Abstract
How follicular helper T cells (TFH cells) differentiate to regulate B cell immunity is critical for effective protein vaccination. Here we define three transcription factor T-bet-expressing antigen-specific effector helper T cell subsets with distinguishable function, migratory properties and developmental programming in vivo. Expression of the transcriptional repressor Blimp-1 distinguished T zone ‘lymphoid’ effector helper T cells (CD62LhiCCR7hi) from CXCR5lo ‘emigrant’ effector helper T cells and CXCR5hi ‘resident’ TFH cells expressing the transcriptional repressor Bcl-6 (CD62LloCCR7lo). We then show by adoptive transfer and intact polyclonal responses that helper T cells with the highest specific binding of peptide-major histocompatibility complex class II and the most restricted T cell antigen receptor junctional diversity ‘preferentially’ developed into the antigen-specific effector TFH compartment. Our studies demonstrate a central function for differences in the binding strength of the T cell antigen receptor in the antigen-specific mechanisms that ‘program’ specialized effector TFH function in vivo.
Vaccination with protein induces high-affinity B cell memory for long-term protection against infectious disease. Antigen-specific helper T cells with many effector functions are the central cognate regulators of this B cell immune response in vivo1,2. Follicular helper T cells (TFH cells) have emerged as another lineage of helper T cells functionally subspecialized to regulate many facets of the development of effector and memory B cells3,4. Understanding the antigen-specific mechanisms that control the differentiation of effector TFH cells is critical for the design of future vaccines but remains poorly resolved in vivo.
The appropriate placement of antigen-specific TFH cells during immune responses is important for the cognate delivery of effector function to antigen-primed B cells5,6. Loss of the chemokine receptor CCR7 and expression of the chemokine receptor CXCR5 relocate effector TFH cells to the B cell areas of secondary lymphoid tissue7,8. In these locations, expression of the costimulatory molecules CD40L9, ICOS10,11, PD-1 (ref. 12) and OX40 (ref. 13) helps modify antigen-specific contact with B cells expressing peptide-major histocompatibility complex (MHC) class II (pMHCII). The cytokines interleukin 4 (IL-4; A001262), IL-10 (ref. 6) and interferon-γ (IFN-γ)12 can also serve an important regulatory function at this cognate interface to affect subsequent development and function of antigen-specific B cells. Early studies of the differentiation program of TFH cells indicated involvement of expression of the transcriptional repressor Bcl-6 (A000369)13, local lymphoid priming14, expression of the ICOS ligand on B cells, IL-6, IL-21 (A001258), the transcription factor STAT3 (ref. 15), the strength of T cell antigen receptor (TCR) signaling through greater abundance of the Vavl guanine nucleotide-exchange factor16 and the duration of stable T cell-B cell contacts controlled by signaling lymphocytic activation molecule-associated proteins17. To delineate the cognate mechanisms of effector TFH differentiation, it remains necessary to define the dynamics of the development of antigen-specific effector TFH cells and distinguish those cells from the many other antigen-specific effector helper T cells that may arise after local protein vaccination.
The strength of TCR signaling affects lineage commitment during thymic development, and in the periphery, it is important for the establishment of selection thresholds for fitness and survival in vitro18,19 and clonal expansion after antigen priming in vivo20–22. It has also been shown that the strength of the pMHCII signal to the TCR influences helper T cell function differently both in vitro23 and in vivo with TCR-transgenic helper T cells24,25. Helper T cells with strong pMHCII binding ‘preferentially’ emerge in the draining lymph nodes and persist locally as a unique compartment of antigen-specific memory TFH cells14. However, it remains unclear whether the distribution of TCRs with substantial pMHCII binding strength is a consequence of local lymphoid priming or whether intrinsic TCR signal strength causally directs the effector TFH developmental program.
To address those issues, it is important to connect the functional development of antigen-specific helper T cells with TCR binding strength and repertoire diversity. The I-Ek-restricted helper T cell response of B10.BR mice to pigeon cytochrome c (PCC) is the only tractable protein vaccination model available at present with this capacity26,27. Antigen-specific helper T cells can be detected with antibodies to expressed variable (V) regions such as Vαll and Vβ3, as well as pMHCII tetramers presenting the dominant peptide epitope. Responding pMHCII-specific helper T cells express a restricted TCR repertoire with demonstrable clonal diversity26,27 and a range of TCR-binding properties28,29. In this model, antigen-specific effector helper T cells have many effector functions30 and changes in TCR triggering associated with the response to antigen31. After vaccination with protein, intrinsic TCR affinity thresholds regulate clonal selection and the composition of pMHCII-specific clones in the local draining lymphoid tissues28,29. Hence, it is critical to determine whether similar TCR affinity-based mechanisms exist for the regulation of the function of antigen-specific TFH cells.
