Abstract
We consider the problem of extracting smooth, low-dimensional neural trajectories that summarize the activity recorded simultaneously from many neurons on individual experimental trials. Beyond the benefit of visualizing the high-dimensional, noisy spiking activity in a compact form, such trajectories can offer insight into the dynamics of the neural circuitry underlying the recorded activity. Current methods for extracting neural trajectories involve a two-stage process: the spike trains are first smoothed over time, then a static dimensionality-reduction technique is applied. We first describe extensions of the two-stage methods that allow the degree of smoothing to be chosen in a principled way and that account for spiking variability, which may vary both across neurons and across time. We then present a novel method for extracting neural trajectories—Gaussian-process factor analysis (GPFA)—which unifies the smoothing and dimensionality-reduction operations in a common probabilistic framework. We applied these methods to the activity of 61 neurons recorded simultaneously in macaque premotor and motor cortices during reach planning and execution. By adopting a goodness-of-fit metric that measures how well the activity of each neuron can be predicted by all other recorded neurons, we found that the proposed extensions improved the predictive ability of the two-stage methods. The predictive ability was further improved by going to GPFA. From the extracted trajectories, we directly observed a convergence in neural state during motor planning, an effect that was shown indirectly by previous studies. We then show how such methods can be a powerful tool for relating the spiking activity across a neural population to the subject's behavior on a single-trial basis. Finally, to assess how well the proposed methods characterize neural population activity when the underlying time course is known, we performed simulations that revealed that GPFA performed tens of percent better than the best two-stage method.
INTRODUCTION
Motivation for single-trial analysis of neural population activity
Neural responses are typically studied by averaging noisy spiking activity across multiple experimental trials to obtain firing rates that vary smoothly over time. However, if the neural responses are more a reflection of internal processing rather than external stimulus drive, the time course of the neural responses may differ on nominally identical trials. This is particularly true of behavioral tasks involving perception, decision making, attention, or motor planning. In such settings, it is critical that the neural data not be averaged across trials, but instead be analyzed on a trial-by-trial basis (Arieli et al. 1996; Briggman et al. 2006; Churchland et al. 2007; Czanner et al. 2008; Horwitz and Newsome 2001; Jones et al. 2007; Nawrot et al. 1999; Ventura et al. 2005; Yu et al. 2006).
The importance of single-trial analyses can be simply illustrated by considering a classic perceptual decision-making study by Newsome and colleagues (Horwitz and Newsome 2001). In this study, they trained monkeys to report the direction of coherent motion in a stochastic random-dot display. Especially in low-coherence conditions, they observed that neurons in the superior colliculus appeared to jump between low and high firing-rate states, suggesting that the subject may have vascillated between the two possible directional choices. For the same random-dot stimulus, the times at which the firing rates jumped appeared to differ from one trial to the next. Such vascillations may also underlie other perceptual and decision-making tasks, including binocular rivalry (Leopold and Logothetis 1996), structure-from-motion (Bradley et al. 1998; Dodd et al. 2001), somatosensory discrimination (de Lafuente and Romo 2005), and action selection (Cisek and Kalaska 2005). Most of these studies provide indirect evidence that the time course of the subject's percept or decision differed on nominally identical trials. Such trial-to-trial differences cannot be eliminated by additional monkey training, since the stimuli are designed to be ambiguous and/or operate near the subject's perceptual threshold.
In the dot-discrimination (Horwitz and Newsome 2001) and binocular rivalry (Leopold and Logothetis 1996) studies, the authors attempted to segment single spike trains based on periods of high and low firing rates. In general, it is very difficult to accurately estimate the time or rate at which the firing rate changes based on a single spike train. If one is able to simultaneously record from multiple neurons and activities of these neurons all reflect a common neural process (e.g., the subject's percept or choice), then one might be able to more accurately estimate the time course of the subject's percept or choice on a single trial. Indeed, developments in multielectrode (Kipke et al. 2008) and optical imaging (Kerr and Denk 2008) technologies are making this a real possibility. However, it is currently unclear how to best leverage the statistical power afforded by simultaneously recorded neurons (Brown et al. 2004) to extract behaviorally relevant quantities of interest (e.g., the time course of the subject's percept or internal decision variable) on a single-trial basis.
In this work, we develop analytical techniques for extracting single-trial neural time courses by leveraging the simultaneous monitoring of large populations of neurons. The approach adopted by recent studies is to consider each neuron being recorded as a noisy sensor reflecting the time evolution of an underlying neural process (Bathellier et al. 2008; Briggman et al. 2005; Broome et al. 2006; Brown et al. 2005; Carrillo-Reid et al. 2008; Levi et al. 2005; Mazor and Laurent 2005; Sasaki et al. 2007; Smith and Brown 2003; Stopfer et al. 2003; Yu et al. 2006). The goal is to uncover this underlying process by extracting a smooth, low-dimensional neural trajectory from the noisy, high-dimensional recorded activity. The activity of each neuron tends to vary significantly between trials, even when experimental conditions are held constant. Some of this variability is due to processes internal to the neuron, such as channel noise in membranes and biochemical noise at synapses (Faisal et al. 2008). However, some portion of the variability reflects trial-to-trial differences in the time evolution of the network state, which may in turn represent different computational paths and may lead to different behavioral outcomes. Because it reflects the network state, we expect this component of the variability to be shared among many (or all) of the neurons that make up the network. The techniques that we develop here seek to embody this shared activity in a neural trajectory, which represents our best estimate of the time evolution of the neural state. The neural trajectory provides a compact representation of the high-dimensional recorded activity as it evolves over time, thereby facilitating data visualization and studies of neural dynamics under different experimental conditions. In principle, relative to the high-dimensional recorded activity, such a parsimonious description should bear clearer and stronger relationships with other experimentally imposed or measurable quantities (e.g., the presented stimulus or the subject's behavior; see the “bouncing ball analogy” in Yu et al. 2006).
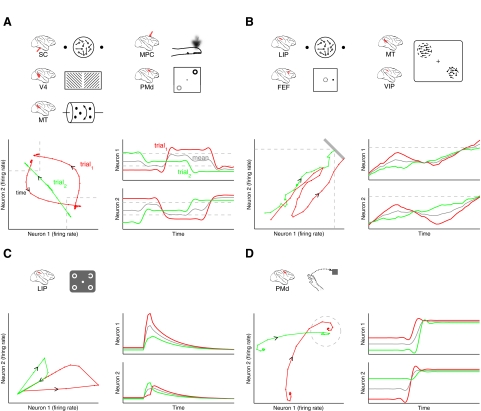
Figure 1 illustrates how such an approach may provide insights into the neural mechanisms underlying perception, decision making, attention, and motor planning. Figure 1A considers the perceptual and decision tasks described earlier, in which there are two possible percepts or choices. Applying the analytical methods developed in this work to the activity of multiple neurons recorded simultaneously may reveal different switching time courses on different trials. In this example (Fig. 1A, bottom left), on trial 1, the subject's percept switched from one choice to another, then back to the first. On trial 2, the percept began to switch, stopped between the two choices, then completed its switch. These switching time courses can be viewed in terms of single-neuron firing rates (Fig. 1A, bottom right), where the two neurons are shown to have anticorrelated firing rates. Note that these firing-rate profiles would be estimated by leveraging the simultaneously recorded spike trains across a neural population on a single-trial basis. In this case, the time course obtained by averaging neural responses across trials (gray) is not representative of the time course on any individual trial (red and green traces). Beyond relating the extracted trajectory (Fig. 1A, bottom left) to the subject's perceptual report or decision on a trial-by-trial basis, such trajectories allow us to ask questions about the dynamics of switching percepts across the neural population. For example, how long does it take to switch between one percept and another? Does it take longer to switch in one direction than the other? Does switching in one direction follow the same path as switching in the other direction? Can regions in firing-rate space be defined corresponding to each percept or choice? If so, what is the shape of these regions?
FIG. 1.
Conceptual illustration showing how the analytical methods presented in this work can be applied to different behavioral tasks, including those involving perception, decision making, attention, and motor planning. The neural mechanisms underlying these behavioral tasks may involve A: switching between two possible percepts or decisions, B: rising to threshold, C: decaying along a single slow mode, D: or converging to an attractor. Each panel includes icons of the relevant behavioral tasks and brain areas (top), single-trial neural trajectories in the firing-rate space of 2 neurons (bottom left), and corresponding firing-rate profiles (both single-trial and trial-averaged) for each neuron (bottom right).
Figure 1B considers a different class of dynamics: rise-to-threshold. Shadlen and colleagues (Roitman and Shadlen 2002) previously showed that single neurons in lateral intraparietal (LIP) cortex appear to integrate sensory evidence until a threshold is reached, at which time a decision is made. By grouping trials based on reaction time, they found that the firing rates approached threshold more quickly on trials with short reaction times than on trials with long reaction times. Similar effects were found in frontal eye field prior to saccade initiation (Hanes and Schall 1996) and middle temporal and ventral intraparietal areas during motion detection (Cook and Maunsell 2002). In other words, the time course of the neural response differed on nominally identical trials in all of these studies. To investigate trial-to-trial differences, correlations were identified between single-trial estimates of firing rate and reaction time (Cook and Maunsell 2002; Roitman and Shadlen 2002). However, due to the limited statistical power in a single spike train, most analyses in these previous studies relied on grouping trials with similar reaction times. If one is able to simultaneously record from multiple neurons, one can then leverage the statistical power across the neural population to more accurately estimate single-trial response time courses. This could potentially uncover even stronger relationships between neural activity and behavioral measurements, such as reaction time. Figure 1B shows two trials in which the decision variable crosses threshold at similar times; thus, the subject would be expected to show similar reaction times on these two trials. However, the time course of the decision variable was quite different on each trial. On trial 1, the decision variable rose quickly toward threshold, then headed back toward baseline (perhaps due to contrary evidence) before finally rising to threshold. On trial 2, the decision variable rose slowly, but steadily, toward threshold. Such subtle differences between trials would be difficult to see based on single spike trains and would be washed out had the neural activity been averaged across trials (Fig. 1B, bottom right, gray trace). By leveraging simultaneous recordings across a neural population, we may be able to uncover such effects on a single-trial basis and gain further insight into the dynamics of decision processes.1
Another potential application of the methods developed in this work is to behavioral tasks that involve attention, which is typically not tied to observable quantities in the outside world. Using a GO/NOGO memory saccade task with visual distractors, Goldberg and colleagues (Bisley and Goldberg 2003) showed that neural activity (averaged across trials and neurons) in LIP indicates the attentionally advantaged part of the visual field. For the same stimulus (target, distractor, and probe), the subject showed different behavioral responses (GO or NOGO) on different trials, where the proportion of correct responses depended on the time, location, and contrast of the probe. If multiple neurons could be monitored simultaneously in LIP, the methods developed in this work could be used to track the instantaneous state of the subject's attention on a single-trial basis, which in turn could be related to the subject's behavioral response. For example, it may be that the same distractor kicks the neural state out farther on some trials (Fig. 1C, bottom left, red trace) than others (green trace), thereby conferring a longer-lasting attentional advantage at the distractor (see Fig. 2 in Ganguli et al. 2008, for a detailed explanation of the state space trajectories shown in Fig. 1C, bottom left). Might it be possible to map out the probability of a correct behavioral response (GO or NOGO) for different neural states at the time of the probe? Such an approach could shed light on the dynamics of attentional shifts between different visual locations and how it influences behavior.
Finally, we consider the arm movement system, which serves as our experimental testbed for exploring analytical methods for extracting single-trial neural time courses. We previously showed that the across-trial variability of neural responses in premotor cortex drops during motor preparation (Churchland et al. 2006). This finding suggested that single-trial neural trajectories might converge during motor preparation, in attractor-like fashion, as illustrated in Fig. 1D (bottom left). Although we previously hypothesized such a convergence of trajectories (see Fig. 1 in Churchland et al. 2006), we have not been able to directly view this effect due to a lack of appropriate analytical methods for extracting trajectories on a single-trial basis. By studying how the neural state evolves from an initially variable baseline state toward a consistent planning state, and relating aspects of the trajectory to the subject's behavior, we can gain insight into how movements are prepared and executed. Although the analytical methods developed here are potentially applicable to many different experimental settings, as exemplified in Fig. 1, we demonstrate the utility of the developed methods in the context of motor preparation and execution in this work.
For each of the examples shown in Fig. 1, there are ways to detect (or indirectly view) trial-to-trial differences in the neural responses, including computing streak indices (Horwitz and Newsome 2001), estimating firing rates from a single spike train (Cook and Maunsell 2002; Roitman and Shadlen 2002), and measuring the across-trial variability of neural responses (Churchland et al. 2006). In all of these cases, however, what one really wants is a direct view of the time evolution of the neural response on single trials. In this report, we present analytical methods that can extract such single-trial time courses from neural population activity.
Existing methods for extracting neural trajectories
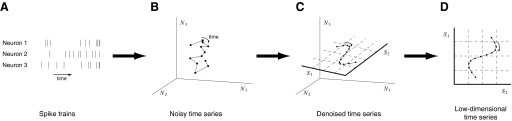
Figure 2 shows conceptually how a neural trajectory relates to a set of simultaneously recorded spike trains. Suppose that we are simultaneously recording from three neurons, whose spike trains are shown in Fig. 2A. Although the following ideas hold for larger numbers of neurons, we use only three neurons here for illustrative purposes. We define a high-dimensional space, where each axis measures the instantaneous firing rate of a neuron being monitored (Fig. 2B). At any given time, the activity of the neural population is characterized by a single point in this space. As the activity of the neural population evolves over time, a noisy trajectory is traced out. The goal is to extract a corresponding smooth neural trajectory that embodies only the shared fluctuations (termed shared variability) in firing rate across the neural population (Fig. 2C). Discarded in this process are fluctuations particular to individual neurons (termed independent variability), which presumably reflect noise processes involved in spike generation that are internal to the neuron.2 Due to the correlated activity across the neural population, the neural trajectory may not explore the entire high-dimensional space; in other words, the neural system may be using fewer degrees of freedom than the number of neurons at play. If this is true, then we would seek to identify a lower-dimensional space (shown as a two-dimensional plane denoted by grid lines in Fig. 2C) within which the neural trajectory lies. The neural trajectory can then be directly visualized in the low-dimensional space and be referred to equivalently using its high-dimensional (N1, N2, N3) or low-dimensional (S1, S2) coordinates (Fig. 2D).
FIG. 2.
Extracting a neural trajectory from multiple spike trains. For clarity, the activity of only 3 neurons is considered in this illustration. A: spike trains recorded simultaneously from 3 neurons. B: the time evolution of the recorded neural activity plotted in a 3-dimensional space, where each axis measures the instantaneous firing rate of a neuron (e.g., N1 refers to neuron 1). C: the neural trajectory (a “denoised” version of the trajectory in B) is shown to lie within a 2-dimensional space with coordinates S1 and S2. D: the neural trajectory can be directly visualized in the low-dimensional space and be referred to using its low-dimensional coordinates (S1, S2).
