianion. For the phosphocysteine intermediate, the instantaneous positions of two MM waters are shown to demonstrate hydrogen bonding to the leaving group. All other atoms belong to the QM subsystem. Important distances (in Å) are labeled.
Product state - pNPP dianion
For the pNPP dianion system, the P-Sγ distance in the phosphocysteinyl structure is slightly shorter than in the monoanion system, at 2.22 Å (Figure 3). The hydrogen bonds between the nonbridge oxygens of the phosphate and Arg479 are 1.63 and 2.04 Å, respectively. Unlike the pNPP monoanion reaction, no QM water molecule was included in the simulation of the pNPP dianion reaction. Upon P-Olg dissociation and formation of the phosphocysteine intermediate, the p-nitrophenoxide leaving group makes hydrogen bonds with two to three TIP3P water molecules.
Although the leaving group does not require protonation for P-O dissociation to occur based on its pKa of 7.0, possible protonation sources for the leaving group should be considered. Following the formation of the phosphocysteine intermediate, a water molecule could attack the phosphocysteine to liberate phosphate and regenerate the active form of the enzyme. The excess proton originating from this water molecule would then be available to protonate the leaving group. Because the phosphocysteine hydrolysis step is not rate-limiting, we did not consider this process in our simulations.
Free energy profile - pNPP monoanion
The computed free energy profile for the reaction of the pNPP monoanion with Cdc25B is shown in Figure 2. The reaction proceeds through three steps. In the first step, the reactant state undergoes a conformational change such that the phosphate hydroxyl group breaks its hydrogen bond with Sγ of Cys473 and forms a new hydrogen bond with a water molecule at the solvent interface (intermediate 1). This conformational rearrangement is high in energy partly because the strongly stabilizing hydrogen bond has been broken and direction of the O-H dipole has been reversed.
The point with the highest free energy on the path, Transition state 2 (Fig. 2), was obtained by QSM optimization followed by transition state optimization. However, the other approximate transition states (transition states 1 and 3) were obtained using QSM optimization only. Thus, the computed free energy barrier for steps 1 and 3 are overestimated and represent upper-bounds to the true free energy barriers.
Free energy profile - pNPP dianion
The computed free energy profile for the reaction of the pNPP dianion with Cdc25B is shown in Figure 3. In this mechanism, only one step is required to convert the reactant state to the phosphocysteinyl structure. The simulation yielded an extremely low barrier process that was somewhat exoergic. The high energy of the reactant state relative to the product suggests that the reactant is unstable due to the electrostatic repulsion in the active site, although the reaction would be tolerated by the dianionic substrate (as evidenced by the extremely low barrier) if binding of the dianion could occur (see below).
For the pNPP dianion reaction, a transition state optimization was not performed. We simply used the QSM estimate, which is an upper-bound to the true activation free energy. As previously stated, the P-Olg bond of pNPP is nearly dissociated (1.93 Å) in the Michaelis complex for the dianion reaction.
The free energy barrier for the pNPP dianion reaction is significantly lower than the experimental value. In fact, the process is nearly barrierless at ~1 kcal mol−1. From classical MD simulations with the substrate phenyl phosphate (see Supporting Information), we observed that the dianionic form dissociated from the active site, whereas the monoanionic form remained bound. For the more typical PTPases, S. cerevisiae low molecular weight protein tyrosine phosphatase and bovine heart phosphotyrosyl phosphatase, the phenyl phosphate dianion remained stably bound for more than 2 ns in each case. That Cdc25 appears incapable of binding small molecule phosphate dianions is likely a result of the relatively high concentration of negative charge in the active site and its inability to stabilize it sufficiently. Thus, although the computed free energy barrier for the dianion was significantly lower than for the monoanion, we suggest that Cdc25B does not bind the pNPP dianion.
Approximations
All QM calculations in this study were performed at the B3LYP/6-31G(d) level of theory. It is entirely possible that another DFT functional or ab initio method would yield better performance for the Cdc25B/pNPP system. However, because there is little thermochemical data with which to compare, and because of the computational cost of the QM/MM-MFEP calculations, we opted to make a compromise between cost and accuracy. Also, our goal was not necessarily to achieve chemical accuracy, but instead to be able to discern between two potentially competing mechanisms and suggest a possible mechanism. Relative free energy differences are often sufficient to accomplish this task. Although we have not quantitatively reproduced the experimental activation free energy of 18.5 kcal mol−1, our predicted transition state structure agrees in many respects (see below) with previous experimental data from kinetic isotope experiments and pH-rate profiles[18, 20], and the computed free energy barrier for the pNPP monoanion reaction is reasonable considering the approximations used in this work.