Here we subcategorized the total antigen-specific helper T cell compartment into three subsets of CD44hiICOShi effector helper T cells. All antigen-specific subsets expressed similar amounts of IL-2, IL-10, IFN-γ and the transcription factor T-bet. Antigen-specific ‘lymphoid’ effector helper T cells expressing Blimp-1 (A003268) retained expression of the chemokine CD62L and CCR7, confining their effector activity to the T cell zones of responding lymph nodes. In contrast, most antigen-specific helper T cells that developed locally exited the lymph nodes as CD62LloCCR7lo ‘emigrant’ effector helper T cells. There were also many Bcl-6-expressing CXCR5hi TFH cells that remained ‘resident’ in lymph nodes with the highest expression of IL-4, IL-21, PD-1 and unique expression of CD69 and OX40 in the B cell zones to regulate antigen-primed B cells. Naive antigen-specific helper T cells with TCR of higher affinity ‘preferentially’ skewed into the CXCR5hl ‘resident’ TFH compartment. Increasing doses of agonist peptide over 3 d in vitro induced maximum IL-21 expression and lower Blimp-1 expression without affecting Bcl-6 expression. In the polyclonal model, TCRs with the strongest pMHCII binding and more restricted junctional diversity ‘preferentially’ skewed into the antigen-specific effector TFH compartment. Furthermore, vaccine adjuvants that promoted responders of higher affinity also induced the largest number of antigen-specific TFH cells in vivo. Thus, we demonstrate that antigen-specific mechanisms for ‘programming’ the differentiation of effector TFH cells in vivo are causally related to the strength of TCR binding and the diversity of the responding helper T cell population. Manipulating these mechanisms in vivo will underpin the formulation of the next generation of protein vaccines.
RESULTS
Three subsets of antigen-specific effector helper T cells
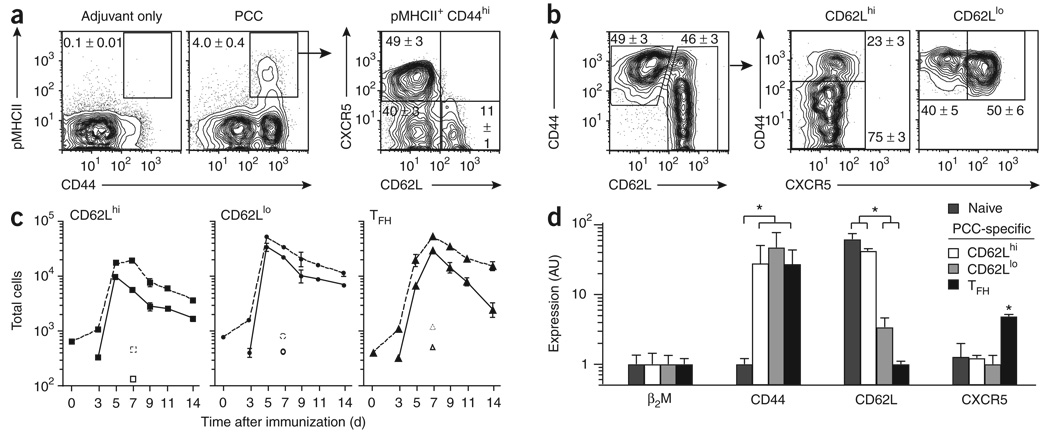
We immunized mice subcutaneously at the base of tail with whole protein antigen in the monophosphoryl lipid A-based adjuvant Ribi. Maximum accumulation of PCC-specific effector helper T cells in draining lymph nodes was reached by the end of the first week after subcutaneous vaccination of B10.BR mice with protein26. Using pMHCII binding (Fig. 1a) and upregulation of CD44 on Vα11+Vβ3+ helper T cells (Fig. 1b), we separated PCC-specific helper T cells into three distinct subsets on the basis of their expression of CXCR5 and CD62L. Two CXCR5lo subsets reached peak numbers by day 5 (Fig. 1c) and could be distinguished by their abundance of CD62L protein and mRNA (Fig. 1d). CXCR5 expression was present in the third major subset of antigen-specific helper T cells that had lost CD62L expression and reached peak numbers by day 7. Quantitative evaluation of mRNA expression in vivo correlated well with protein expression for CD44, CD62L and CXCR5 (Fig. 1d). Thus, after protein vaccination, CXCR5hi TFH cells constitute only half of the antigen-specific helper T cells that accumulate in the draining lymph node.