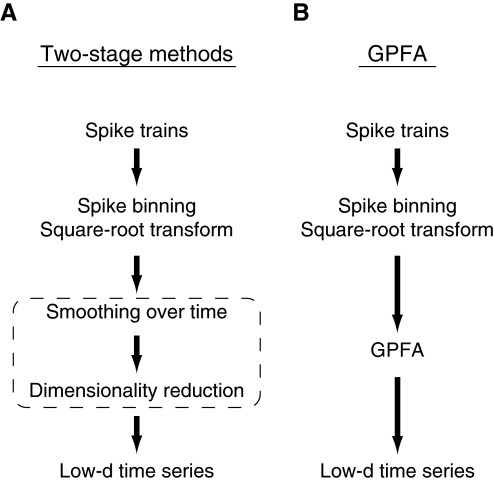
A simple way to extract neural trajectories is to first estimate a smooth firing-rate profile for each neuron on a single trial (e.g., by convolving each spike train with a Gaussian kernel), then apply a static dimensionality-reduction technique (e.g., principal components analysis [PCA]) (Levi et al. 2005; Nicolelis et al. 1995). The signal flow diagram for these so-called two-stage methods is shown in Fig. 3A. Smooth firing-rate profiles may also be obtained by averaging across a small number of trials (if the neural time courses are believed to be similar on different trials) (Broome et al. 2006; Brown et al. 2005; Mazor and Laurent 2005; Stopfer et al. 2003) or by applying more advanced statistical methods for estimating firing-rate profiles from single spike trains (Cunningham et al. 2008b; DiMatteo et al. 2001; Ventura et al. 2005). Numerous linear and nonlinear dimensionality-reduction techniques exist, but to our knowledge only PCA (Levi et al. 2005; Mazor and Laurent 2005; Nicolelis et al. 1995) and locally linear embedding (LLE) (Broome et al. 2006; Brown et al. 2005; Roweis and Saul 2000; Stopfer et al. 2003) have been used to extract neural trajectories. Smoothed firing-rate trajectories based on pairs of simultaneously recorded neurons without dimensionality reduction have also been studied (Aksay et al. 2003).
FIG. 3.
Signal flow diagram of how neural trajectories are extracted from spike trains using A: two-stage methods and B: Gaussian-process factor analysis (GPFA). Whereas the smoothing and dimensionality-reduction operations are performed sequentially with the two-stage methods (dotted box), they are performed simultaneously using GPFA. For the two-stage methods, smoothing can be performed either directly on spike trains or on binned spike counts. The spike trains must be binned if one desires to apply the square-root transform, which operates only on count data.
The two-stage methods have been fruitfully applied in studies of the olfactory system, where the presentation of an odor sets off a time course of neural activity across the recorded population. To understand how the population response varies under different experimental conditions (e.g., different presented odors), one could attempt to directly compare the recorded spike trains. However, this quickly becomes unmanageable as the number of neurons and the number of experimental conditions increase. Instead, a neural trajectory can be extracted for each trial condition (typically averaged across a small number of trials) and compared in a low-dimensional space. This approach has been adopted to study the population response across different odor identities (e.g., Brown et al. 2005), concentrations (Stopfer et al. 2003), durations (Mazor and Laurent 2005), and sequences (Broome et al. 2006). Dynamical behaviors resembling fixed points (Mazor and Laurent 2005) and limit cycles (Bathellier et al. 2008) have also been identified. In these studies, hypotheses were generated based on the visualized trajectories, then tested using the high-dimensional recorded activity. Without the low-dimensional visualizations, many of these hypotheses would have remained unposed and thus untested.
Methodological advances proposed here
Although two-stage methods have provided informative low-dimensional views of neural population activity, there are several aspects that can be improved. 1) Because the smoothing and dimensionality reduction are performed sequentially, there is no way for the dimensionality-reduction algorithm to influence the degree or form of smoothing used. This is relevant both to the identification of the low-dimensional space and to the extraction of single-trial neural trajectories. 2) PCA and LLE have no explicit noise model and thus have difficulty distinguishing between changes in the underlying neural state (i.e., shared variability) and spiking noise (i.e., independent variability). 3) For kernel smoothing, the degree of smoothness is often arbitrarily chosen. We instead seek to learn the appropriate degree of smoothness from the data. With probabilistic methods, a principled approach would be to ask what is the degree of smoothness that maximizes the probability of having observed the data at hand. Unfortunately, kernel smoothing, PCA, and LLE are all nonprobabilistic methods, so such standard parameter-learning techniques are not applicable. One may try to get around this problem by applying kernel smoothing, followed by a probabilistic dimensionality-reduction technique (e.g., probabilistic PCA [PPCA]; Roweis and Ghahramani 1999; Tipping and Bishop 1999), which does assign probabilities to data. However, the problem with this scheme is that these probabilities correspond to the smoothed data (the input to the probabilistic dimensionality-reduction technique), rather than the unsmoothed data (the input to the kernel smoother). Because the smoothed data change depending on the degree of smoothness chosen, the resulting probabilities are not comparable. 4) The same kernel width is typically used for all spike trains across the neural population, which implicitly assumes that the population activity evolves with a single timescale. Because we do not know a priori how many timescales are needed to best characterize the data at hand, we seek to allow for multiple timescales.
In this work, we first propose extensions of the two-stage methods that can help to address issues 2) and 3) cited earlier. We summarize these extensions here; details can be found in methods. For 2) we explore dimensionality-reduction algorithms possessing different explicit noise models and consider the implications of the different noise assumptions. We find that an effective way to combat spiking noise (whose variance may vary both across neurons and across time) is to use the square-root transform (Kihlberg et al. 1972) in tandem with factor analysis (FA) (Everitt 1984). Taking the square root of the spike counts serves to approximately stabilize the spiking noise variance. FA is a dimensionality-reduction technique related to PCA that, importantly, allows different neurons to have different noise variances. Although nonlinear dimensionality-reduction techniques with explicit noise models have been developed (for a probabilistic LLE-inspired algorithm, see Teh and Roweis 2003), we consider only linear mappings between the low-dimensional neural state space and the high-dimensional space of recorded activity in this work for mathematical tractability. For 3), we adopt a goodness-of-fit metric that measures how well the activity of each neuron can be predicted by the activity of all other recorded neurons, based on data not used for model fitting. This metric can be used to compare different smoothing kernels and allows for the degree of smoothness to be chosen in a principled way. An advantage of this metric is that it can be applied in both probabilistic and nonprobabilistic settings. In results, we will use this as a common metric by which different methods for extracting neural trajectories are compared.
Next, we develop Gaussian-process factor analysis (GPFA), which unifies the smoothing and dimensionality-reduction operations in a common probabilistic framework. GPFA takes steps toward addressing all of the issues (1–4) described earlier and is shown in results to provide a better characterization of the recorded population activity than the two-stage methods. Because GPFA simultaneously performs the smoothing and dimensionality-reduction operations (Fig. 3B), rather than sequentially (Fig. 3A), the degree of smoothness and the relationship between the low-dimensional neural trajectory and the high-dimensional recorded activity can be jointly optimized. GPFA allows for multiple timescales, whose optimal values can be found automatically by fitting the GPFA model to the recorded activity. Unlike the two-stage methods, GPFA assigns probabilities to the unsmoothed data, which allows the timescale parameters to be optimized using standard maximum likelihood techniques. As with FA, GPFA specifies an explicit noise model that allows different neurons to have different noise variances. The time series model involves Gaussian processes (GPs), which require only the specification of a parameterized correlation structure of the neural state over time.
A critical assumption when attempting to extract a low-dimensional neural trajectory is that the recorded activity evolves within a low-dimensional manifold. Previous studies have typically assumed that the neural trajectories lie in a three-dimensional space for ease of visualization. In this work, we investigate whether this low-dimensional assumption is justified in the context of reach planning and execution. If so, we will attempt to identify the appropriate dimensionality. Furthermore, we will systematically compare different analytical methods for extracting neural trajectories.
We first detail the two-stage methods, GPFA, and dynamical system approaches to extracting neural trajectories. Next, the behavioral task and the neural recordings in premotor and motor cortices are described. We then apply the different extraction techniques to study the dimensionality and time course of the recorded activity during reach planning and execution.
Preliminary versions of this work were previously published (Yu et al. 2008, 2009).
METHODS
Two-stage methods
The two-stage methods involve first estimating a smooth firing-rate profile for each neuron on a single trial, then applying a static dimensionality-reduction technique. For the simplest two-stage method, the firing-rate estimates are obtained by convolving each spike train with a Gaussian kernel. These firing-rate estimates, taken across all simultaneously recorded neurons, define a trajectory in the high-dimensional space of recorded activity (Fig. 2B). This trajectory is represented by a series of data points (dots in Fig. 2B). In the simplest case, the data points of many such trajectories are then passed to PCA, which identifies the directions of greatest variance in the high-dimensional space. The high-dimensional data points are then projected into the low-dimensional space defined by the principal component axes (conceptualized by the S1S2 plane shown in Fig. 2C). The projected data points can then be strung back together over time to obtain a low-dimensional neural trajectory (Fig. 2D).
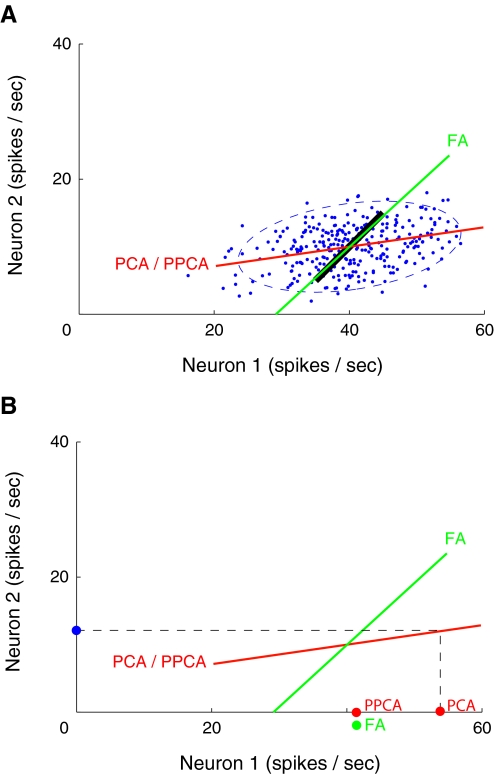
Although PCA is widely used and simple to apply, it is problematic when applied to neural data because neurons with higher firing rates are known to show higher count variability (i.e., their Poisson-like behavior) (Dayan and Abbott 2001). Because PCA finds directions in the high-dimensional space of greatest variance, these directions tend to be dominated by the neurons with the highest firing rates. This is illustrated in Fig. 4 A using two neurons. In this simulation, the underlying firing rates of the two neurons are perfectly correlated (black line), representing the ground truth. What we are able to observe, however, are noise-corrupted versions (blue dots) of the underlying firing rates, where the noise is assumed to be independent for each neuron. These blue dots are analogous to the dots in Fig. 2B. The goal is to recover the true relationship between the activity of the two neurons (black line) using only the noise-corrupted data points (blue dots). Once this relationship is identified (i.e., an estimate of the black line), the data points are then projected onto the estimated line, yielding “denoised” firing-rate estimates for the two neurons. In this case, identifying the true one-dimensional relationship between the two neurons provides a succinct account of the noisy recorded activity.
FIG. 4.
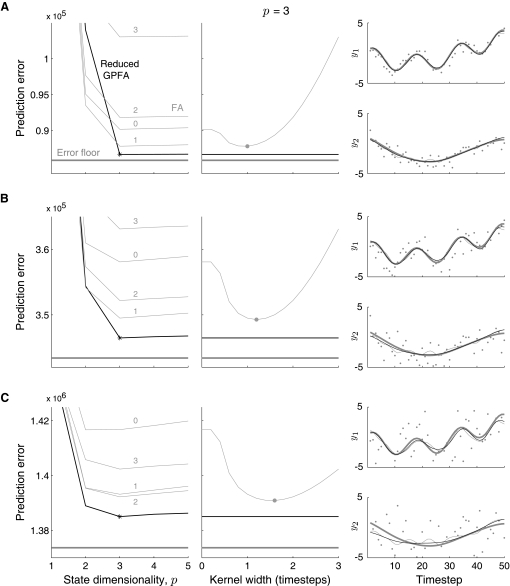
Simulation comparing principal components analysis (PCA), probabilistic PCA (PPCA), and factor analysis (FA) in the two-neuron case. A: FA (green line) is better able to uncover the true firing-rate relationship (black line) between the two neurons than PCA/PPCA (red line). The noise-corrupted observations (blue dots) and two SD covariance ellipse (dashed blue) are shown. B: leave-neuron-out model prediction for PCA (red dot labeled “PCA”), PPCA (red dot labeled “PPCA”), and FA (green dot). Each model predicts the activity of neuron 1, given the activity of neuron 2 (blue dot).
Compared with neuron 2, neuron 1 has a higher mean firing rate and correspondingly higher firing-rate variability in Fig. 4A. The higher variability leads to an elongation of the covariance ellipse (dashed blue) along the horizontal direction. When PCA is applied to the data points, the direction of highest variance (red line) is identified. In a comparison of the red and black lines, it is apparent that PCA provides a poor estimate of the true firing-rate relationship between the two neurons. The reason for the mismatch is that PCA implicitly assumes that the noise variance is isotropic (i.e., the same for all neurons regardless of mean firing rate). PCA erroneously identifies a direction that is biased in the direction of high noise variance, in this case along the horizontal axis corresponding to neuron 1. The same incorrect direction would be found by probabilistic PCA (PPCA) (Roweis and Ghahramani 1999; Tipping and Bishop 1999), which explicitly assumes isotropic noise.3
Here we propose to relax the assumption of isotropic noise by applying factor analysis (FA) (Everitt 1984) instead of PCA/PPCA. The only difference between FA and PPCA is that FA allows for each neuron to have a different noise variance that is learned from the data. Figure 4A shows that the direction recovered by FA (green line) is much closer to the true relationship (black line) than that recovered by PCA/PPCA (red line). The reason is that FA does not simply seek directions of greatest variance; rather, it seeks directions of greatest covariance while allowing for different noise variances along the different observed dimensions.
Although FA better estimates the true firing-rate relationship compared to PCA/PPCA, there remains a problem that is common to all three techniques. In Fig. 4A, the underlying firing rates (black line) of the two neurons lie within a relatively small range of 10 spikes/s. However, neurons can change their firing rate by many tens of spikes/s, for example, in response to a stimulus or during movement preparation and execution. As described earlier, the noise variance can therefore also change drastically over time for a given neuron. This is problematic because PCA, PPCA, and FA all assume that the noise variance of each neuron is fixed over time, regardless of how the neuron's underlying firing rate fluctuates. A possible solution is to replace the Gaussian observation noise model of PPCA and FA with a point-process (Smith and Brown 2003; Truccolo et al. 2005; Yu et al. 2006) likelihood model. Such an extension is challenging due to issues of mathematical tractability and computational complexity (Cunningham et al. 2008b). In this work, we consider a simpler approach based on discrete time steps. The square-root transform is known to stabilize the variance of Poisson-distributed counts (Kihlberg et al. 1972). By stabilizing the noise variance, dimensionality-reduction techniques that assume stationary noise variance (such as PCA, PPCA, and FA) can then be applied. Because the square-root transform operates only on count data, we propose performing the following sequence of preprocessing operations in lieu of kernel-smoothing the spike trains directly: 1) spike counts are taken in nonoverlapping time bins, 2) the counts are square-root transformed,4 and 3) the transformed counts are kernel-smoothed over time. The resulting data points are then passed to PCA, PPCA, or FA.