It was found that including Arg479 and one water molecule in the QM subsystem lowered the free energy barrier by approximately 3 kcal mol−1 (data not shown). It is possible that the addition of other residues or solvent molecules to the QM subsystem would lower the barrier further and bring the results closer to the experimental value. It should be noted that we did not perform frequency calculations to obtain thermal or zero-point energy corrections in this work. Inclusion of zero-point energies would most likely lower the free energy barrier by approximately 1–3 kcal mol−1.[47–49] A higher level of theory and/or more complete basis set may result in a more accurate transition state structure and activation free energy, although this was not investigated.
Solvent-exposed active site
From our simulations, it is clear that the p-nitrophenyl group of the substrate protrudes into the solvent because the active site is located at the solvent interface, not in a buried, hydrophobic pocket. The shallow, somewhat hydrophilic nature of the active site likely contributes to the lowered activity observed with small molecule substrates relative to the natural protein substrate.
With the natural Cdk2/CycA substrate, the active site is likely to be characterized by a large protein-protein interface that excludes bulk solvent. As a result, it would be expected to possess a lower effective dielectric constant. Such a modified dielectric environment may result in perturbed pKas that promote acid catalysis. In that case, it is possible that Cdc25 may bind the dianionic form of the substrate, as evidenced by previous mechanistic studies.[16, 19] Favorable electrostatic interactions between residues on Cdc25 and Cdk2/CycA may impart charge complementarity capable of stabilizing the additional negative charge of the physiologically more prevalent dianionic substrate.
Substrate binding
Because the computed free energy barrier for the pNPP dianion reaction is significantly lower (nearly barrierless) than that of the pNPP monoanion, here we address why we believe the dianionic reaction is not the preferred pathway. Most likely, the dianion is not able to bind effectively due to the concentration of negative charge in the active site. It has been noted previously that Cdc25 is the only known PTPase that lacks a Ser/Thr residue immediately following the catalytic motif in the active site (See Ref. 20 and references therein). From the crystal structures of other PTPases such as PTP1B and PTEN, it appears that the Ser/Thr residue forms a stabilizing interaction with the thiolate of the catalytic Cys. For VHR, Denu et al. found that the Ser hydroxyl does not significantly affect substrate binding or formation of the phosphocysteine intermediate, but instead acts to facilitate phosphocysteine hydrolysis by stabilizing the thiolate leaving group.[50] However, in Cdc25B no Ser/Thr is present, yet the phosphocysteine hydrolysis step is rapid compared to the first step of the reaction.
Due to its lack of an analogous stabilizing residue, the active site of Cdc25 would be burdened with additional negative charge character if the dianionic substrate were bound. On the other hand, if the substrate is in the monoanionic form the role of charge stabilization of the thiolate may be performed by the phosphate hydroxyl group. The pKa of the nonbridging phosphate oxygen of pNPP has been experimentally determined to be around 5.0 [51, 52], suggesting that only a small fraction of the phosphate would be present in the monoanionic form at physiologically relevant pH values. Although the relative abundance of the monoanion is quite low, it is presumably the only form that is able to bind. In our simulations, binding is not a part of the rate (or free energy) determination, which is calculated using transition state theory from kcat, not kcat/KM.
Another scenario that must be addressed is the possibility that the enzyme binds the dianionic substrate poorly, but upon binding the reaction proceeds rapidly. However, an experimental investigation of the temperature dependence of binding of Cdc25B with pNPP does not support this notion. Sohn and Rudolph [53] found that the binding free energy of pNPP with Cdc25B was −8 kcal mol−1, suggesting that although binding of phosphate at the active site is somewhat weak, the formation of the Michaelis complex is thermodynamically favorable. As discussed previously, our classical MD simulations of Cdc25 and two other PTPases provide evidence to support the unique preference of Cdc25 for the monoanionic form of small molecule substrates such as pNPP.