Figure 1. Three subsets of antigen-specific effector helper T cells emerge in draining lymph nodes.
(a,b) Expression of CD62L and CXCR5 (a) or CD44 and CXCR5 (b) on the surface of cells from draining lymph nodes (inguinal and periaortic) of B10.BR mice (n ≥ 6) 7 d after priming with PCC (a,b) or adjuvant only (a). (a) PCC-specific effector helper T cells (propidium iodide-negative (PI−), B220−CD8−CD11b−Vα11+pMHCII+CD44hi). (b) Helper T cells (PI−B220−CD8−CD11b−Vα11+Vβ3+, and either CD62Lhi or CD62Llo). Numbers adjacent to outlined areas or in quadrants indicate percent cells in each (mean ± s.e.m.). (c) Total PCC-specific Vα11+Vβ3+CD44hi (dashed lines) or Vα11+pMHCII+CD44hi (solid lines) effector helper T cells over time in draining lymph nodes after PCC priming (n ≥ 3 mice for each time point). Open symbols indicate estimate of total cells from B10.BR mice 7 d after adjuvant-only priming. (d) Expression of mRNA immediately after the isolation of 2 × 103 naive helper T cells (Vα11+Vβ3+CD44loCD62Lhi) or PCC-specific helper T cells (Vα11+Vβ3+CD44hi) from lymph nodes on day 7 after subcutaneous priming (n = 3 mice). Estimates are presented in arbitrary units (AU) relative to β2-microglobulin mRNA (β2M), set as 1. PCC-specific: CD62Lhi, CD62LhiCXCR5lo; CD62Llo, CD62LloCXCR5lo; TFH, CD62LloCXCR5hi. *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of at least three experiments.
Blimp-1-expressing ‘lymphoid’ effector helper T cells
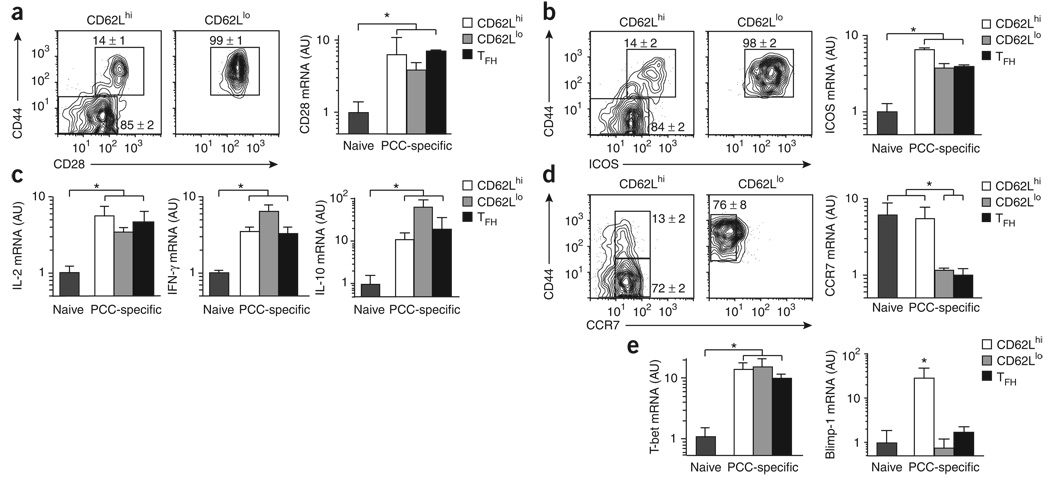
All three antigen-specific helper T cells had the qualities of effector cells in vivo by day 7. All subsets had higher in vivo surface expression of CD28 protein and mRNA (Fig. 2a) and ICOS protein and mRNA (Fig. 2b). There was much higher expression of IL-2, IFN-γ and IL-10 mRNA without in vitro restimulation (Fig. 2c), which indicated similar effector cell potential for all subsets in vivo. The CD62Lhi subset also retained expression of CCR7 similar to that of naive helper T cells (Fig. 2d), which indicated a capacity for lymphoid recirculation and placement in the T cell zones, even during an active immune response. As for the transcriptional regulation of effector cell differentiation, all subsets had similarly higher expression of T-bet than that of naive helper T cells, but only the CD62LhiCXCR7hi subset expressed Blimp-1 at this stage of the immune response (Fig. 2e). On the basis of the cell surface phenotype, we refer to the Blimp-1-expressing subset as ‘lymphoid’ effector helper T cells.