If the spike counts were indeed Poisson-distributed and if the square-root transform were able to perfectly stabilize the variance of Poisson-distributed counts, then the use of PCA/PPCA would be justified, since the spiking noise (in the space of smoothed, square-rooted counts) would be isotropic across different neurons and time points. However, spike counts of real neurons are known to deviate from a Poisson distribution (e.g., Churchland et al. 2006; Tolhurst et al. 1983) and the square-root transform only approximately stabilizes the variance of Poisson-distributed counts (Kihlberg et al. 1972). This is the case both across neurons and across time points. To compensate for unequal variances across neurons, we apply FA rather than PCA/PPCA. In results, we show that two-stage methods based on FA outperform those based on PCA/PPCA.
Leave-neuron-out prediction error
There are several modeling choices to be made when extracting neural trajectories. First, for the two-stage methods involving kernel smoothing, we seek to find the appropriate degree of smoothness by comparing different smoothing kernel widths. Second, for the two-stage methods, we seek to compare different dimensionality-reduction techniques (in this work, PCA, PPCA, and FA). Third, we seek to compare the two-stage methods with GPFA. Fourth, for all two-stage methods and GPFA, we seek to compare different dimensionalities of the low-dimensional state space. Such a comparison would help to determine whether the high-dimensional recorded activity can indeed be succinctly summarized by a low-dimensional neural trajectory and help to select the appropriate dimensionality of the low-dimensional space.
Such modeling choices are typically made either by comparing cross-validated prediction errors (Hastie et al. 2001) or likelihoods, or by comparing Bayesian marginal likelihoods (MacKay 2003), which are often approximated using the Akaike information criterion or the Bayesian information criterion. There are two reasons why the likelihood approaches are not applicable here. First, most of the two-stage methods are partially or entirely nonprobabilistic. In particular, kernel smoothing and PCA are nonprobabilistic operations. Second, even if a probabilistic dimensionality-reduction technique (e.g., PPCA or FA) is used, the likelihoods obtained are based on the smoothed data. When the data are altered by different presmoothing operations (or not, in the case of GPFA), the likelihoods are no longer comparable. Here, we introduce a goodness-of-fit metric by which all of the comparisons listed earlier can be made.
We describe the basic idea of the metric in this section; the mathematical details are given in the appendix. First, we select a particular method for extracting neural trajectories for which we want to evaluate goodness-of-fit. For the two-stage methods using kernel smoothing, this involves specifying the smoothing kernel width and the dimensionality-reduction technique (e.g., PCA, PPCA, or FA) to be used. Next, the model parameters are fit to the training data. For example, for the PCA-based two-stage method, the model parameters are the principal directions and data mean found by applying PCA to the smoothed square-rooted spike counts. Then, based on data not used for model fitting, we leave out one neuron at a time and ask how well the fitted model is able to predict the activity of that neuron, given the activity of all other recorded neurons.
This leave-neuron-out model prediction is illustrated in Fig. 4B. Consider the same situation with two neurons as in Fig. 4A. Here, we leave out neuron 1 and ask each dimensionality-reduction technique (PCA, PPCA, FA) to predict the activity of neuron 1 based only on the activity of neuron 2 (blue dot). For PCA, this is a simple geometric projection, yielding the red dot labeled “PCA.” Although PPCA finds the same principal direction as PCA (as shown in Fig. 4A), it yields a different model prediction (red dot labeled “PPCA”). The reason is that PPCA has an explicit noise model, which allows deviations of neuron 2's activity away from its mean to be attributed partially to noise, rather than entirely to changes in the low-dimensional state (i.e., movement along the red line). Thus, the PPCA model prediction is more robust to observation noise than the PCA model prediction. One can use the PPCA intuition to understand the FA model prediction (green dot). The only difference is that FA allows different neurons to have different noise variances. Although PPCA and FA are shown to give nearly identical model predictions in the two-neuron case in Fig. 4B, their model predictions are generally different for larger numbers of neurons (cf. Fig. 5A). The same ideas can be applied to compute the model prediction for GPFA, which incorporates the concept of time.
FIG. 5.
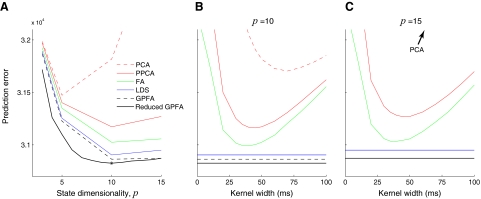
Prediction errors of two-stage methods (PCA, dotted red; PPCA, solid red; FA, green), linear dynamical system (LDS, blue), GPFA (dashed black), and reduced GPFA (solid black), computed using 4-fold cross-validation. A: prediction errors for different state dimensionalities. For two-stage methods, prediction errors shown for 50-ms kernel width (SD of Gaussian kernel). For reduced GPFA, the horizontal axis corresponds to p̃ orthonormalized dimensions of a GPFA model fit with p = 15. Star indicates minimum of solid black curve. Denser sampling of kernel widths shown for B: p = 10 and C: p = 15. Note that the dashed and solid black lines are overlaid in C, by definition. Analyses in this figure are based on 56 trials and q = 61 units for the reach target at distance 100 mm and direction 45°, Experiment G20040123.
For all methods considered in this work, the model prediction can be computed analytically because all variables involved are jointly Gaussian, as detailed in the appendix. We compute a prediction error, defined as the sum-of-squared differences between the model prediction and the observed square-rooted spike counts across all neurons and time points. This prediction error can be computed for the two-stage methods (using various smoothing kernel widths and dimensionality-reduction techniques) and GPFA, across different choices of the state space dimensionality. The comparisons listed at the beginning of this section can then be made by comparing the prediction errors.
Gaussian-process factor analysis
In this section, we provide motivation for GPFA before describing it mathematically. Then, we detail how to fit the GPFA model to neural data. Finally, we show how the extracted trajectories can be intuitively visualized using an orthonormalization procedure and describe how this gives rise to a “reduced” GPFA model with fewer state dimensions than timescales.
MOTIVATION FOR GPFA.
PCA, PPCA, and FA are all static dimensionality-reduction techniques. In other words, none of them takes into account time labels when applied to time series data; the measurements are simply treated as a collection of data points. In the two-stage methods, the temporal relationships among the data points are taken into account during kernel smoothing. There is then no explicit use of time label information during dimensionality reduction.
Here we propose an extension of FA that performs smoothing and dimensionality reduction in a common probabilistic framework, which we term Gaussian-process factor analysis (GPFA). Unlike FA, GPFA leverages the time label information to provide more powerful dimensionality reduction for time series data. The GPFA model is simply a set of factor analyzers (one per time point, each with identical parameters) that are linked together in the low-dimensional state space by a Gaussian process (GP) (Rasmussen and Williams 2006) prior. Introducing the GP allows for the specification of a correlation structure across the low-dimensional states at different time points. For example, if the system underlying the time series data is believed to evolve smoothly over time, we can specify that the system's state should be more similar between nearby time points than between faraway time points. Extracting a smooth low-dimensional neural trajectory can therefore be viewed as a compromise between the low-dimensional projection of each data point found by FA and the desire to string them together using a smooth function over time.
MATHEMATICAL DESCRIPTION OF GPFA.
As with the two-stage methods, spike counts are first taken in nonoverlapping time bins and square-rooted. However, unlike the two-stage methods, there is no presmoothing of the square-rooted spike counts for GPFA, since the smoothing and dimensionality reduction are performed together. Let y:,t ∈ ℝq×1 be the high-dimensional vector of square-rooted spike counts recorded at time point t = 1, …, T, where q is the number of neurons being recorded simultaneously. We seek to extract a corresponding low-dimensional latent neural state x:,t ∈ ℝp×1 at each time point, where p is the dimensionality of the state space (p < q). For notational convenience,5 we group the neural states from all time points into a neural trajectory denoted by the matrix X = [x:,1, …, x:,T] ∈ ℝp×T. Similarly, the observations can be grouped into a matrix Y = [y:,1, …, y:,T] ∈ ℝq×T. We define a linear-Gaussian relationship between the observations y:,t and neural states x:,t
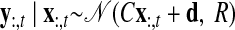 |
(1) |
where C ∈ ℝq×p, d ∈ ℝq×1, and R ∈ ℝq×q are model parameters to be learned. As in FA, we constrain the covariance matrix R to be diagonal, where the diagonal elements are the independent noise variances of each neuron. In general, different neurons can have different independent noise variances. Although a Gaussian is not strictly a distribution on square-rooted counts, its use in Eq. 1 preserves computational tractability (e.g., Wu et al. 2006).
The neural states x:,t at different time points are related through Gaussian processes, which embody the notion that the neural trajectories should be smooth. We define a separate GP for each dimension of the state space indexed by i = 1, …, p
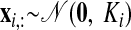 |
(2) |
where xi,: ∈ ℝ1×T is the ith row of X and Ki ∈ ℝT×T is the covariance matrix for the ith GP. The form of the GP covariance can be chosen to provide different smoothing properties on the neural trajectories. In this work, we chose the commonly used squared exponential (SE) covariance function
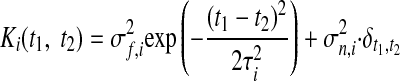 |
(3) |
where Ki(t1, t2) denotes the (t1, t2)th entry of Ki and t1, t2 = 1, …, T. The SE covariance is defined by its signal variance σf,i2 ∈ ℝ+, characteristic timescale τi ∈ ℝ+, and GP noise variance σn,i2 ∈ ℝ+. The Kroneker delta δt1,t2 equals 1 if t1 = t2 and 0, otherwise. The SE is an example of a stationary covariance; other stationary and nonstationary GP covariances (Rasmussen and Williams 2006) can be applied in a seamless way.
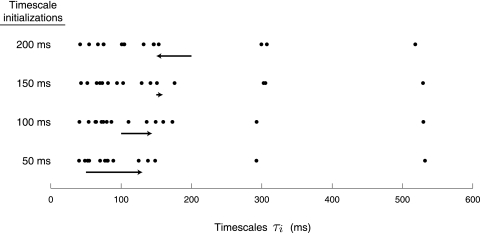
Because the neural trajectories X are hidden and must be inferred from the recorded activity Y, the scale of X (defined by the Ki in Eq. 2) is arbitrary. In other words, any scaling of X can be compensated by appropriately scaling C (which maps the neural trajectory into the space of recorded activity) such that the scale of Y remains unchanged.6 To remove this model redundancy without changing the expressive power of the model, we fix the scale of X and allow C to be learned without constraints. By direct analogy to FA, we set the prior distribution of the neural state x:,t at each time point t to be 𝒩(0, I) by fixing Ki(t, t) = 1 (however, note that the x:,t are still correlated across different t). This can be achieved by setting σf,i2 = 1 − σn,i2, where 0 < σn,i2 ≤ 1.7 Because we seek to extract smooth neural trajectories, we fixed σn,i2 to a small value (10−3), as is often done for GPs (Rasmussen and Williams 2006). In the appendix, we consider learning σn,i2 from the data. For all analyses described in results, the timescale τi is the only parameter of the SE covariance that is learned.
FITTING THE GPFA MODEL.
The parameters of the GPFA model (Eqs. 1 and 2) can be fit using the commonly used expectation-maximization (EM) algorithm (Dempster et al. 1977). The EM algorithm seeks the model parameters θ = {C, d, R, τ1, …, τp} that maximize the probability of the observed data Y. In the appendix, we derive the EM update equations for the GPFA model. Because the neural trajectories and model parameters are both unknown, the EM algorithm iteratively updates the neural trajectories (in the E-step) and the model parameters (in the M-step), while the other remains fixed. This algorithm is guaranteed to converge to a local optimum. The E-step involves using the most recent parameter updates to evaluate the relative probabilities of all possible neural trajectories given the observed spikes. This Gaussian posterior distribution P(X|Y) can be computed exactly because the x:,t and y:,t across all time points are jointly Gaussian, by definition. In the M-step, the model parameters are updated using the distribution P(X|Y) over neural trajectories found in the E-step. The updates for C, d, and R can be expressed in closed form and are analogous to the parameter updates in FA. The characteristic timescales τi can be updated using any gradient optimization technique. Note that the degree of smoothness (defined by the timescales) and the relationship between the low-dimensional neural trajectory and the high-dimensional recorded activity (defined by C) are jointly optimized. Furthermore, a different timescale is learned for each state dimension indexed by i.
VISUALIZING TRAJECTORIES VIA ORTHONORMALIZATION.
Once the GPFA model is learned, we can use it to extract neural trajectories E [X|Y] (Eq. A6) from the observed activity Y. These low-dimensional neural trajectories can be related to the high-dimensional observed activity using Eq. 1, which defines a linear mapping C between the two spaces. The following is one way to understand this mapping. Each column of C defines an axis in the high-dimensional space. The ith element of x:,t (i = 1, …, p) specifies “how far to go” along the axis defined by the ith column of C. The location in the high-dimensional space corresponding to the neural state x:,t is given by the summed contributions along each of the p aforementioned axes, plus a constant offset d.
Although the relationship between the low- and high-dimensional spaces is mathematically well defined, it is difficult to picture this relationship without knowing the direction and scale of the axes defined by the columns of C. For example, any point in two-dimensional space can be represented as a linear combination of arbitrary two-dimensional vectors w1 and w2, provided that the two vectors are not scaled versions of each other. If w1 and w2 are neither orthogonal nor of unit length, understanding how the two vectors are linearly combined to form different points can be nonintuitive. Thus points in two-dimensional space are typically referred to using their (x, y) Cartesian coordinates. These coordinates specify how unit vectors pointing along the x and y axes (i.e., orthonormal vectors) should be linearly combined to obtain different points in the two-dimensional space. This provides an intuitive specification of points in the two-dimensional space. In GPFA, the columns of C are not orthonormal, akin to the arbitrary w1 and w2. The following paragraphs describe how to orthonormalize the columns of C in GPFA to make visualization more intuitive. This is akin to expressing two-dimensional points in terms of their (x, y) Cartesian coordinates.
In the case of PCA, the identified principal directions are orthonormal, by definition. It is this orthonormal property that yields the intuitive low-dimensional visualization—i.e., the intuitive mapping between the low-dimensional principal components space and high-dimensional data space. Although the columns of C are not constrained to be orthonormal for GPFA, we can still obtain an intuitive “PCA-like” mapping between the two spaces for ease of visualization. The basic idea is to find a set of orthonormal basis vectors spanning the same space as the columns of C. This is akin to finding the unit vectors that point along the two Cartesian axes from the arbitrary w1 and w2, in the preceding example. This orthonormalization procedure does not alter the GPFA model-fitting procedure nor the extracted neural trajectories; it simply offers a more intuitive way of visualizing the extracted trajectories.
The orthonormalization procedure involves applying the singular value decomposition (Strang 1988) to the learned C. This yields C = UDV′, where U ∈ ℝq×p and V ∈ ℝp×p each have orthonormal columns and D ∈ ℝp×p is diagonal. Thus, we can write Cx:,t = U(DV′x:,t) = Ux̃:,t, where x̃:,t = DV′x:,t ∈ ℝp×1 is the orthonormalized neural state at time point t. Note that x̃:,t is a linear transformation of x:,t. The orthonormalized neural trajectory extracted from the observed activity Y is thus DV′ E [X|Y]. Since U has orthonormal columns, we can now intuitively visualize the trajectories extracted by GPFA, in much the same spirit as for PCA.