The enzymatic reaction with the pNPP substrate is quite slow, as demonstrated by the rather high activation free energy of 18.5 kcal mol−1 for the reaction. Available experimental data for the reaction of Cdc25 with its natural protein substrate [19] suggest a preference for the dianionic substrate by Cdc25B. Taken together, experiments and our simulations imply that the mechanisms for the protein substrate and various small molecule substrates are different. Presumably, there is some degree of flexibility in the types of reaction, specifically the protonation states of the phosphate and leaving group pKas, that may be accommodated in certain circumstances by Cdc25B. In fact, experimental evidence [20] suggests that the closely related Cdc25A uses different mechanisms for small molecule substrates based on the pKa of the substrate leaving group. As a result, it is difficult to extract mechanistic details for the natural protein substrate from the current study.
Rate-limiting step for the Cdc25 reaction
We have suggested that the first free energy barrier of the monoanion reaction, corresponding to transition state 1 in Figure 2, is an upper bound to the true barrier. Because the Olg-P-O-H dihedral may rotate either in a clockwise or counterclockwise direction, there are multiple paths by which the reactant may be converted to intermediate 1. This will result in a favorable entropic contribution to the free energy barrier associated with transition state 1. It is also possible that tunneling may further contribute to lowering the barrier as the process essentially involves the movement of hydrogen over a barrier, although this was not investigated. Additionally, we did not perform a true transition state optimization of this structure. Thus, the activation free energy of the first step has most likely been significantly overestimated.
Because the equilibrium constant between the reactant state and intermediate 1 heavily favors the reactant state based on the free energy difference of more than 13 kcal mol−1 between these two structures, the enzyme is not expected to bind intermediate 1 directly. Thus, we conclude that the rate-limiting step of the reaction involves the formation of transition state 2, the structure on the path with the overall highest free energy.
Although a proton was ultimately transferred from the phosphate to the leaving group in our simulation of the monoanion mechanism, this does not imply acid catalysis because the protonation occurred after the rate-limiting step. However, for other substrates with higher leaving group pKas, protonation, and therefore acid catalysis, becomes necessary to enable P-Olg dissociation.[20]
McCain et al. provided isotope effect data for Cdc25A with pNPP at pH 7.2.[18] It is encouraging that our predicted structure agrees quite well with the reported values of 15N and 18Obridge, which suggest a dissociative transition state without protonation of the leaving group. However, if the substrate is actually a monoanion, the 18Ononbridge kinetic isotope effect must be corrected for the equilibrium isotope effect resulting from protonation, which would change the KIE value from slightly inverse (0.9988 at pH 7.2) to a significantly normal effect (1.014). A normal 18Ononbridge effect would imply that deprotonation of the phosphate group occurs at the transition state. However, our calculations at the B3LYP/6-31G(d) level of theory predict transfer of the phosphoryl proton after the transition state. Although the KIE data may be interpreted to support a monoanionic substrate, it should be noted that McCain et al. suggested that their results favor a dianionic substrate.
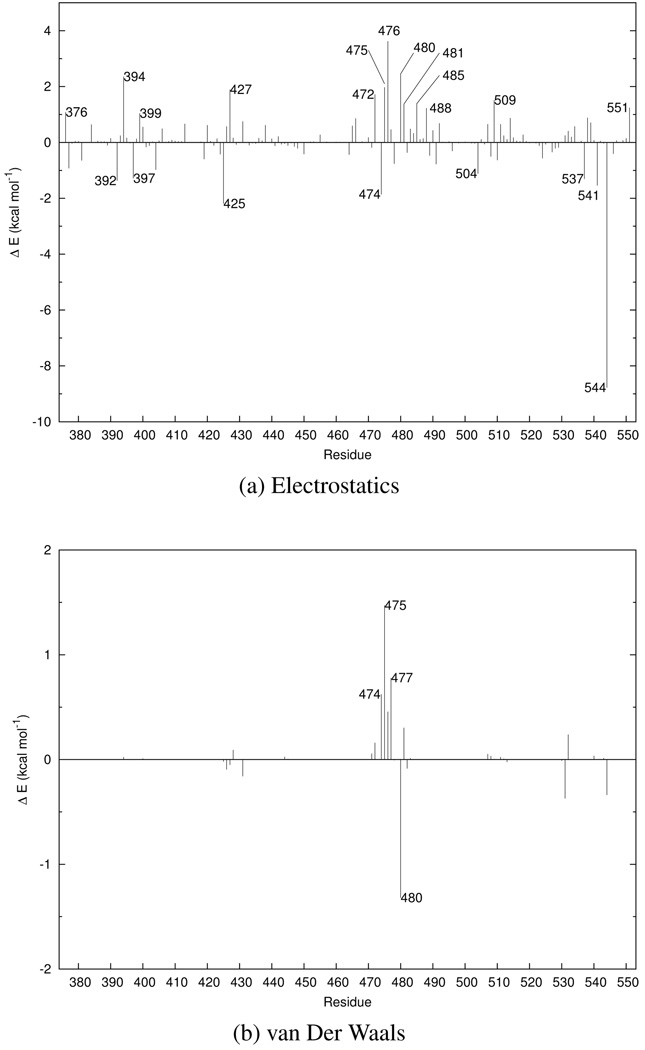
Residue analysis
In order to understand the contributions of individual residues to catalysis, we performed a nonbonded energy analysis in which we computed the electrostatic and van derWaals interaction energies between the QM subsystem as a whole and each protein residue. The QM subsystem was held fixed and its electrostatic potential was represented by ESP charges. The electrostatic and van der Waals interaction energies were averaged over a period of 160 ps. The energies are defined as
where i represents a protein residue in the MM subsystem, represents either the electrostatic or van der Waals interaction energy between the QM subsystem and residue i, and the brackets indicate ensemble averaging over the MM degrees of freedom in which the QM conformation is fixed at either the transition state or reactant state structure. Negative values indicate that the residue lowers the free energy barrier, whereas positive values indicate contributions that increase the free energy barrier.