Figure 2. Lymphoid effector helper T cells express Blimp-1.
(a,b) Left, expression of CD44 and either CD28 (a) or ICOS (b) on the surface of cells (PI−CD8−CD11b−B220−Vα11+Vβ3+, and either CD62Lhi or CD62Llo), as described in Figure 1b from B10.BR mice on day 7 after PCC priming (n ≥ 4 mice). Numbers adjacent to outlined areas indicate percent cells in each (mean ± s.e.m.). Far right, expression CD28 mRNA (a) or ICOS mRNA (b) immediately after isolation of 2 × 103 naive helper T cells or PCC-specific helper T cell subsets (as described in Fig. 1d) from lymph nodes on day 7 after priming. (c) Expression of IL-2, IFN-γ and IL-10 mRNA, analyzed as described in Figure 1d. (d) Expression of CD44 and CCR7, analyzed as described in Figure 1d. (e) Expression of T-bet and Blimp-1 mRNA, analyzed as described in Figure 1d. Estimates for mRNA are presented in arbitrary units (mean and s.e.m.) relative to results obtained for naive cells, set as 1 (n = 3 mice). *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of three experiments.
Most helper T cells are ‘emigrant’ effector helper T cells
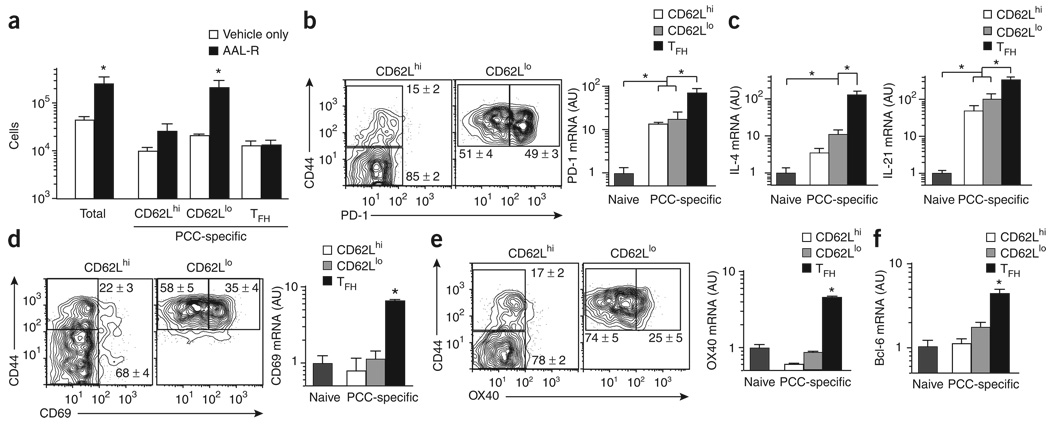
Sphingosine 1-phosphate receptor agonists such as FTY720 and its chiral analog AAL-R (2-amino-4-(4-heptyloxyphenyl)-2-methylbutanol) inhibit the migration of and promote the lymph node retention of lymphocytes32,33. We found many more antigen-specific helper T cells in draining lymph nodes after 7 d of treatment with AAL-R than after treatment with vehicle only (Fig. 3a and Supplementary Fig. 1 online). There were no changes in the number of antigen-specific TFH cells, which indicated the ‘resident’ nature of this effector helper T cell compartment. The ‘lymphoid’ effector helper T cells also remained mostly unchanged, which suggests that this T zone-localized effector compartment delivers its function locally during this acute phase of the immune response. In contrast, there were over ten times more CD62LloCCR7loCXCR5lo effector helper T cells after AAL-R treatment (Fig. 3a), which suggested that most of these effector helper T cells developed locally but exited the draining lymph node during this first week after priming. Furthermore, there were considerably fewer antigen-specific helper T cells in the spleen after AAL-R treatment, which could be accounted for mostly by the lower number of CD62LloCXCR5lo effector helper T cells than in untreated mice (Supplementary Fig. 2 online). Thus, we refer to this CD62LloCXCR5lo antigen-specific compartment as ‘emigrant’ effector helper T cells to emphasize their most distinguishing cellular attribute in vivo.