There is one other important advantage of the orthonormalization procedure. Whereas the elements of x:,t (and the corresponding columns of C) have no particular order, the elements of x̃:,t (and the corresponding columns of U) are ordered by the amount of data covariance explained, analogous to PCA. Especially when the number of state dimensions p is large, the ordering facilitates the identification and visualization of the dimensions of the orthonormalized neural trajectory that are most important for explaining the recorded activity. The ordering is made possible by the singular value decomposition, which specifies the scaling of each of the columns of U in the diagonal entries of D (i.e., the singular values). If these diagonal entries are arranged in decreasing order, then the columns of U specify directions in the high-dimensional space in order of decreasing data covariance explained. Overall, the orthonormalization procedure allows us to view the neural trajectories extracted by GPFA using PCA-like intuition.8 In particular, the low-dimensional axes are ordered and can be easily pictured in the high-dimensional space. These concepts are illustrated in Fig. 2, C and D, where S1 and S2 correspond to the first two dimensions of the orthonormalized neural state x̃:,t.
REDUCED GPFA.
According to Eq. 2, each neural state dimension indexed by i has its own characteristic timescale τi. This implies that a GPFA model with a p-dimensional neural state possesses a total of p timescales. However, there may be cases where the number of timescales needed to describe the data exceeds the number of state dimensions. For example, it may be that a system uses only two degrees of freedom (i.e., two state dimensions), but evolves over time with a wide range of different speeds that cannot be well captured using only two timescales. Here, we describe a way to obtain a GPFA model whose number of timescales p exceeds the effective state dimensionality p̃. First, a GPFA model with state dimensionality p is fit using the EM algorithm. Next, the orthonormalization procedure described earlier is applied, yielding the orthonormalized neural state x̃:,t ∈ ℝp×1. Note that, although each dimension of x:,t possesses a single characteristic timescale, each dimension of x̃:,t represents a mixture of p timescales. Because the dimensions of x̃:,t are ordered by the amount of data covariance explained, we can choose to retain only the top p̃ dimensions of x̃:,t (p̃ = 1, …, p) and to discard the remaining lowest dimensions. This yields a p̃-dimensional neural trajectory for the reduced GPFA model.
Dynamical systems approaches
Another way to extract neural trajectories is by defining a parametric dynamical model that describes how the low-dimensional neural state evolves over time. A hidden Markov model (HMM) is a dynamical model in which the state jumps among a set of discrete values. HMMs have been fruitfully applied to study single-trial neural population activity in monkey frontal cortex (Abeles et al. 1995; Gat et al. 1997; Seidemann et al. 1996), rat gustatory cortex (Jones et al. 2007), monkey premotor cortex (Kemere et al. 2008), and songbird premotor areas (Danóczy and Hahnloser 2006; Weber and Hahnloser 2007). In many experimental contexts, it is desirable to allow for a continuous-valued state, rather than one that jumps among a set of discrete values. Even in settings where the experimental paradigm defines discrete states (e.g., Fig. 1A, one state per percept or decision), there are advantages to using continuous-valued states. Whereas a HMM would indicate when the switches occur in Fig. 1A, a dynamical model with continuous-valued states would allow one to study the details of how the switching is carried out—in particular, along what path and how quickly. Although it is always possible to define the HMM with a larger number of discrete states to approximate a model with continuous-valued states, such an approach is prone to overfitting and requires appropriate regularization (Beal et al. 2002).
A commonly used dynamical model with continuous-valued state is a first-order linear autoregressive (AR) model (Kulkarni and Paninski 2007; Smith and Brown 2003), which captures linear Markovian dynamics. Such a model can be expressed as a Gaussian process, since the state variables are jointly Gaussian. This can be shown by defining a separate first-order AR model for each state dimension indexed by i ∈ {1, …, p}
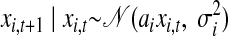 |
(4) |
Given enough time (t → ∞) and |ai| < 1, the model will settle into a stationary state that is equivalent to Eq. 2 with
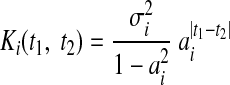 |
(5) |
as derived elsewhere (Turner and Sahani 2007). The first-order AR model described by Eqs. 2 and 5, coupled with the linear-Gaussian observation model Eq. 1, will henceforth be referred to as “LDS” (linear dynamical system). Different covariance structures Ki can be obtained by going from a first-order to an nth-order AR model. One drawback of this approach is that it is usually not easy to construct an nth-order AR model with a specified covariance structure. In contrast, the GP approach requires only the specification of the covariance structure, thus allowing different smoothing properties to be applied in a seamless way. AR models are generally less computationally demanding than those based on GP, but this advantage shrinks as the order of the AR model grows. Another difference is that Eq. 5 does not contain an independent noise term σn,i2·δt1,t2 as in Eq. 3. The innovations noise σi2 in Eq. 4 is involved in setting the smoothness of the time series, as shown in Eq. 5. Thus, Eq. 4 would need to be augmented to explicitly capture departures from the AR model.
One may also consider defining a nonlinear dynamical model (Yu et al. 2006), which typically has a richer set of dynamical behaviors than that of linear models. The identification of the model parameters provides insight into the dynamical rules governing the time evolution of the system under study. However, especially in exploratory data analyses, it may be unclear what form this model should take. Even if an appropriate nonlinear model can be identified, using it to extract neural trajectories may require computationally intensive approximations and yield unstable model-fitting algorithms (Yu et al. 2006). In contrast, the model-fitting algorithm for GPFA is stable, approximation-free, and straightforward to implement. The use of GPFA can be viewed as a practical way of going beyond a first-order linear AR model without having to commit to a particular nonlinear system, while retaining computational tractability.
Relative to previously proposed models in the machine learning literature, GPFA is most similar to the semiparametric latent factor model (Teh et al. 2005), where the GPFA model can be obtained by letting time indices play the role of inputs. Although GPFA involves Gaussian processes and latent variables, it is quite different from the Gaussian process latent variable model (GP-LVM) (Lawrence 2005). The GP-LVM uses Gaussian processes to define a nonlinear relationship between the latent and the observed variables. In that case, the GP smoothing is defined by how close two points are in the latent space. In contrast, GPFA defines a linear mapping between the latent and observed variables; the GP smoothing is defined by how close two points are in time (Lawrence and Moore 2007). GPFA is also quite different from Gaussian process dynamical models (GPDM) (Wang et al. 2006). Whereas GPDM extends Markovian linear AR models to the nonlinear regime (while remaining Markovian), GPFA extends to the non-Markovian regime (while remaining linear) with arbitrary temporal covariance structures. As with GP-LVM, GPDM defines a nonlinear relationship between the latent and observed variables.
Delayed-reach task and neural recordings
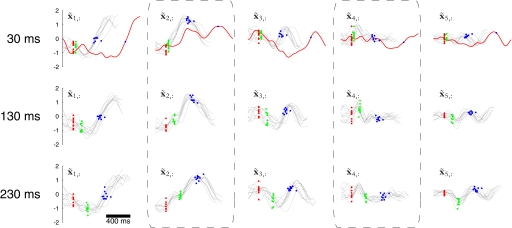
Animal protocols were approved by the Stanford University Institutional Animal Care and Use Committee. We trained an adult male monkey (Macaca mulatta, monkey G) to perform delayed center-out reaches for juice rewards. Visual targets were back-projected onto a frontoparallel screen about 30 cm in front of the monkey. The monkey touched a central target and fixated his eyes on a crosshair at the upper right corner of the central target. After a center hold period of 1,000 ms, a pseudorandomly chosen peripheral reach target was presented at one of 14 possible locations (directions: 0, 45, 90, 135, 180, 225, 315°; distances: 60, 100 mm).9 After a randomly chosen instructed delay period, the “go” cue (signaled by both the enlargement of the reach target and the disappearance of the central target) was given and the monkey reached to the target. In the present work, we analyzed data from two experiments that differ only in the distribution of delay periods used. Whereas experiment G20040123 used delay periods in the range 200–700 ms, experiment G20040124 used three discrete delay periods of 30, 130, and 230 ms. Eye fixation at the crosshair was enforced throughout the delay period. After a hold time of 200 ms at the reach target, the monkey received a liquid reward.
During experiments, monkeys sat in a custom chair (Crist Instruments, Hagerstown, MD) with the head braced and the nonreaching arm strapped to the chair. The presentation of the visual targets was controlled using the Tempo software package (Reflective Computing, St. Louis, MO). A custom photodetector recorded the timing of the video frames with 1-ms resolution. The position of the hand was measured in three dimensions using the Polaris optical tracking system (Northern Digital, Waterloo, Ontario, Canada; 60 Hz, 0.35-mm accuracy), whereby a passive marker taped to the monkey's fingertip reflected infrared light back to the position sensor. Eye position was tracked using an overhead infrared camera (Iscan, Burlington, MA; 240 Hz, estimated accuracy of 1°).
A 96-channel silicon electrode array (Cyberkinetics, Foxborough, MA) was implanted straddling dorsal premotor (PMd) and motor (M1) cortex in the right hemisphere, contralateral to the reaching arm. Surgical procedures have been described previously (Churchland et al. 2006). An intraoperative photo showing the exact location of array implantation can be found in Batista et al. (2007). We manually discriminated spike waveforms at the start of each session using two time–amplitude window discriminators on each channel. Isolations were tagged as either single unit or multiunit based on visual inspection of their quality during the experiment. On this particular electrode array, we found several groups of electrodes that yielded nearly identical (or highly similar) spike trains. Although the source of this electrode “cross talk” is currently unclear, we speculate that it may be due to faulty electrical isolation among the channels either in the pedestal connectorization, in the wire bundle leading out of the array, or in the array itself. It is unlikely that two adjacent electrodes recorded from the same neuron(s), given the distance between adjacent electrodes (400 μm). We have observed such cross talk on a few different electrode arrays. For prosthetic decoding (e.g., of arm trajectories), this is typically not a major concern, since it simply gives the repeated unit(s) a greater influence on the decoded result. For extracting neural trajectories, this is a major problem, since the goal is to identify structure in the correlated activity across the neural population. If two units have identical (or nearly identical) activity, one of the dimensions of the neural trajectory is likely to be dedicated to describing this spurious, strong correlation between the pair of units. Thus, before analyzing the data, we checked all pairs of the 96 electrodes for cross talk by computing the percentage of coincident spikes (allowing for ±1-ms jitter) for each pair. Across all pairs, this yielded a clear, bimodal distribution. On this electrode array, we found cross talk in three electrode pairs, two triplets, and one quadruplet. We thus removed 10 of the 96 channels before any of the analyses described in this paper were performed.
The analyses in the present work are concerned primarily with the neural activity during the delay period. However, many of our isolations showed the strongest modulation during the movement period and/or showed weakly modulated delay-period activity. A single unit or multiunit was thus included in our analyses only if 1) it possessed tuned (P < 0.1) delay-period activity with reasonable modulation (≥5 spikes/s difference between the most and least responsive conditions) and 2) the delay-period firing rate averaged across all conditions was ≥20% of the movement-period firing rate averaged across all conditions. For these assessments, the delay-period firing rate was computed in a 200-ms window starting 150 ms after reach target presentation, whereas the movement-period firing rate was computed in a 300-ms window starting 100 ms this paper movement onset.
In total, we analyzed 784 (910) trials for experiment G20040123 (G20040124), comprising 18 (18) single units and 43 (44) multiunits. The distribution of reaction times, defined as the time between the go cue and movement onset, had mean ± SD of 293 ± 48 ms (329 ± 54 ms). The arm movement durations were 269 ± 40 ms (280 ± 44 ms). Both data sets have previously appeared (Churchland et al. 2006). We have explicitly chosen to analyze the same data sets here to uncover the single-trial substrates of the trial-averaged effects reported in our previous studies.
RESULTS
We considered neural data for one reach target at a time, ranging from 200 ms before reach target onset to movement end. This period comprised the randomly chosen delay period following reach target onset, the monkey's reaction time, and the duration of the arm reach. Spike counts were taken in nonoverlapping 20-ms bins, then square-rooted.10 For the two-stage methods, these square-rooted counts were first smoothed over time using a Gaussian kernel before being passed to a static dimensionality-reduction technique: PCA, PPCA, or FA. LDS and GPFA were given the square-rooted spike counts with no kernel presmoothing.
Using the goodness-of-fit metric described in methods, we can compare different degrees of smoothness, dimensionality-reduction techniques (PCA, PPCA, and FA), and state dimensionalities for the two-stage methods. Figure 5A shows the prediction error for PCA (dashed red), PPCA (solid red), and FA (green) across different state dimensionalities (p = 3, 5, 10, 15) with a kernel width of 50 ms. Alternatively, we can fix the state dimensionality (p = 10 in Fig. 5B; p = 15 in Fig. 5C) and vary the kernel width. There are two primary findings for the two-stage methods. First, PCA, PPCA, and FA yielded progressively lower prediction error (Wilcoxon paired-sample test, P <0.001). Statistical significance was assessed by looking across the 14 reach targets; for each reach target, we obtained the prediction error for each method at its optimal state dimensionality and kernel width. FA outperforms PCA and PPCA because it allows different neurons to have different noise variances. Recall that prediction errors were evaluated based on data not used for model-fitting (i.e., cross-validated), so this result cannot simply be due to FA having more parameters. PCA has the worst performance because it has no explicit noise model and is thus unable to distinguish between changes in the underlying neural state and spiking noise. Second, for these data, the optimal smoothing kernel width was approximately 40 ms for both PPCA and FA, as indicated by Fig. 5, B and C.
The same metric can be used to compare the two-stage methods with LDS and GPFA. As indicated in Fig. 5, LDS (blue) yielded lower prediction error than the two-stage methods (Wilcoxon paired-sample test, P <0.001). Furthermore, GPFA (dashed black) outperformed LDS and the two-stage methods (Wilcoxon paired-sample test, P <0.001). As before, statistical significance was assessed by looking across the 14 reach targets; for each reach target, we obtained minimum prediction error for each method (LDS and GPFA) at its optimal state dimensionality. The prediction error was further reduced by taking only the top p̃ orthonormalized state dimensions of a GPFA model fit with p = 15 (reduced GPFA, solid black). Among the methods for extracting neural trajectories compared in this work,11 reduced GPFA produced the lowest prediction error (Wilcoxon paired-sample test, P <0.001). Further insight regarding the performance of the reduced GPFA model is provided in the following text.
Based only on Fig. 5, it is difficult to assess the benefit of GPFA relative to competing methods in terms of percentage improvement in prediction error. The reason is that we do not know what the theoretical lower limit on the prediction error is for real neural data. It would be incorrect to compute the percentage improvement in terms of distance from zero error. Thus, we performed a simulation (described in the appendix and Fig. A2) in which the error floor can be computed. Based on this error floor (which was far above zero), we found that GPFA provided tens of percent improvement in prediction error relative to that of the best two-stage method. This suggests that GPFA may have a similar percentage improvement for the real neural data shown in Fig. 5.
FIG. A2.
Simulated data with known error floor. Each row corresponds to a different independent noise variance: A: 0.5, B: 2, C: 8. Left column: prediction errors for two-stage method with FA (green) and reduced GPFA (black), along with error floor (orange), at different state dimensionalities. Each green curve corresponds to a different kernel width (numbers of time steps are labeled). Star indicates minimum of black curve. Middle column: denser sampling of kernel widths for p = 3. Minimum of green curved denoted by green dot. Right column: each panel corresponds to an observed dimension. The same 2 observed dimensions are used in A, B, and C. Shown are the activity level of each neuron before noise was added (orange curves), noisy observations (orange dots), leave-neuron-out prediction using best two-stage method (green), and leave-neuron-out prediction using reduced GPFA (black).