The results from the electrostatic analysis are shown in Figure 4a. Glu474 and Glu478 contribute about −1.75 and −0.85 toward stabilizing the transition state, and are the only active site residues (excluding Cys473 and Arg479) that provide electrostatic stabilization of the transition state. Arg544 exhibits quite favorable electrostatic interactions with the QM subsystem. This residue, located more than 10 Å away from Cys473 in the active site, interacts strongly with the nitro group of pNPP in our simulations.
FIG. 4.
Electrostatic and van der Waals contributions to catalysis for the pNPP monoanion reaction. Negative values indicate stabilizing interactions.
Interestingly, many of the residues in the active site impart unfavorable electrostatic interactions on the QM subsystem. His472 and Phe475 contribute around 1.75 kcal mol−1, Gly480 contributes 2.4 kcal mol−1, and Ser476 contributes 3.5 kcal mol−1 of destabilization in the transition state relative to the reactant state.
As shown in Figure 4b, the van der Waals energetic analysis yields results that are similar to the electrostatic analysis. All active site residues except one impart relatively small but unfavorable contributions. The exception is Gly480, which lowers the barrier by slightly more than 1 kcal mol−1.
Taken together, our results suggest that the active site is not optimized for the dephosphorylation of pNPP. In fact, several residues in the active site contribute net destabilizing effects on the transition state. Often, simulations are performed with enzymes and their natural substrates, so residues in the active site are expected to yield an overall favorable catalytic effect at the transition state relative to the reactant state. However, because pNPP is not a natural substrate for Cdc25, it should come as no surprise that the active site is not optimized to perform the hydrolysis of pNPP.
IV. CONCLUSIONS
We have performed QM/MM-MFEP calculations in an effort to understand the enzymatic reaction mechanism of Cdc25B with the small molecule substrate p-nitrophenyl phosphate. Our simulations support a mechanism in which the monoanionic form of the substrate is dephosphorylated rather than the dianion. Our computed transition state structure is largely consistent with available experimental data which indicates that the transition state is dissociative and no catalytic acid is required for catalysis. Because Cdc25 lacks a Ser/Thr residue immediately following the HCX5R catalytic motif that would otherwise stabilize the Cys473 thiolate, it may be unable to effectively bind the dianionic substrate. Our analysis of the electrostatic and van derWaals contributions to the free energy barrier reveals that Cdc25B is not optimized for the hydrolysis of the small molecule substrate pNPP, further reinforcing the notion that pNPP is a poor substrate for Cdc25. To the best of our knowledge, the proposed mechanism is in contrast to that of all other known PTPases. The Cdc25 phosphatases appear to represent a unique, fundamentally different type of dual-specificity phosphatase as Cdc25 is the only PTPase that does not utilize acid catalysis for pNPP hydrolysis.