Figure 3. ‘Resident’ effector TFH cells express Bcl-6.
(a) Total PCC-specific effector helper T cells (Vα11+Vβ3+CD44hi) and PCC-specific effector helper T cell subsets (horizontal axis, as described for Fig. 1d key) from lymph nodes on day 7 after priming of mice (n = 3) treated with AAL-R or vehicle only (mean and s.e.m.). (b) Left, expression of CD44 and PD-1 on the surface of lymph node cells (CD8−CD11b−B220−PI−Vα11+Vβ3+ and either CD62Lhi or CD62Llo) from B10.BR mice on day 7 after PCC priming (n ≥ 4 mice). Far right, expression PD-1 mRNA immediately after isolation of 2 × 103 naive helper T cells or PCC-specific helper T cell subsets (as described in Fig. 1d) from lymph nodes on day 7 after priming, (c) Expression of IL-4 and IL-21 mRNA, analyzed as described in Figure 1d. (d,e) Expression of CD44 and either CD69 (d) or 0X40 (e) on the surface of lymph node cells (left), and CD69 mRNA (d, right) or 0X40 mRNA (e, right), analyzed as described in Figure 1d. (f) Expression of Bcl-6 mRNA, analyzed as described in Figure 1d. Numbers adjacent to outlined areas (b,d,e) indicate percent cells in each (mean ± s.e.m.). Results for mRNA (b,d,e, far right; c,f) are presented in arbitrary units (mean and s.e.m.) relative to results obtained for naive cells, set as 1 (n ≥ 3 mice). *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of at least three experiments.
Bcl-6-expressing ‘resident’ effector TFH cells
In addition to CXCR5, the negative regulator of T cell activation PD-1 has been reported to be present on a germinal center subset of TFH cells34. We found that similar fractions of CD44hiCD62Llo antigen-specific helper T cells and cells of the CXCR5+ TFH subset expressed PD-1 protein and mRNA (Fig. 3b). Although all three populations of effector helper T cells expressed more PD-1 mRNA than did naive helper T cells, the effector TFH compartment had significantly higher expression than did the other two subsets. Cytokines IL-4 and IL-21, which are also associated with the generation and function of effector TFH cells1–4, were expressed by all three effector helper T cell subsets, but the effector TFH compartment had significantly higher expression (Fig. 3c). In contrast, high expression of CD69 (Fig. 3d) and OX40 (Fig. 3e) ‘assorted’ uniquely into the effector TFH compartment. As for the transcriptional control of effector compartments, Bcl-6 was also uniquely expressed by the effector TFH compartment and was not present in the other two subsets at this stage of the acute response (Fig. 3f). Therefore, higher expression of PD-1, IL-4 and IL-21 and unique expression of CD69, OX40 and Bcl-6 distinguish the ‘resident’ effector TFH compartment that is ‘placed’ in the B cell zones of lymph nodes to deliver these activities to antigen-primed B cells.