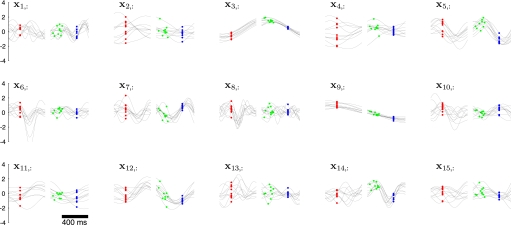
Figure 6 shows the neural trajectories E[X|Y] (Eq. A5) extracted by GPFA with p = 15. Each panel corresponds to a different neural state dimension, which evolves over time according to its own characteristic timescale τi that is learned from the data. For example, the timescales for the first five dimensions in Fig. 6 are 54, 160, 293, 173, and 111 ms. Although we have obtained a substantial reduction in dimensionality in going from the 61-dimensional recorded neural responses to the 15-dimensional neural trajectories, it is still difficult to gain intuition about how the neural responses are evolving over time based solely on Fig. 6, for two reasons. First, the dimensions of the neural state are not ordered; thus, we do not know whether certain state dimensions are more important than others for explaining the activity across the neural population. Second, although each state dimension corresponds to a column of C, one cannot readily picture how the low-dimensional neural trajectories would appear if mapped out into the high-dimensional space using Eq. 1. The reason is that the columns of the learned C may have different scalings and are not guaranteed to be mutually orthogonal.
FIG. 6.
Neural trajectories for GPFA with p = 15. Each panel corresponds to one of the 15 dimensions of the neural state, which is plotted vs. time. The neural trajectory for one trial comprises one black trace from each panel. Dots indicate time of reach target onset (red), go cue (green), and movement onset (blue). Due to differing trial lengths, the traces on the left/right half of each panel are aligned on target/movement onset for clarity. However, the GPFA model was fit using entire trials with no gaps. Note that the polarity of these traces is arbitrary, as long as it is consistent with the polarity of C. Each trajectory corresponds to planning and executing a reach to the target at distance 100 mm and direction 45°. For clarity, only 10 randomly chosen trials with delay periods >400 ms are plotted. Experiment G20040123, q = 61 units.
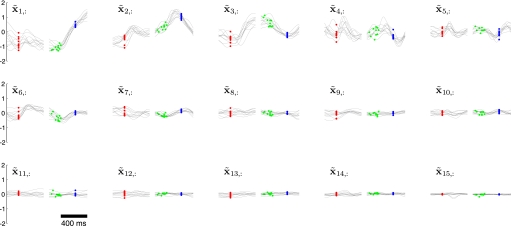
These difficulties can be overcome by applying the orthonormalization procedure described in methods based on the singular value decomposition of C. The resulting orthonormalized neural trajectories are shown in Fig. 7, where each panel corresponds to an orthonormalized state dimension and involves a mixture of timescales. Importantly, the panels are arranged in decreasing order of data covariance explained. This ordering is apparent in Fig. 7 if one considers the range of values explored by the orthonormalized neural trajectory along each of its dimensions. The top orthonormalized dimensions indicate fluctuations in the recorded population activity shortly after target onset (red dots) and again after the go cue (green dots). Furthermore, the neural trajectories around the time of the arm movement are well aligned on movement onset. These observations are consistent with previous analyses of the same data (Churchland et al. 2006), as well as other studies of neural activity collected during similar tasks in the same cortical areas. Note that the neural trajectories in Fig. 7 are remarkably similar (but not identical) on different trials, even though 1) the spike timing differs across repeated trials and 2) there is no constraint built into GPFA requiring that neural trajectories should trace out similar paths on different trials. The orthonormalized dimensions x̃1,: and x̃2,: are analogous to S1 and S2, respectively, in Fig. 2. Unlike in Fig. 2D, where the trajectory is plotted in the space of S1 versus S2, each orthonormalized dimension is plotted versus time in Fig. 7 to show more than just the top two (or three) dimensions.
FIG. 7.
Orthonormalized neural trajectories for GPFA with p = 15. These are the same 10 trials shown in Fig. 6. Each panel corresponds to one of the 15 dimensions of the orthonormalized neural state, which is plotted vs. time. The orthonormalized neural trajectory for one trial comprises one black trace from each panel. Note that the polarity of these traces is arbitrary, as long as it is consistent with the polarity of U. Figure conventions identical to those in Fig. 6.
The range of values explored by the trajectories in each orthonormalized dimension is analogous to the variance explained by each principal component in PCA. A common way to estimate the data dimensionality with PCA is to look for an “elbow” in the residual variance curve. Such an “elbow,” if it exists, is typically considered to separate the signal dimensions from the noise dimensions. Similarly, we can obtain a rough estimate of the data dimensionality with GPFA by counting the number of top orthonormalized dimensions showing “meaningful” time-varying structure in Fig. 7. Although the top six dimensions show strong temporal structure, it is unclear by eye whether the lower dimensions are needed to describe the population response. The number of “meaningful” dimensions can be rigorously quantified by computing the prediction error based only on the top p̃ orthonormalized dimensions (reduced GPFA), as described in methods. In Fig. 5A (solid black), we found that the prediction error continued to decrease as more orthonormalized dimensions (x̃1,:, x̃2,:,) were included, up to x̃10,:. This indicates that dimensions x̃1,: to x̃10,: contain meaningful structure for explaining the population response. Beyond x̃10,:, adding additional dimensions increased the prediction error, indicating that the weak temporal structure seen in these lowest orthonormalized dimensions is primarily “noise.” Thus, the solid black line reaches its minimum at p̃ = 10 (referred to as p*). By definition, the solid and dashed black lines coincide at p̃ = 15.
Figure 5A also shows that prediction error using only the top 10 orthonormalized dimensions (solid black, p̃ = 10) is lower than that obtained by directly fitting a GPFA model with a 10-dimensional neural state (dashed black, p = 10). This can be understood by recalling that each panel in Fig. 7 represents a mixture of 15 characteristic timescales. Thus, the top 10 orthonormalized dimensions can make use of up to 15 timescales. In contrast, a GPFA model fit with p = 10 can have at most 10 timescales. By fitting a GPFA model with a large number of state dimensions p (each with its own timescale) and taking only the top p̃ = p* orthonormalized dimensions, we can obtain neural trajectories whose effective dimensionality is smaller than the number of timescales at play.
Based on the solid black line in Fig. 5A we consider the effective dimensionality of the recorded population activity to be p* = 10. In other words, the linear subspace within which the recorded activity evolved during reach planning and execution for this particular target was 10-dimensional. Across the 14 reach targets, each considered separately, the effective dimensionality ranged from 8 to 12, with a mode of 10. All major trends seen in Fig. 5 were preserved across all reach targets.
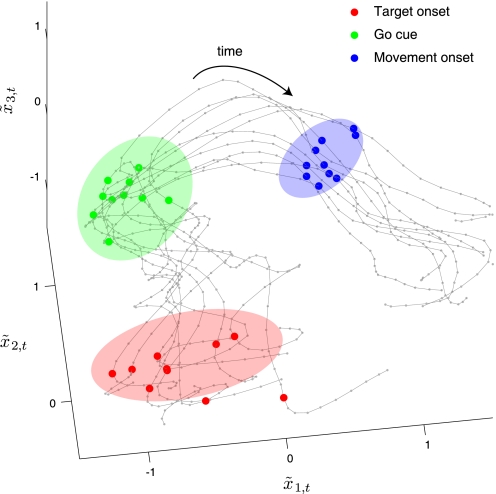
Having developed a method for extracting low-dimensional neural trajectories that yields lower prediction error than existing methods, we sought to apply it to study neural population activity on a trial-by-trial basis. We previously showed that the across-trial neural variability decreased during reach planning (Churchland et al. 2006), which led to the conception that the underlying neural trajectories (indexing the process of motor planning) may be converging over time. However, this effect could only be inferred indirectly by collapsing over many neurons and trials. Using the methods described in the present work, we can now track the progress of motor planning on single trials and directly view their convergence over time. Figure 8 shows neural trajectories plotted in the space of the top three orthonormalized state dimensions (corresponding to the first three panels of Fig. 7). The extent to which these trajectories converged during reach planning can be quantified by comparing the spread of neural states at target onset (red dots) to that at the go cue (green dots). These spreads are described by the covariance ellipsoids about the scatter of neural states at each of these time points, shown as shaded ellipses in Fig. 8. Formally, we computed the volume of the covariance ellipsoid, defined by the square root of the determinant of the covariance matrix. To compare the spreads at two different time points, we took the ratio of volumes of the two covariance ellipsoids. We computed the ratio of volumes using the top p* orthonormalized dimensions (in this case, 10), rather than just the top three orthonormalized dimensions shown in Fig. 8. It is essential to compute volumes (and perform other analyses) in the space of optimal dimensionality p*, since important features of the trajectories can be lost by using only a subset of its dimensions. To compare this result across different reach targets that may have different p*, we then took the p* th root of this ratio to obtain a “ratio per dimension.” Only trials with delay periods >400 ms, for which there is enough time for the motor planning process to come to completion, were included in this analysis.
FIG. 8.
Top 3 dimensions of orthonormalized neural trajectories for GPFA with p = 15. Each gray trace corresponds to a single trial (same 10 trials as in Figs. 6 and 7). Small gray dots are time points separated by 20 ms. Larger dots indicate time of reach target onset (red), go cue (green), and movement onset (blue). Ellipses (two SD around mean) indicate the across-trial variability of neural state at reach target onset (red shading), go cue (green shading), and movement onset (blue shading). These ellipses can be obtained equivalently in two ways. One can either first project the neural states from the optimal 10-dimensional space into the 3-dimensional space shown, then compute the covariance ellipsoids in the 3-dimensional space; or, one can first compute the covariance ellipsoids in the 10-dimensional space, then project the ellipsoids into the 3-dimensional space. The covariance ellipsoids were computed based on all 45 trials with delay periods >400 ms for this reach target.
For the reach target considered in Figs. 7 and 8, the ratio per dimension from target onset to the go cue was 1.34.12 Across the 14 reach targets, the ratio per dimension was 1.31 ± 0.13 (mean ± SD). The mean of this distribution was greater than unity (one-sided t-test, P <0.001), indicating that the neural state converged during reach planning. From the go cue (green dots) to movement onset (blue dots), the neural state further converged (one-sided t-test, P <0.001), a finding that is also consistent with that of Churchland et al. (2006). In this case, the ratio per dimension was 1.17 ± 0.14 (mean ± SD) across the 14 reach targets. Since the columns of U are orthonormal, the same volumes can be obtained by first mapping the neural trajectories into the high-dimensional space using U (yielding denoised high-dimensional data) and computing the volumes there. Because firing rates and the associated spiking noise variances tend to rise after target onset (Churchland et al. 2006), the spread of raw spike counts (with no smoothing or dimensionality reduction) in the high-dimensional space at the time of the go cue would be larger than that at the time of target onset.
Previous reports have shown that reaction times tend to be shorter on trials with longer delay periods, suggesting that some time-consuming motor preparatory process is given a head start during the delay (Churchland et al. 2006; Crammond and Kalaska 2000; Riehle and Requin 1989). In these studies, evidence is provided by the trial-averaged response of single neurons or a one-dimensional time course (e.g., the average firing rate or the Fano factor) collapsed across the neural population. The methods presented in this work allow us to view such effects in a multidimensional neural state space on single trials. We applied GPFA to a data set with three discrete delay periods of 30, 130, and 230 ms. With these short delay periods, we can visualize the effect of the go cue arriving at different times during the early stages of motor preparation. The GPFA model with p = 15 was fit to trials of all delays periods together. Figure 9 shows the extracted orthonormalized neural trajectories with trials grouped by delay period. Recall that the orthonormalized dimensions are ordered; within each row, the panels are arranged in decreasing order of data covariance explained. The panels in the first column (x̃1,:) appear to be largely capturing the movement-related neural activity (the ramp to the right of the green dots). The panels in the second column (x̃2,:, left dotted box) suggest that, prior to movement onset, the orthonormalized neural state must move from a baseline state (red dots) to a state appropriate for movement (blue dots) along this dimension. With a 30-ms delay period, nearly the entire traversal from baseline state to movement state occurs after the go cue (green dots). In contrast, with a 230-ms delay, the neural state performs part of the traversal during the delay period and appears to hold while waiting for the go cue. When the go cue arrives, the remainder of the traversal is carried out. If there is a limit on how quickly firing rates (and therefore the neural state) can change over time, then one would expect the reaction times (i.e., the time between the green and blue dots) to be longer for the 30-ms delays than for the 230-ms delays. Indeed, we found that the reaction times for the 30-ms delays were greater than those for the 230-ms delays (P <0.01, t-test) (Churchland et al. 2006). Comparing the panels in the fourth column (x̃4,:, right dotted box), the neural state appears to trace out a similar path along that orthonormalized dimension following target onset, regardless of when the go cue arrives. Kalaska and colleagues (Crammond and Kalaska 2000) previously reported single PMd neurons with similar response properties, whereby a phasic response was emitted only after the first signal with instructional value in reaction-time (analogous to 30-ms delay) and instructed-delay (analogous to 230-ms delay) reach trials. The phasic response was interpreted as information processing that would not need to occur after the go cue if given enough time to be carried out during the delay period. Although we cannot rule out that the “phasic response” seen in the fourth column of Fig. 9 is primarily a sensory response (to the appearance of the reach target) rather than motor processing, such visualizations provide invaluable intuition for the recorded activity and suggest tantalizing hypotheses that can be further investigated in future studies.
FIG. 9.
Orthonormalized neural trajectories for trials with discrete delay periods of 30 ms (top row), 130 ms (middle row), and 230 ms (bottom row). The red traces in the top row correspond to a single trial with an outlying reaction time (reaction time: 844 ms, trial ID 68). The top 5 orthonormalized dimensions of a GPFA model fit with p = 15 are shown for each delay period; the remaining orthonormalized dimensions are qualitatively similar to dimensions 6 to 15 in Fig. 7. Dotted boxes highlight the 2nd and 4th orthonormalized dimensions, which are referred to in results. For clarity, only 10 randomly chosen trials of each delay period are plotted, aligned on target onset. All trials shown correspond to the reach target located at distance 100 mm and direction 45°. Figure conventions are otherwise identical to those in Fig. 6. There is a small amount of temporal jitter in the green points due to the refresh rate of the visual display projector. Experiment G20040124, q = 62 units.
The methods developed here provide a concise summary of the activity recorded across a neural population on a single trial. By extracting such a summary (i.e., the neural trajectory) for each trial, we can readily compare how the neural activity observed on one trial differs from that observed on other trials and possibly link such differences to the subject's behavior. Such a comparison would be onerous based solely on the raw spike trains recorded simultaneously from tens to hundreds of neurons. The power of this approach is illustrated in Fig. 9. Among the trials with 30-ms delay, one particular trial was readily identified as an outlier, whose neural trajectory (red traces) appeared very different from the trajectories on other trials. Note that the visualization is extracted from neural activity alone, with no experimental timing or behavioral information provided. Can we relate this outlying trajectory to the subject's behavior? Indeed, when we labeled the neural trajectories with experimental timing markers (red, green, and blue dots), it became clear that the reaction time (i.e., the time between the green and blue dots) on the outlying trial was much longer than that on the other trials. However, the neural activity around the time of the arm movement on the outlying trial matched well with that on the other trials (seen by aligning the trajectories in time based on the blue dots). This neural activity is presumably related to generating the arm movement and it is thus sensible that it is time-locked to movement onset (blue dots). Such visualizations are invaluable when screening large volumes of neural data and during exploratory data analyses. While this outlying trial provides a particularly illustrative example of how differences in the neural trajectories can be indicative of differences in single-trial behavior, we hope to relate more subtle properties of the neural trajectories to the subject's behavior in future studies.