Although Cdc25B exhibits a unique mechanism with pNPP, it is important to realize that the mechanism with the native protein substrate is most likely quite different. Comparison between pNPP and native protein substrate is difficult, and one should not speculate too much given the huge differences in reactivity and the fact that the protein substrate involves extensive protein-protein interactions around the active upon substrate binding. Interfacial interactions and conformational changes could presumably overcome any “minor" charge differences by inducing changes in local dipoles and modifying the dielectric environment of the active site, thus allowing dianionic phosphate to bind readily. In the case of pNPP, there are no surrounding groups or residues capable of offsetting or attenuating the repulsive electrostatics of a dianionic substrate. As a result, small molecule substrates such as pNPP may not be effective mechanistic probes for understanding the reaction of Cdc25 with its natural protein substrate. However, our results may prove useful in the design of improved, specific inhibitors of Cdc25. For example, phosphate mimetics, which are often thought to be a good starting point for the design of protein phosphatase inhibitors, may require a charge of −1, rather than −2, for effective binding and inhibition in the case of Cdc25.
Supplementary Material
Complete Refs. 27 and 33, overview of the QM/MMMFEP method, model calculations and simulation details. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The authors thank the National Institutes of Health (NIH R01-GM-061870) for financial support. J.M.P. would like to thank Steven K. Burger for helpful discussions.
References
- 1.Fersht A. Structure and Mechanism in Protein Science. A Guide to Enzyme Catalysis and Protein Folding. New York: W. H. Freeman and Co.; 1999. [Google Scholar]
- 2.Jackson MD, Denu JM. Chem. Rev. 2001;101:2313–2340. doi: 10.1021/cr000247e. [DOI] [PubMed] [Google Scholar]
- 3.Boutros R, Lobjois V, Ducommun B. Nat. Rev. Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 4.Lyon MA, Ducruet AP, Wipf P, Lazo JS. Nat. Rev. Drug Discov. 2002;1:961–975. doi: 10.1038/nrd963. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph J. Biochemistry. 2007;46:3595–3604. doi: 10.1021/bi700026j. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph J. Biochemistry. 2002;41:14613–14623. doi: 10.1021/bi0263513. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Burke SP. J. Biol. Chem. 1996;271:5118–5124. doi: 10.1074/jbc.271.9.5118. [DOI] [PubMed] [Google Scholar]
- 8.Kolmodin K, Nordlund P, Åqvist J. Chem. Comm. 1997;15:1431–1432. [Google Scholar]
- 9.Alhambra C, Wu L, Zhang Z-Y, Gao J. J. Am. Chem. Soc. 1998;120:3858–3866. [Google Scholar]
- 10.Kolmodin K, Nordlund P, Åqvist J. Proteins: Struct. Func. Genet. 1999;36:370–379. [PubMed] [Google Scholar]
- 11.Dillet V, Etten RLV, Bashford D. J. Phys. Chem. B. 2000;104:11321–11333. [Google Scholar]
- 12.Asthagiri D, Dillet V, Liu T, Noodleman L, Etten RLV, Bashford D. J. Am. Chem. Soc. 2002;124:10225–10235. doi: 10.1021/ja020046n. [DOI] [PubMed] [Google Scholar]
- 13.Asthagiri D, Liu T, Noodleman L, Etten RLV, Bashford D. J. Am. Chem. Soc. 2004;126:12677–12684. doi: 10.1021/ja048638o. [DOI] [PubMed] [Google Scholar]
- 14.Arantes GM. Phys. Chem. Chem. Phys. 2006;8:347–353. doi: 10.1039/b511805k. [DOI] [PubMed] [Google Scholar]
- 15.Arantes GM. Biochem. J. 2006;399:343–350. doi: 10.1042/BJ20060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Wilborn M, Rudolph J. Biochemistry. 2000;39:10781–10789. doi: 10.1021/bi000909u. [DOI] [PubMed] [Google Scholar]
- 17.Sohn J, Parks JM, Buhrman G, Brown P, Kristjánsdóttir K, Safi A, Edelsbrunner H, Yang W, Rudolph J. Biochemistry. 2005;44:16563–16573. doi: 10.1021/bi0516879. [DOI] [PubMed] [Google Scholar]
- 18.McCain DF, Grzyska PK, Wu L, Hengge AC, yin Zhang Z. Biochemistry. 2004;43:8256–8264. doi: 10.1021/bi0496182. [DOI] [PubMed] [Google Scholar]
- 19.Rudolph J. Bioorg. Chem. 2005;33:264–273. doi: 10.1016/j.bioorg.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 20.McCain DF, Catrina IE, Hengge AC, Zhang Z-Y. J. Biol. Chem. 2002;277:11190–11200. doi: 10.1074/jbc.M109636200. [DOI] [PubMed] [Google Scholar]