High-affinity precursors develop into ‘resident’ TFH cells
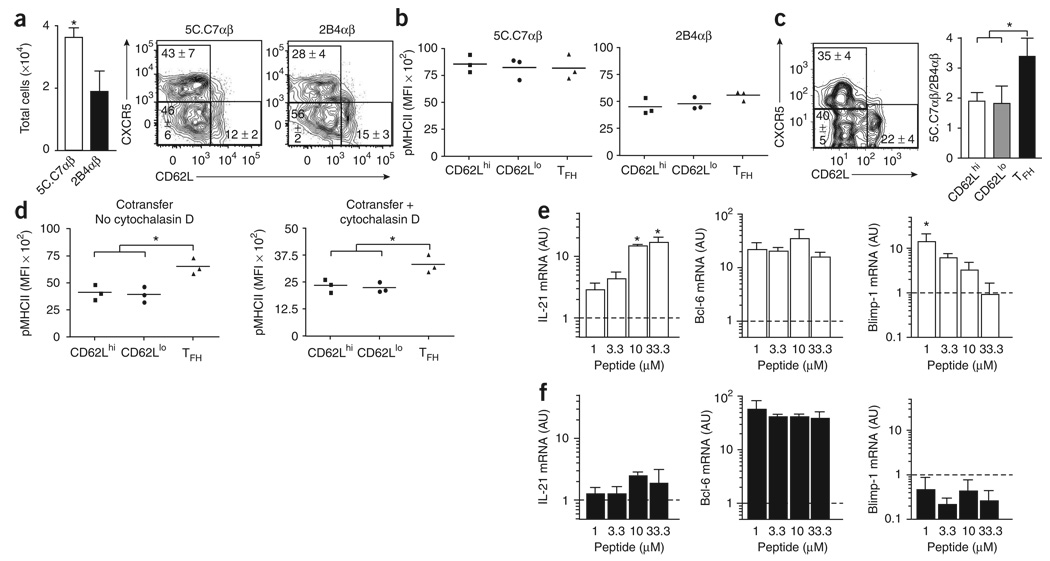
To begin to assess the involvement of TCR affinity in the regulation of effector TFH function, we used the 5C.C7 (higher affinity) and 2B4 (lower affinity) PCC-specific TCRαβ-transgenic helper T cell populations in adoptive-transfer experiments22,28. We transferred small numbers of CD90.2-expressing transgenic cells into otherwise replete unmanipulated recipient mice, which allowed the emergence of endogenous CD90.1-expressing PCC-specific responses (Supplementary Fig. 3 online). The frequency of recruited 5C.C7 helper T cells differed from that of the 2B4 cells with no indication of the differences in the dynamics of clonal expansion or contraction expected in these vaccination conditions28,29. Nevertheless, a significantly larger fraction of high-affinity responders than low-affinity responders developed into CXCR5+ ‘resident’ TFH cells (Fig. 4a). Even after 7 d of activation and effector cell differentiation in vivo, the differences in mean fluorescence intensity (MFI) for pMHCII tetramer binding correlated with the preimmune differences in TCR affinity for the helper T cell populations (Fig. 4b). All subsets had equivalent expression of Vα11 (data not shown). With both transgenic populations together in a cotransfer experimental design, significantly more higher affinity 5C.C7 helper T cells than lower affinity 2B4 helper T cells developed into CXCR5+ ‘resident’ TFH cells (Fig. 4c). These data show that monoclonal TCR-expressing helper T cells can develop into all three subsets, yet precursors with a high-affinity TCR ‘preferentially’ develop into CXCR5+ ‘resident’ TFH cells in vivo.
Figure 4. Precursors with high-affinity TCR ‘preferentially’ develop into ‘resident’ TFH cells.
(a) Left, total PCC-specific transgenic effector helper T cells (PI−B220−CD8−CD11b−Vβ3+CD90.2+CD44hi) in CD90.1 hosts given 1 × 104 syngeneic naive PCC-specific 2B4 or 5C.C7 TCRαβ-transgenic helper T splenocytes (CD4+Vα11+Vβ3+CD44loCD62Lhi) and then immunized with PCC in Ribi (mean and s.e.m.; n = 3 mice). Right, CXCR5 and CD62L staining on the surface of PCC-specific transgenic effector helper T cells. Numbers in quadrants indicate percent cells in each (mean ± s.e.m.; n = 3 mice), (b) Staining of pMHCII tetramers for each transgenic effector helper T cell subset in a (all CD90.1−Vα11+pMHC11+CD44hi; horizontal axis, as described for Fig. 1d key). Each symbol represents an individual mouse; small horizontal bars indicate the mean. (c) Left, CXCR5 and CD62L staining on the surface of PCC-specific transgenic effector helper T cells (PI−B220−CD8−CD11b−Vβ3+CD90.22+CD44hi) from CD90.1 hosts given a mixture of 1 × 104 syngeneic naive PCC-specific 2B4 and 5C.C7 TCRαβ-transgenic helper T splenocytes and then immunized with PCC in Ribi; cells are from lymph nodes obtained on day 7 after immunization. Numbers in quadrants indicate percent cells in each (mean ± s.e.m.; n = 3 mice). Right, single-cell RT-PCR analysis of the cell origin of 2B4αβ and 5C.C7β helper T cells among antigen-experienced PCC-specific transgenic helper T cells (all Vβ3+CD44hi CD90.2+; horizontal axis, as described for Fig. 1d key), assessed with Jβ2.5- and Jβ1.2-specific primers and presented as the ratio of total 5C.C7αβ-transgenic cells participating in the PCC response to 2B4αβ-transgenic cells in each effector helper T cell subset (mean and s.e.m.; n = 3). (d) Staining of pMHCII tetramers for each transgenic effector helper T cell subset (as described in b), assessed in the presence or absence of cytochalasin D. (e,f) Quantitative PCR analysis of IL-21, Bcl-6 and Blimp-1 mRNA from 5 × 104 naive PCC-specific, 5C.C7 (e) or 2B4 (f) TCRαβ-transgenic helper T splenocytes (CD4+Vα11+Vβ3+CD44loCD62Lhi) cultured for 3 d in vitro in the presence of 5 × 103 CD11c+ antigen-presenting cells loaded with various concentrations of MCC(88–103) (Peptide; horizontal axis). Results are presented in arbitrary units (mean and s.e.m.) relative to results obtained for naive cells (dashed lines), set as 1 (n = 3 mice for each peptide concentration). *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of three experiments.