DISCUSSION
In this work, we have extended existing two-stage methods and developed a new method (GPFA) for extracting single-trial neural trajectories from neural population activity. For the two-stage methods, we introduced 1) dimensionality-reduction techniques PPCA and FA, which explicitly account for spiking noise; 2) the square-root transform, which approximately stabilizes the spiking noise variance across neurons and across time; and 3) a goodness-of-fit metric, which allows for the degree of smoothing to be chosen in a principled way and for different extraction methods to be objectively compared. We then presented GPFA, which unifies the smoothing and dimensionality-reduction operations in a common probabilistic framework without any loss in predictive power compared to the best two-stage method. We applied these methods to neural activity recorded during a delayed-reach task in premotor and motor cortices. We found that 1) the 61-dimensional recorded activity could be succinctly captured by neural trajectories that evolve within a far lower dimensional (8- to 12-dimensional) space; 2) the single-trial trajectories converged over time during motor planning, an effect that was shown indirectly by previous studies; and 3) properties of the trajectories could be related to the subject's behavior on a single-trial basis.
One of the advantages of GPFA over the two-stage methods is that the degree of smoothing (defined by the characteristic timescales τi) and the relationship between the low-dimensional neural trajectory and the high-dimensional recorded activity (defined by C in Eq. 1) can be jointly optimized. For the two-stage methods, the relationship between the low- and high-dimensional spaces is optimized given that the neural data have already been presmoothed in some way (e.g., using a Gaussian kernel with a predetermined kernel width). This suggests a “brute-force” approach to joint optimization by presmoothing the neural data in different ways, then optimizing the relationship between the two spaces in each case. However, the brute-force approach can be carried out only if the search space of different ways to presmooth the neural data is not too large. For example, allowing each neuron to have its own smoothing kernel width would only be tractable for small numbers of neurons. In Fig. 5, B and C (red and green lines), we were able to carry out this brute-force search for the two-stage methods by assuming that all neurons have the same smoothing kernel width, effectively collapsing 61 parameters down to one parameter. Despite this brute-force approach to joint optimization and restricting all neurons to the same smoothing kernel width, the best two-stage method (FA with a 40-ms smoothing kernel width) was able to extract neural trajectories that look qualitatively similar to those extracted by GPFA, shown in Figs. 7–9. It is reassuring that different methods produce similar trajectories when applied to the same data. However, when compared quantitatively, the best two-stage method still yielded higher prediction error than that of GPFA (Fig. 5). It remains to be seen how important this difference in prediction error is in terms of one's ability to relate features of the neural trajectories to the subject's behavior. The leave-neuron-out prediction error is a general and fundamental criterion for measuring how well a neural trajectory captures the correlated firing rates across a neural population. Depending on the goals of the visualization and scientific questions being asked, there may be other reasonable criteria for comparing different methods for extracting neural trajectories.
It is tempting to try to relate the smoothing kernel width (40 ms) of the best two-stage method to the timescales τi learned by GPFA, since the SE covariance has the same shape as the Gaussian smoothing kernel. However, as shown in Fig. A1, nearly all of the timescales learned by GPFA are >40 ms. This apparent mismatch can be understood by considering the equivalent kernel of the SE covariance (Sollich and Williams 2005), which takes on a sinc-like13 shape whose main lobe is generally far narrower than a Gaussian kernel with the same width parameter. It is therefore reasonable that the timescales learned by GPFA are larger than the optimal smoothing kernel width.
Because only the GP covariance structure needs to be specified, GPFA is particularly attractive for exploratory data analyses, where the rules governing the dynamics of the system under study are unknown. Based on the trajectories obtained by GPFA, one can then attempt to define an appropriate dynamical model that describes how the neural state evolves over time. Such an approach will allow us to reexamine, and potentially advance, the dynamical systems approach we previously proposed (Yu et al. 2006). Compared with the two-stage methods, the choice of GP covariance allows for more explicit specification of the smoothing properties of the low-dimensional trajectories. This is important when investigating (possibly subtle) properties of the system dynamics. For example, one may seek to ask whether the system exhibits second-order dynamics by examining the extracted trajectories. In this case, it is critical that second-order effects not be built-in by the smoothness assumptions used to extract the trajectories. With GPFA, it is possible to select a triangular GP covariance that assumes smoothness in position, but not in velocity. In contrast, it is unclear how to choose the shape of the smoothing kernel to achieve this in the two-stage methods.
Whether a two-stage method or GPFA is used to extract neural trajectories, one should critically evaluate the assumptions made by the extraction method before using it to answer scientific questions. No method is assumption-free and one must verify that the assumptions made by the method are not trivially producing the observed effect (e.g., when studying second-order dynamics). This often requires looking at the same data with related methods that apply different assumptions to see whether the observed effect holds up. Even with the same data, different scientific questions may call for the use of different methods. Examples of such assumptions include the choice of smoothing kernel or GP covariance, the use of the square-root transform, the observation noise model, the linear mapping between the low- and high-dimensional spaces,14 and edge effects when estimating finite-duration neural trajectories. To avoid possible artifacts introduced by the extraction method, one may consider first generating hypotheses by visualizing the low-dimensional neural trajectories, then testing the hypotheses using the raw high-dimensional recorded activity (e.g., Mazor and Laurent 2005). Although this approach is in principle “safer,” the high-dimensional recorded activity is noisy and may mask subtle relationships that are revealed only in the (denoised) low-dimensional neural trajectories.
While being mindful of these caveats, based on our findings described in this report, we believe that the GPFA framework offers better single-trial characterization of population activity and greater flexibility for testing different scientific hypotheses relative to competing methods. Given a new data set with neural activity recorded simultaneously across a neural population, we suggest taking the following steps to extract and visualize single-trial neural trajectories.
Signal conditioning: identify and remove electrode channels with cross talk (see methods), then spike sort remaining channels.
Apply square-root transform to binned spike counts.
Fit the parameters of the GPFA model using the EM algorithm, as detailed in the appendix.
Using these parameters, extract neural trajectories E [X|Y] (Eq. A6) from the observed activity Y.
Apply the orthonormalization procedure described in methods to the neural trajectories. This step is critical for visualization because it orders the dimensions of the low-dimensional trajectory by the amount of data covariance explained.
Plot each dimension of the orthonormalized neural trajectory versus time, as in Fig. 7. These time courses should be inspected for qualitative agreement with prior analyses of the same or related data sets. For example, one may expect firing rates, and thus the neural state, to change shortly after stimulus presentation. A rough estimate of the data dimensionality can be obtained by counting the number of orthonormalized dimensions showing time-varying structure; the data dimensionality can be formally computed using the leave-neuron-out prediction error described in methods.
Plot the top three (or any three) dimensions of the orthonormalized neural trajectories in a three-dimensional state space, as in Fig. 8.
Taken together, we consider steps 2 through 7 to be part of the GPFA framework for extracting and visualizing neural trajectories. Step 1 is necessary “best practices” when asking scientific questions about electrode array data.
For visualization in three dimensions, Fig. 5 shows that it is still better to fit a GPFA model with a large number of state dimensions (in this case, p = 15) and take the top three orthonormalized dimensions, rather than to fit a GPFA model directly with p = 3. This allows the neural trajectories to make use of a large number of timescales, rather than just three timescales. Although such a visualization is intuitively appealing, it is able to show only three selected dimensions and thus may be missing important structure contained in the dimensions not plotted. This can be partially overcome by plotting different sets of three dimensions, but we are seeking better ways to visualize higher-dimensional trajectories.
The ability of the methods developed here to concisely summarize the neural events on a single trial offers a powerful tool for studying the time course of neural population activity. We intend to apply these methods to data recorded during other behavioral tasks and in other brain areas, as schematized in Fig. 1. This is enabled by the development and increasing adoption of large-scale neural recording technologies, including multielectrode arrays and optical imaging techniques. Such analyses should provide insights into the neural mechanisms underlying cognitive processes (such as perception, decision making, attention, and motor planning), which are not directly yoked to observable quantities in the outside world and whose time course may differ substantially from trial to trial. More generally, these methods can be applied in experimental settings with no trial structure, such as in freely behaving animals (Chestek et al. 2009; Eliades and Wang 2008; Jackson et al. 2007; Santhanam et al. 2007). In such settings, traditional data analysis methods relying on trial-averaging are not applicable. Instead, if large-scale neural recordings are available, the methods presented here can be applied to track the subject's instantaneous neural state during a recording session.15 Another potential application of the developed methods is in studies of learning. Whereas analyzing the activity of single neurons can detect the presence of learning in neural activity, it is often unclear how the activity across a neural population is changing during the learning process and why such changes might be advantageous. By tracking the subject's instantaneous neural state using the methods developed here, we may be able to further our understanding of the neural mechanisms underlying learning.
Several extensions to the GPFA methodology can be envisaged. It may be possible to 1) couple the covariance structure of the one-dimensional GPs, which would provide for a richer description of the multidimensional neural state x:,t evolving over time; 2) apply nonstationary GP covariances, since the neural activity can be nonstationary; 3) allow for nonlinear relationships between the low- and high-dimensional spaces; and 4) incorporate point-process likelihood models (Truccolo et al. 2005) with appropriate stimulus and spike history dependence.
GRANTS
This work was supported by National Institute of Neurological Disorders and Stroke/Collaborative Research in Computational Neuroscience Grant 5-R01-NS-054283, National Defense Science and Engineering Graduate Fellowships, National Science Foundation (NSF) Graduate Research Fellowships, Gatsby Charitable Foundation, Michael Flynn Stanford Graduate Fellowship, Christopher and Dana Reeve Foundation, Burroughs Wellcome Fund Career Award in the Biomedical Sciences, Stanford Center for Integrated Systems, NSF Center for Neuromorphic Systems Engineering at Caltech, Office of Naval Research, Sloan Foundation, and Whitaker Foundation.
Acknowledgments
We thank S. Eisensee for administrative assistance, M. Howard for surgical assistance and veterinary care, D. Haven for computing support, and Dr. Mark Churchland for experimental guidance and discussions regarding analyses.
APPENDIX
GPFA model fitting
This section details how the parameters of the GPFA model are fit using the EM algorithm, as well as the associated computational requirements.
E-STEP.
The E-step computes the relative probabilities P(X|Y) of all possible neural trajectories X given the observed activity Y, using the most recent parameter estimates. We will first find the joint distribution of X and Y, which is Gaussian by definition. The desired conditional distribution P(X|Y) is therefore also Gaussian and can then be obtained using the basic result of conditioning for jointly Gaussian random variables.
Equations 1 and 2 can be reexpressed as
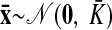 |
(A1) |
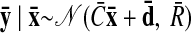 |
(A2) |
where x̄ = [x′:,1 x′:,T]′ ∈ ℝpT×1 is a concatenation of the columns of X, and ȳ = [y′:,1 y′:,T]′ ∈ ℝqT×1 is a concatenation of the columns of Y. The block diagonal matrices C̄ ∈ ℝqT×pT and R̄ ∈ ℝqT×qT comprise T blocks of C and R, respectively. The vector d̄ ∈ ℝqT×1 is a concatenation of T copies of d. The covariance matrix
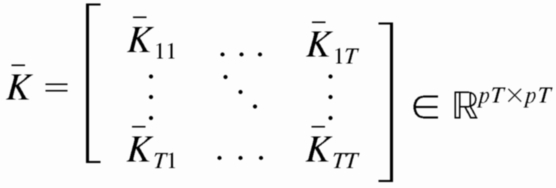 |
(A3) |
comprises blocks K̄t1t2 = diag {K1(t1, t2), …, Kp(t1, t2)} ∈ ℝp×p, where the diag operator returns a diagonal matrix whose nonzero elements are given by its arguments, K̄i(t1, t2) is defined in Eq. 3, and t1, t2 = 1, …, T. One can interpret K̄t1t2 as the covariance of the neural states at time points t1 and t2. From Eqs. A1 and A2, the joint distribution of x̄ and ȳ can be written
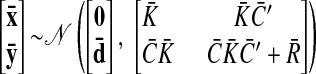 |
(A4) |
Using the basic result of conditioning for jointly Gaussian random variables16
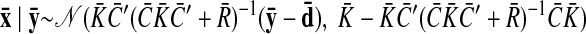 |
(A5) |
Thus, the extracted neural trajectory is
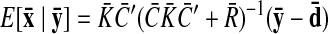 |
(A6) |
From Eq. A4, the data likelihood P(Y) can be easily computed since
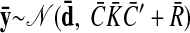 |
(A7) |
M-STEP.
The M-step involves maximizing ℰ(θ) = E [log P(X, Y|θ)] with respect to the parameters θ = {C, d, R, τ1, …, τp}. The expectation in ℰ(θ) is taken with respect to the distribution P(X|Y) found in the E-step, given in Eq. A5. Although this is a joint optimization with respect to all parameters in θ, it turns out that their optimal values are dependent on only a few or none of the other parameters, as shown in the following text. For clarity, we first define the following notation
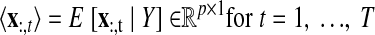 |
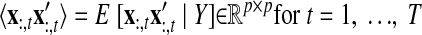 |
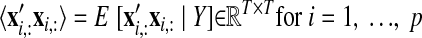 |
where these expectations can be obtained from Eq. A5.
Maximizing ℰ(θ) with respect to C and d yields
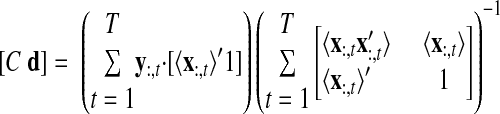 |
(A8) |
which does not depend on any of the other parameters. The update for R is
 |
(A9) |
where the diag operator zeros all off-diagonal elements of its argument. The new values of C and d found in Eq. A8 should be used in Eq. A9. Note that the updates for C, d, and R have the same analytic form as for FA, except that the sums here are taken over different time points rather than different data points in the case of FA.
Although there is no analytic form for the timescale updates, they can be obtained using any gradient optimization technique. The gradient of ℰ(θ) with respect to τi (i = 1, …, p) is
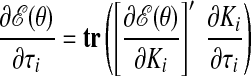 |
(A10) |
where
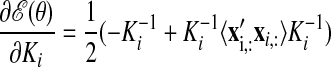 |
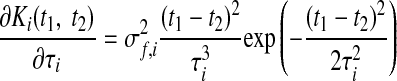 |
As in methods, Ki(t1, t2) denotes the (t1, t2)th entry of Ki and t1, t2 = 1, …, T. Note that Eq. A10 does not depend on the other p − 1 timescales, nor on the other model parameters. Thus, each of the timescales can be optimized individually. Because the timescales must be nonnegative, the optimization should be performed under the constraint that τi ≥ 0. This constrained optimization problem can be converted into an equivalent unconstrained optimization problem by optimizing with respect to log τi (which can be positive or negative) rather than τi using a change of variable.