- 21.Hu H, Lu Z, Parks JM, Burger SK, Yang W. J. Chem. Phys. 2008;128:034105. doi: 10.1063/1.2816557. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds RA, Yem AW, Wolfe CL, Deibel MR, Jr, Chidester CG, Watenpaugh KD. J. Mol. Bio. 1999;293:559–568. doi: 10.1006/jmbi.1999.3168. [DOI] [PubMed] [Google Scholar]
- 23.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins: Struct. Func. Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 24.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucl. Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bashford D, Karplus M. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 26.Myers J, Grothaus G, Narayana S, Onufriev A. Proteins: Struct. Func. Bioinf. 2006;63:928–938. doi: 10.1002/prot.20922. [DOI] [PubMed] [Google Scholar]
- 27.MacKerell AD, Jr, et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Zhang Y, Lee T-S, Yang W. J. Chem. Phys. 1999;110:46–54. [Google Scholar]
- 30.Mann G, Yun RH, Nyland L, Prins J, Board J, Hermans J. The Sigma MD program and a generic interface applicable to multi-functional programs with complex, hierarchical command structure. Computational Methods for Macromolecules: Challenges and Applications; Proceedings of the 3rd International Workshop on Algorithms for Macromolecular Modelling; New York. 2002. pp. 129–145. [Google Scholar]
- 31.Hu H, Elstner M, Hermans J. Proteins: Struct. Func. Genet. 2003;3:451–463. doi: 10.1002/prot.10279. [DOI] [PubMed] [Google Scholar]
- 32.Hu H, Yang W. J. Chem. Phys. 2005;123:041102. doi: 10.1063/1.1990113. [DOI] [PubMed] [Google Scholar]
- 33.Frisch MJ, et al. Gaussian 03, Revision C.02. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 34.Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 35.Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 36.Singh UC, Kollman PA. J. Comput. Chem. 1984;5:129–145. [Google Scholar]
- 37.Berendsen H, Postma J, van Gunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 38.Zhang Y, Liu H, Yang W. J. Chem. Phys. 2000;112:3484–3492. [Google Scholar]
- 39.Cisneros GA, Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2003;125:10384–10393. doi: 10.1021/ja029672a. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Lu Z, Cisneros GA, Yang W. J. Chem. Phys. 2004;121:697–706. doi: 10.1063/1.1759318. [DOI] [PubMed] [Google Scholar]
- 41.Cisneros GA, Wang M, Silinski P, Fitzgerald MC, Yang W. Biochemistry. 2004;43:6885–6892. doi: 10.1021/bi049943p. [DOI] [PubMed] [Google Scholar]
- 42.Hu H, Lu Z, Yang W. J. Chem. Theory Comput. 2007;3:390–406. doi: 10.1021/ct600240y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu H, Boone A, Yang W. J. Am. Chem. Soc. 2008;130:14493–14503. doi: 10.1021/ja801202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senn HM, Thiel W. Top. Curr. Chem. 2007;268:173–290. [Google Scholar]
- 45.Hu H, Yang W. Ann. Rev. Phys. Chem. 2008;59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burger SK, Yang W. J. Chem. Phys. 2006;124:054109. doi: 10.1063/1.2163875. [DOI] [PubMed] [Google Scholar]
- 47.Cui Q, Karplus M. J. Phys. Chem. B. 2002;106:7927–7947. [Google Scholar]
- 48.Zhang X, Bruice TC. Proc. Natl. Acad. Sci. U.S.A. 2006;44:16141–16146. doi: 10.1073/pnas.0607503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claeyssens F, Harvey JN, Manby FR, Mata RA, Mulholland AJ, Ranaghan KE, Schütz M, Thiel S, Thiel W, Werner H-J. Angew. Chem. Int. Ed. 2006;45:6856–6859. doi: 10.1002/anie.200602711. [DOI] [PubMed] [Google Scholar]
- 50.Denu JM, Dixon JE. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5910–5914. doi: 10.1073/pnas.92.13.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourne N, Williams A. J. Org. Chem. 1984;49:1200–1204. [Google Scholar]
- 52.Zhang Z-Y, Malachowski WP, Etten RLV, Dixon JE. J. Biol. Chem. 1994;269:8140–8145. [PubMed] [Google Scholar]
- 53.Sohn J, Rudolph J. Biophys. Chem. 2007;125:549–555. doi: 10.1016/j.bpc.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete Refs. 27 and 33, overview of the QM/MMMFEP method, model calculations and simulation details. This material is available free of charge via the Internet at http://pubs.acs.org.