In the cotransfer experiment, we detected more binding of pMHCII in the effector TFH compartment than in the other effector compartments, consistent with the higher ratio of 5C.C7 helper T cells to 2B4 helper T cells in the TFH compartment. We obtained similar results using cytochalasin D (Fig. 4d), which disrupts cytoskeletal activity needed for aggregation of TCRs at the cell surface. These results suggest that the greater intensity of pMHCII binding in the TFH compartment was due more to TCR repertoire differences than to physiological changes associated with effector cell differentiation in vivo. The endogenous PCC-specific response in this cotransfer experiment showed trends for pMHCII binding similar to those obtained with the transgenic helper T cells and also remained in the presence of cytochalasin D (Supplementary Fig. 4 online). Consistent with those findings, adoptive transfer of 5C.C7 TCRβ chain-transgenic helper T cells into unmanipulated mice followed by immunization with PCC also produced a CXCR5+ TFH compartment with the highest pMHCII tetramer binding of the three phenotypically defined helper T cell subsets that emerged (Supplementary Fig. 5 online). These data collectively indicate a causal connection between the TCR affinity of antigen-specific precursors and their differentiation into various effector helper T cell compartments.
Greater TCR signal strength induces IL-21 in vitro
We next addressed the function of TCR signal strength with peptide stimulation of transgenic helper T cells in vitro. All doses of moth cytochrome c (MCC) agonist peptide induced maximum clonal expansion of naive 5C.C7 helper T cells, as assessed by the resultant abundance of β2-microglobulin mRNA after the use of various stimuli (Supplementary Fig. 5a). In contrast, expression of IL-21 mRNA required tenfold higher concentrations of MCC peptide, with 10 µM MCC inducing maximum amounts of IL-21 in vitro (Fig. 4e). Furthermore, all peptide doses induced similar Bcl-6 expression, but Blimp-1 expression was inversely correlated with IL-21 induction. The lower affinity 2B4 clonotype induced less maximum clonal accumulation at all doses of MCC peptide (Supplementary Fig. 6a online) but failed to induce substantial expression of IL-21 mRNA above the baseline expression detected in vitro (Fig. 4f). Unlike the results obtained with 5C.C7 cells, expression of both Bcl-6 and Blimp-1 remained unchanged in 2B4 cells at all peptide doses tested (Fig. 4f). We used the peptide MCC(102S)22 to create a ligand of lower affinity for the 5C.C7 αβ TCR and again found less total clonal expansion at all peptide concentrations in vitro (Supplementary Fig. 6a). In these conditions, we noted higher IL-21 expression only at the highest peptide concentration; this also correlated with higher Bcl-6 expression. However, in these conditions, Blimp-1 expression was much higher at all doses of peptide (Supplementary Fig. 6b). Thus, it is clear that maximum IL-21 expression can be induced by stronger TCR signals in vitro with an effect on the relative balance in the expression of Bcl-6 and Blimp-1, even in the absence of B cells.