The derivations in this section assume a single time series (corresponding to a single experimental trial) with T time points. We typically want to learn the model parameters θ based on multiple time series, each with a possibly different T. The preceding parameter update equations can be extended in a straightforward way to accommodate multiple time series. Instead of optimizing ℰ(θ) for a single time series, we optimize their sum ∑n ℰn(θ) across all time series indexed by n. Equations analogous to Eqs. A8–A10 can be derived by considering ∂[∑n ℰn(θ)]/∂θ rather than ∂ℰ(θ)/∂θ. This assumes that the time series are independent, given the model parameters. In other words, there is no constraint built into the model that similar neural trajectories should be obtained on different trials. However, the neural trajectories are assumed to lie within the same low-dimensional state space with the same timescales.
PARAMETER INITIALIZATION AND LOCAL OPTIMA.
Because EM is an iterative algorithm that is guaranteed to converge to a local optimum, the values at which the parameters are initialized are important. Recall that the neural trajectories extracted by GPFA can be viewed as a compromise between the low-dimensional FA projection of each data point and the desire to string them together using a smooth function over time. Under this view, we initialized the parameters C, d, and R using FA, which provides dimensionality reduction, but no smoothness over time. We then allowed GPFA to refine these estimates to obtain smooth neural trajectories. The degree of smoothness is defined by the timescales τi, which also need to be initialized. We fit the GPFA model starting at four different timescales: 50, 100, 150, and 200 ms. In each case, all p = 15 timescales were initialized to the same value. Figure A1 shows the resulting learned timescales for each initialization. Although the learned timescales were initialization dependent, their distributions were similar. In each case, there was one learned timescale around 525 ms, one or two around 300 ms, and the others in the range 40–180 ms. As indicated by the arrows in Fig. A1, the mean of the 15 learned timescales ranged from 125 to 155 ms. Furthermore, the resulting training data likelihoods, as well as the extracted orthonormalized neural trajectories, were very similar in the four cases (results not shown). Unless otherwise specified, all results in this work are based on initializing the timescales to 100 ms and running EM for 500 iterations. We also reran the analysis in Fig. A1 using 2,000 EM iterations to verify that there are true local optima in the space of timescales. Although other parameter initializations are possible (e.g., starting at random values with multiple restarts), we found that FA provided a sensible and effective initialization.
FIG. A1.
Learned GPFA timescales τi (i = 1, …, 15) after 500 EM iterations starting at 4 different initial values: 50, 100, 150, 200 ms. Arrows denote how the mean of the 15 timescales changed between their initial and learned values. These results are based on the same data used in Figs. 5–8. For the 100-ms initialization, each of the learned timescales corresponds to a different panel in Fig. 6.
Because we seek to extract smooth neural trajectories, we fixed the GP noise variances σn,i2 to a small value (10−3) for all results shown in this work. Larger values of σn,i2 generally yield neural trajectories that are less smooth. We also considered learning σn,i2 from the data, where each state dimension indexed by i can have a different GP noise variance. This involves finding the gradient of ℰ(θ) with respect to σn,i2 (similar to Eq. A10) and taking gradient steps in the joint space of σn,i2 and τi for each i during the M-step. If the σn,i2 are initialized to 10−3, their learned values (after 2,000 EM iterations) remain on the order of 10−3, yielding a similar training data likelihood and prediction error as when the σn,i2 are fixed to 10−3.
COMPUTATIONAL REQUIREMENTS.
This section details the computation time required to fit the GPFA model parameters and to extract neural trajectories. The computation time is a function of the state dimensionality p, the number of neurons q, the number of trials, the number of time steps T in each trial, and the number of EM iterations. Although fitting a GPFA model is generally computationally demanding, extracting a single neural trajectory can be very fast, as described in the following text.
Table A1 lists the computation time required for fitting a GPFA model for different state dimensionalities p and time bin widths. The values are based on q = 61 neurons, 56 trials, and 500 EM iterations. Because the absolute time duration of each trial is fixed, a larger time bin width means a smaller number of time steps T, whose range across the 56 trials is shown in Table A1. A naïve implementation scales as O(q3T3) due to the matrix inversion in Eq. A5. The matrix inversion lemma can be applied to reduce the computational load to O(p3T3). Overall, the most costly operations in fitting the GPFA model are the matrix inversion and multiplications in Eq. A5, as well as the iterative gradient optimization (Eq. A10) of the timescales.
TABLE A1.
Time required for fitting GPFA model
|
Time Bin Width |
||
|---|---|---|
| 20 ms | 50 ms | |
| p = 3 | 50 min | 8 min |
| p = 15 | 9 h | 45 min |
| T range | 47–71 | 19–28 |
Results were obtained on a 2006-era Linux (FC4) 64-bit workstation with 2–4 GB of RAM running MATLAB (R14sp3, BLAS ATLAS 3.2.1 on AMD processor).
There are several ways in which computation time can be reduced for the same state dimensionality p, number of neurons q, and number of trials. First, if different trials have the same T, the costly matrix inversion and multiplications in Eq. A5 can be reused. The computation time can be drastically reduced if all or many trials have the same T. In Table A1, many trials had different T and thus did not take full advantage of this savings. Second, depending on how crucial time resolution is, one might consider using a larger time bin width, thereby reducing T. Increasing the time bin width has the additional benefit that it increases the number of trials with the same T. As shown in Table A1, increasing the time bin width from 20 to 50 ms reduced the computation time from 9 h to 45 min for p = 15. Third, depending on how quickly the data likelihood Eq. A7 converges, one may need fewer (or more) than 500 EM iterations. The computation time scales linearly with the number of EM iterations. Fourth, approximate techniques can be applied to reduce computation time (Cunningham et al. 2008a; Teh et al. 2005). In this work, we perform all computations exactly, without the potential speedups of approximate techniques.
Once the GPFA model parameters are learned, extracting a single neural trajectory (Eq. A6) can be very fast, given the appropriate precomputation. In particular, the expensive matrix inversion and multiplications in Eq. A6 can be precomputed for each T. Depending on the values of p and T, the time required for this precomputation ranges from a few milliseconds to a few seconds for each T. Once the precomputation is finished, extracting a single neural trajectory takes 2.5 ms for p = 15 and T = 71 (the most computationally demanding trajectory in our data set), and less time for smaller values of p and T. It is readily possible to envision having single-trial, low-dimensional visualizations (as extracted by GPFA) appear during the inter-trial interval (<1 s) of behaving animal experiments using standard PC hardware and Matlab.
Computing prediction error
For GPFA, we first fit the model parameters θ = {C, d, R, τ1, …, τp} using the EM algorithm to the training data. We show here how to evaluate model goodness-of-fit by applying these learned parameters to data not used for model fitting. As described in methods, we seek to predict the activity of a neuron given the activity of all other (q − 1) recorded neurons. Let ȳj ∈ ℝT×1 be the activity of neuron j and ȳ−j ∈ ℝ(q−1)T×1 be the activity of the other (q − 1) neurons across all T time points, where j = 1, …, q. In other words, ȳj is equal to the transpose of the jth row of Y, whereas ȳ−j comprises all but the jth row of Y. The model prediction ŷj ∈ ℝT×1 for neuron j is defined as E [ȳj | ȳ−j]. Because ȳj and ȳ−j are jointly Gaussian by definition, the model prediction can be computed analytically.
We first define sparse binary matrices Bj ∈ {0, 1}T×qT and B−j ∈ {0, 1}(q−1)T×qT such that ȳj = Bjȳ and ȳ−j = B−jȳ. Multiplication by Bj and B−j can be viewed as picking out the elements in ȳ corresponding to neuron j and to all other neurons, respectively. Using Eq. A7
 |
(A11) |
where Σ = C̄K̄C̄′ + R̄ is introduced for notational clarity. Applying the basic result of conditioning for jointly Gaussian random variables
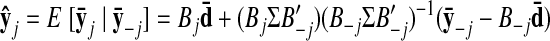 |
(A12) |
The prediction error is defined as the sum-of-squared differences between the model prediction and the observed square-rooted spike counts across all neurons and time points
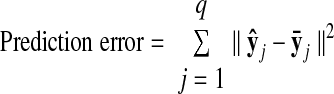 |
(A13) |
For the two-stage methods using PCA, PPCA, or FA, the model prediction is analogous to Eq. A12, but has a simpler form because these static dimensionality-reduction techniques have no concept of time. It is important to note that the training data and the data used to compute the model prediction must be presmoothed in the same way (e.g., using the same kernel) for the two-stage methods. However, when evaluating the prediction error, the model prediction must be compared to unsmoothed square-rooted spike counts, as in Eq. A13 for GPFA. Thus, for both the two-stage methods and GPFA, a smooth model prediction is compared to unsmoothed square-rooted spike counts.
To compute the model prediction for the reduced GPFA model, we cannot simply apply Eq. A12. Instead, we must consider an alternate approach to computing the model prediction via the orthonormalized state space. The basic idea is that a p-dimensional orthonormalized neural trajectory is first estimated using all but the jth neuron. Then, the activity of the jth neuron is predicted using the only the top p̃ dimensions (p̃ = 1, …, p) of the orthonormalized neural trajectory. The following equations formalize these concepts
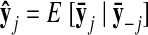 |
(A14) |
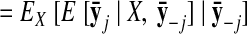 |
(A15) |
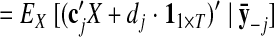 |
(A16) |
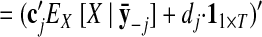 |
(A17) |
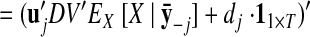 |
(A18) |
Equation A15 is obtained from Eq. A14 by conditioning on X. In Eq. A16, we use the fact that ȳj is independent of ȳ−j conditioned on X. Furthermore, c′j ∈ ℝ1×p is the jth row of C, dj ∈ ℝ is the jth element of d, and 11×T is a 1 × T vector of all ones. Equation A18 uses the singular value decomposition of C = UDV′, as described in methods. Note that u′j ∈ ℝ1×p is the jth row of U, and that DV′ EX [X | ȳ−j] is the orthonormalized neural trajectory estimated using all but the jth neuron.
Equation A18 is an alternate approach to computing the GPFA model prediction and yields the same result as Eq. A12. Equation A18 says that the model prediction for neuron j can be obtained by projecting the orthonormalized neural trajectory estimated using all but the jth neuron onto the jth axis in the high-dimensional space. Although Eq. A18 tends to be more computationally demanding than Eq. A12, it allows us to compute the model prediction for the reduced GPFA model. For the reduced GPFA model, Eq. A18 is computed using only the top p̃ elements of uj (since its elements are ordered due to orthonormalization) and setting all other elements of uj to zero.
Simulation with error floor
In Fig. 5, the benefit of GPFA over competing methods appears to be small (in percentage terms) if measured in terms of distance from zero prediction error. However, zero prediction error is unachievable due to the noise present in the data to be predicted. Thus, we conducted a simulation to determine the benefit of GPFA relative to a known error floor. This must be done in simulation, since the error floor is unknown for real neural data.
The simulated data were generated using a three-dimensional state space, where each state dimension evolved in time according to a sinusoid with a different frequency. These sinusoids were then linearly combined and mapped out into a 61-dimensional observation space according to Eq. 1. The independent noise was assumed to be isotropic and Gaussian across the 61 dimensions. We simulated 56 trials, each with 50 time steps. We then applied the two-stage methods and GPFA using four-fold cross-validation, as we did for the real neural data.
As shown in Fig. A2 (left column), all methods correctly indicated that the data are three-dimensional, since all curves reach their minimum at p = 3. Since the data were generated with isotropic noise, the results for two-stage methods using PPCA and FA were nearly identical. We then more densely sampled the kernel width for p = 3 to find the optimal smoothing kernel width, shown in Fig. A2 (middle column). Depending on the level of independent noise, we found that GPFA (black) yielded prediction errors that were tens of percent (A: 58.5%; B: 47.9%; C: 33.9%) lower than that of the best two-stage method (green dot), relative to the error floor (orange). The error floor was computed based on the level of activity of each neuron before noise was added, shown in Fig. A2 (right column, orange curves). The orange curves provide the theoretical limit for how well the leave-neuron-out model prediction can come to the observed data on average.
Footnotes
Although the concept of a threshold is well defined for a single neuron, it is unclear how it generalizes for a population of neurons. Is the decision made when any one of the neurons in the population reaches threshold (i.e., when the neural trajectory first hits a dotted line in Fig. 1B, bottom left)? Or is it the sum of the activity across the population that matters (i.e., when the neural trajectory first hits the thick gray bar in Fig. 1B, bottom left)? It may be possible to address such questions using the methods developed here.
Although the independent variability indeed feeds back into the network and can affect the aggregate network state, we assume that such effects are small.
PCA is the limiting case of PPCA as the noise variance goes to zero.
Our data sets include multiunit activity, comprising the activity of single neurons whose spike waveforms could not be discriminated through spike sorting. We consider a multiunit spike count to be the summed spike counts of its constituent single neurons. Because the sum of Poisson random variables is Poisson, we apply the square-root transform to both single units and multiunits in our data sets.
A colon in the subscript denotes all elements in a particular row or column. For example, x:,t specifies all elements in the tth column of X, whereas xi,: specifies all elements in the ith row of X.
This can be seen mathematically in Eq. A7, where K̄ defines the scale of X. The scale of Y depends on the product C̄K̄C̄′, not on K̄ and C̄ individually. Thus any scaling on K̄ can be compensated by appropriately scaling C̄.
The GP noise variance σn,i2 must be nonzero to ensure that Ki is invertible. If σn,i2 = 1, there is no correlation across time and GPFA becomes FA.
This orthonormalization procedure is also applicable to PPCA and FA. In fact, it is through this orthonormalization procedure that the principal directions found by PPCA are equated with those found by PCA. In general, the solutions found by PPCA and FA are unique up to an arbitrary rotation of the low-dimensional space. The orthonormalization procedure resolves this ambiguity.
Reach targets were not presented directly below the central target (i.e., direction: 270°) since they would be occluded by the monkey's hand while he is touching the central target.
All major trends in Fig. 5 were preserved without the square-root transform. We also considered smoothing spike trains directly (i.e., without binning) for the two-stage methods, which yielded results nearly identical to those of smoothing (non-square-rooted) spike counts. In the present work, the spikes are binned because this allows the square-root transform to be used, as described in methods.
For comparison, one may also consider computing the prediction error using the trial-averaged neural responses from the training data. However, trial-averaging is possible only if the experimental timing is identical on different trials. For the data being analyzed here, across-trial averaging is not possible because different trials have different delay periods, reaction times, and arm movement durations.
If computed improperly using only the top three orthonormalized dimensions rather than the top p* orthonormalized dimensions, the ratio per dimension would be 1.01. In other words, there is nearly no decrease in volume between the spread of the red dots and that of the green dots in the top three orthonormalized dimensions shown in Fig. 8.
The sinc function is defined as sinc(x) = sin(x)/x.
PCA, PPCA, FA, and GPFA all assume a linear relationship between the low-dimensional state space and the high-dimensional space of square-rooted spike counts. However, because the square-root transform is a nonlinear operation, the identified manifold is nonlinear in the original high-dimensional space of firing rates (or raw spike counts).
Due to computational considerations (cf. the appendix), it may be desirable to segment a recording session into multiple nonoverlapping intervals before applying GPFA. The methods presented here can then be applied unchanged, even though the segments are not multiple realizations of an experimental trial.
Since x̄ is obtained by reshaping X, they contain the same elements. The same is true for ȳ and Y. Thus P(x̄|ȳ) is equivalent to P(X|Y).
REFERENCES
- Abeles M, Bergman H, Gat I, Meilijson I, Seidemann E, Tishby N, Vaadia E. Cortical activity flips among quasi-stationary states. Proc Natl Acad Sci USA 92: 8616–8620, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksay E, Major G, Goldman MS, Baker R, Seung HS, Tank DW. History dependence of rate covariation between neurons during persistent activity in an oculomotor integrator. Cereb Cortex 13: 1173–1184, 2003. [DOI] [PubMed] [Google Scholar]
- Arieli A, Sterkin A, Grinvald A, Aertsen A. Dynamics of ongoing activity: explanation of the large variability in evoked cortical responses. Science 273: 1868–1871, 1996. [DOI] [PubMed] [Google Scholar]
- Bathellier B, Buhl DL, Accolla R, Carleton A. Dynamic ensemble odor coding in the mammalian olfactory bulb: sensory information at different timescales. Neuron 57: 586–598, 2008. [DOI] [PubMed] [Google Scholar]
- Batista AP, Santhanam G, Yu BM, Ryu SI, Afshar A, Shenoy KV. Reference frames for reach planning in macaque dorsal premotor cortex. J Neurophysiol 98: 966–983, 2007. [DOI] [PubMed] [Google Scholar]
- Beal MJ, Ghahramani Z, Rasmussen CE. The infinite hidden Markov model. In: Advances in Neural Information Processing Systems, edited by Dietterich TG, Becker S, Ghahramani Z. Cambridge, MA: MIT Press, 2002, vol. 14, p. 577–585.
- Bisley JW, Goldberg ME. Neuronal activity in the lateral intraparietal area and spatial attention. Science 299: 81–86, 2003. [DOI] [PubMed] [Google Scholar]
- Bradley DC, Chang GC, Andersen RA. Encoding of three-dimensional structure-from-motion by primate area MT neurons. Nature 392: 714–717, 1998. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Abarbanel HDI, Kristan WB Jr. Optical imaging of neuronal populations during decision-making. Science 307: 896–901, 2005. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Abarbanel HDI, Kristan WB Jr. From crawling to cognition: analyzing the dynamical interactions among populations of neurons. Curr Opin Neurobiol 16: 135–144, 2006. [DOI] [PubMed] [Google Scholar]
- Broome BM, Jayaraman V, Laurent G. Encoding and decoding of overlapping odor sequences. Neuron 51: 467–482, 2006. [DOI] [PubMed] [Google Scholar]
- Brown EN, Kass RE, Mitra PP. Multiple neural spike train data analysis: state-of-the-art and future challenges. Nat Neurosci 7: 456–461, 2004. [DOI] [PubMed] [Google Scholar]
- Brown SL, Joseph J, Stopfer M. Encoding a temporally structured stimulus with a temporally structured neural representation. Nat Neurosci 8: 1568–1576, 2005. [DOI] [PubMed] [Google Scholar]
- Carrillo-Reid L, Tecuapetla F, Tapia D, Hernández-Cruz A, Galarraga E, Drucker-Colin R, Bargas J. Encoding network states by striatal cell assemblies. J Neurophysiol 99: 1435–1450, 2008. [DOI] [PubMed] [Google Scholar]
- Chestek CA, Gilja V, Nuyujukian P, Ryu SI, Kier RJ, Solzbacher F, Harrison RR, Shenoy KV. HermesC: RF low-power wireless neural recording system for freely moving primates. In: Proceedings of the IEEE Symposium on Circuits and Systems (ISCAS). Piscataway, NJ: IEEE, 2009, p. 1752–1755. [DOI] [PubMed]
- Churchland MM, Yu BM, Ryu SI, Santhanam G, Shenoy KV. Neural variability in premotor cortex provides a signature of motor preparation. J Neurosci 26: 3697–3712, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland MM, Yu BM, Sahani M, Shenoy KV. Techniques for extracting single-trial activity patterns from large-scale neural recordings. Curr Opin Neurobiol 17: 609–618, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek P, Kalaska JF. Neural correlates of reaching decisions in dorsal premotor cortex: specification of multiple direction choices and final selection of action. Neuron 45: 801–814, 2005. [DOI] [PubMed] [Google Scholar]
- Cook EP, Maunsell JHR. Dynamics of neuronal responses in macaque MT and VIP during motion detection. Nat Neurosci 5: 985–994, 2002. [DOI] [PubMed] [Google Scholar]
- Crammond D, Kalaska J. Prior information in motor and premotor cortex: activity during the delay period and effect on premovement activity. J Neurophysiol 84: 986–1005, 2000. [DOI] [PubMed] [Google Scholar]
- Cunningham JP, Shenoy KV, Sahani M. Fast Gaussian process methods for point process intensity estimation. In: Proceedings of the 25th International Conference on Machine Learning, edited by McCallum A, Roweis S. London: ICML, 2008a, vol. 307, p. 192–199.
- Cunningham JP, Yu BM, Shenoy KV, Sahani M. Inferring neural firing rates from spike trains using Gaussian processes. In: Advances in Neural Information Processing Systems, edited by Platt J, Koller D, Singer Y, Roweis S. Cambridge, MA: MIT Press, 2008b, vol. 20, p. 329–336.
- Czanner G, Eden UT, Wirth S, Yanike M, Suzuki WA, Brown EN. Analysis of between-trial and within-trial neural spiking dynamics. J Neurophysiol 99: 2672–2693, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danóczy M, Hahnloser R. Efficient estimation of hidden state dynamics from spike trains. In: Advances in Neural Information Processing Systems, edited by Weiss Y, Schölkopf B, Platt J. Cambridge, MA: MIT Press, 2006, vol. 18, p. 227–234.
- Dayan P, Abbott LF. Theoretical Neuroscience. Cambridge, MA: MIT Press, 2001.
- de Lafuente V, Romo R. Neuronal correlates of subjective sensory experience. Nat Neurosci 8: 1698–1703, 2005. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm (with discussion). J R Stat Soc Ser B 39: 1–38, 1977. [Google Scholar]
- DiMatteo I, Genovese CR, Kass RE. Bayesian curve-fitting with free-knot splines. Biometrika 88: 1055–1071, 2001. [Google Scholar]
- Dodd JV, Krug K, Cumming BG, Parker AJ. Perceptually bistable three-dimensional figures evoke high choice probabilities in cortical area MT. J Neurosci 21: 4809–4821, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliades SJ, Wang XQ. Chronic multi-electrode neural recording in free-roaming monkeys. J Neurosci Methods 172: 201–214, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BS An Introduction to Latent Variable Models. London: Chapman & Hall, 1984.
- Faisal AA, Selen LPJ, Wolpert DM. Noise in the nervous system. Nat Rev Neurosci 9: 292–303, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli S, Bisley JW, Roitman JD, Shadlen MN, Goldberg ME, Miller KD. One-dimensional dynamics of attention and decision making in LIP. Neuron 58: 15–25, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat I, Tishby N, Abeles M. Hidden Markov modelling of simultaneously recorded cells in the associative cortex of behaving monkeys. Network 8: 297–322, 1997. [Google Scholar]
- Hanes DP, Schall JD. Neural control of voluntary movement initiation. Science 274: 427–430, 1996. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York: Springer-Verlag, 2001.
- Horwitz GD, Newsome WT. Target selection for saccadic eye movements: prelude activity in the superior colliculus during a direction-discrimination task. J Neurophysiol 86: 2543–2558, 2001. [DOI] [PubMed] [Google Scholar]
- Jackson A, Mavoori J, Fetz EE. Correlations between the same motor cortex cells and arm muscles during a trained task, free behavior, and natural sleep in the macaque monkey. J Neurophysiol 97: 360–374, 2007. [DOI] [PubMed] [Google Scholar]
- Jones LM, Fontanini A, Sadacca BF, Miller P, Katz DB. Natural stimuli evoke dynamic sequences of states in sensory cortical ensembles. Proc Natl Acad Sci USA 104: 18772–18777, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemere C, Santhanam G, Yu BM, Afshar A, Ryu SI, Meng TH, Shenoy KV. Detecting neural state transitions using hidden Markov models for motor cortical prostheses. J Neurophysiol 100: 2441–2452, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JND, Denk W. Imaging in vivo: watching the brain in action. Nat Rev Neurosci 9: 195–205, 2008. [DOI] [PubMed] [Google Scholar]
- Kihlberg JK, Herson JH, Schotz WE. Square root transformation revisited. Appl Statist 21: 76–81, 1972. [Google Scholar]
- Kipke DR, Shain W, Buzsáki G, Fetz E, Henderson JM, Hetke JF, Schalk G. Advanced neurotechnologies for chronic neural interfaces: new horizons and clinical opportunities. J Neurosci 28: 11830–11838, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni JE, Paninski L. Common-input models for multiple neural spike-train data. Network 18: 375–407, 2007. [DOI] [PubMed] [Google Scholar]
- Lawrence N Probabilistic non-linear principal component analysis with Gaussian process latent variable models. J Mach Learn Res 6: 1783–1816, 2005. [Google Scholar]
- Lawrence ND, Moore AJ. The hierarchical Gaussian process latent variable model. In: Proceedings of the 24th Annual International Conference on Machine Learning (ICML 2007), edited by Ghahramani Z. East Sussex, UK: Omnipress, 2007, p. 481–488.
- Leopold DA, Logothetis NK. Activity changes in early visual cortex reflect monkeys' percepts during binocular rivalry. Nature 379: 549–553, 1996. [DOI] [PubMed] [Google Scholar]
- Levi R, Varona R, Arshavsky YI, Rabinovich MI, Selverston AI. The role of sensory network dynamics in generating a motor program. J Neurosci 25: 9807–9815, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay D Information Theory, Inference, and Learning Algorithms. Cambridge, UK: Cambridge Univ. Press, 2003.
- Mazor O, Laurent G. Transient dynamics versus fixed points in odor representations by locust antennal lobe projection neurons. Neuron 48: 661–673, 2005. [DOI] [PubMed] [Google Scholar]
- Nawrot M, Aertsen A, Rotter S. Single-trial estimation of neuronal firing rates: from single-neuron spike trains to population activity. J Neurosci Methods 94: 81–92, 1999. [DOI] [PubMed] [Google Scholar]
- Nicolelis MAL, Baccala LA, Lin RCS, Chapin JK. Sensorimotor encoding by synchronous neural ensemble activity at multiple levels of the somatosensory system. Science 268: 1353–1358, 1995. [DOI] [PubMed] [Google Scholar]
- Rasmussen CE, Williams CKI. Gaussian Processes for Machine Learning. Cambridge, MA: MIT Press, 2006.
- Riehle A, Requin J. Monkey primary motor and premotor cortex: single-cell activity related to prior information about direction and extent of an intended movement. J Neurophysiol 61: 534–549, 1989. [DOI] [PubMed] [Google Scholar]
- Roitman JD, Shadlen MN. Response of neurons in the lateral intraparietal area during a combined visual discrimination reaction time task. J Neurosci 22: 9475–9489, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roweis S, Ghahramani Z. A unifying review of linear Gaussian models. Neural Comput 11: 305–345, 1999. [DOI] [PubMed] [Google Scholar]
- Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science 290: 2323–2326, 2000. [DOI] [PubMed] [Google Scholar]
- Santhanam G, Linderman MD, Gilja V, Afshar A, Ryu SI, Meng TH, Shenoy KV. HermesB: a continuous neural recording system for freely behaving primates. IEEE Trans Biomed Eng 54: 2037–2050, 2007. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsuki N, Ikegaya Y. Metastability of active CA3 networks. J Neurosci 27: 517–528, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidemann E, Meilijson I, Abeles M, Bergman H, Vaadia E. Simultaneously recorded single units in the frontal cortex go through sequences of discrete and stable states in monkeys performing a delayed localization task. J Neurosci 16: 752–768, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Brown EN. Estimating a state-space model from point process observations. Neural Comput 15: 965–991, 2003. [DOI] [PubMed] [Google Scholar]
- Sollich P, Williams CKI. Using the equivalent kernel to understand Gaussian process regression. In: Advances in Neural Information Processing Systems, edited by Saul LK, Weiss Y, Bottou L. Cambridge, MA: MIT Press, 2005, vol. 17, p. 1313–1320.
- Stopfer M, Jayaraman V, Laurent G. Intensity versus identity coding in an olfactory system. Neuron 39: 991–1004, 2003. [DOI] [PubMed] [Google Scholar]
- Strang G Linear Algebra and Its Applications. Philadelphia, PA: Elsevier/Saunders, 1988.
- Teh YW, Roweis S. Automatic alignment of local representations. In: Advances in Neural Information Processing Systems, edited by Becker S, Thrun S, Obermayer K. Cambridge, MA: MIT Press, 2003, vol. 15, p. 841–848.
- Teh YW, Seeger M, Jordan MI. Semiparametric latent factor models. In: Proceedings of the Tenth International Workshop on Artificial Intelligence and Statistics (AISTATS), edited by Cowell RG, Ghahramani Z. Hackensack, NJ: Society for Artificial Intelligence and Statistics, 2005, p. 333–340.
- Tipping ME, Bishop CM. Probabilistic principal component analysis. J R Stat Soc Ser B 61: 611–622, 1999. [Google Scholar]
- Tolhurst DJ, Movshon JA, Dean AF. The statistical reliability of signals in single neurons in cat and monkey visual cortex. Vision Res 23: 775–785, 1983. [DOI] [PubMed] [Google Scholar]
- Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. J Neurophysiol 93: 1074–1089, 2005. [DOI] [PubMed] [Google Scholar]
- Turner RE, Sahani M. A maximum-likelihood interpretation for slow feature analysis. Neural Comput 19: 1022–1038, 2007. [DOI] [PubMed] [Google Scholar]
- Ventura V, Cai C, Kass RE. Trial-to-trial variability and its effect on time-varying dependency between two neurons. J Neurophysiol 94: 2928–2939, 2005. [DOI] [PubMed] [Google Scholar]
- Wang J, Fleet D, Hertzmann A. Gaussian process dynamical models. In: Advances in Neural Information Processing Systems, edited by Weiss Y, Schölkopf B, Platt J. Cambridge, MA: MIT Press, 2006, vol. 18, p. 1441–1448.
- Weber AP, Hahnloser RHR. Spike correlations in a songbird agree with a simple Markov population model. PLoS Comput Biol 3: 2520–2531, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Gao Y, Bienenstock E, Donoghue J, Black M. Bayesian population decoding of motor cortical activity using a Kalman filter. Neural Comput 18: 80–118, 2006. [DOI] [PubMed] [Google Scholar]
- Yu BM, Afshar A, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Extracting dynamical structure embedded in neural activity. In: Advances in Neural Information Processing Systems, edited by Weiss Y, Schölkopf B, Platt J. Cambridge, MA: MIT Press, 2006, vol. 18, p. 1545–1552.
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Gaussian process factor analysis for low-dimensional single-trial analysis of neural population activity. Soc Neurosci Abstr 319.9, 2008. [DOI] [PMC free article] [PubMed]
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. In: Advances in Neural Information Processing Systems, edited by Koller D, Schuurmans D, Bengio Y, Bottou L. Cambridge, MA: MIT Press, 2009, vol. 21, p. 1881–1888. [DOI] [PMC free article] [PubMed]