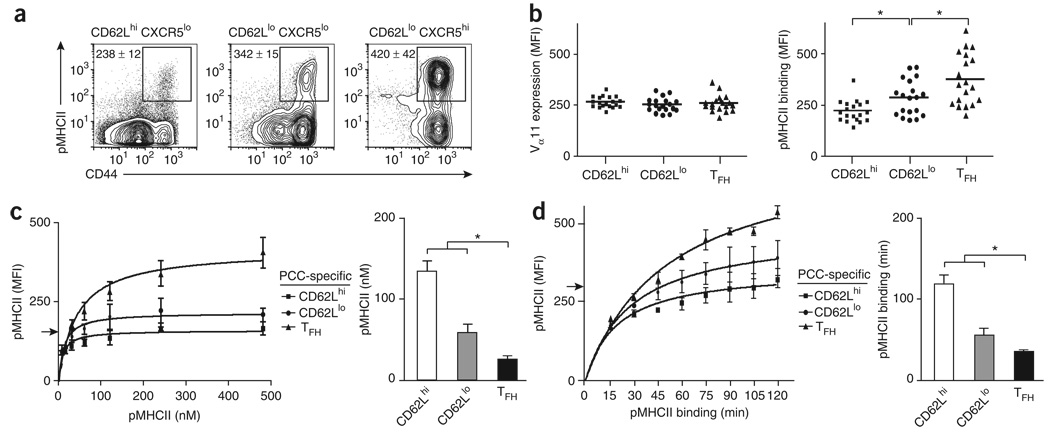
‘Resident’ TFH cells have stronger pMHCII binding
Although adoptive transfer provides direct access to antigen-specific precursors, it was important to address this issue in B10.BR mice with a polyclonal TCR repertoire in which precursor frequencies are more physiologically relevant. We detected considerable differences in pMHCII binding intensity for the three antigen-specific effector helper T cell compartments (Fig. 5a). All subsets expressed similar amounts of TCR, on the basis of Vα11 expression, but had progressively higher MFI values of pMHCII binding (Fig. 5b). ‘Resident’ TFH cells had the greatest pMHCII binding intensity for each mouse tested. At all concentrations of pMHCII, the ‘resident’ TFH compartment bound significantly more pMHCII complex than did the ‘lymphoid’ or ‘emigrant’ effector helper T cell compartment (Fig. 5c and Supplementary Fig. 7 online). Although the ‘emigrant’ effector helper T cells present at day 7 in draining lymph nodes represented a small fraction of this compartment (Fig. 3a and Supplementary Fig. 2), the splenic ‘effector’ helper T cells present from this time point on are considered part of the emigrant subset and have also been shown to have less pMHCII binding14. At optimal concentrations of pMHCII, the ‘resident’ TFH compartment bound more pMHCII more quickly than did the other effector compartments, reaching the maximum tetramer binding detected for the CD62Lhi subset in only 30 min (Fig. 5d and Supplementary Fig. 8 online). These trends in the polyclonal repertoire also indicate a skewing of antigen-specific precursors with stronger pMHCII binding of TCR into the ‘resident’ TFH compartment.
Figure 5. ‘Resident’ TFH cells have stronger pMHCII binding than do other effector helper T cells.
(a) Expression of pMHCII and CD44 on the surface of helper T cell subsets (CD8−CD11b−B220−prVα11+ and CD62LhiCXCR5lo, CD62LloCXCR5lo or CD62LloCXCR5hi) from lymph nodes on 7 d after PCC priming of mice. Numbers adjacent to outlined areas indicate MFI of each (mean ± s.e.m.; n ≥ 4 mice), (b) Vα11 expression and pMHCII tetramer staining for each PCC-specific effector helper T cell subset (all Vα11+pMHCII+CD44h; horizontal axis, as described for Fig. 1d key). Each symbol represents an individual mouse; small horizontal bars indicate the mean. *, P ≤ 0.01 (one-tail paired Student's t-test). (c,d) Left, pMHCII tetramer staining on the surface of PCC-specific helper T cell subsets (as in b; day 7 after priming) after staining with varying amounts of pMHCII tetramer (c) or with optimal pMHCII tetramer concentration but varying times of pMHCII tetramer binding (d). Right, pMHCII tetramer concentration required for each subset to reach an MFI of 150 (arrow at left; c) and time of pMHCII tetramer binding required for each subset to reach an MFI of 300 (arrow at left; d). Results presented as mean and s.e.m. (n = 3 mice for each condition). *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of at least three experiments.
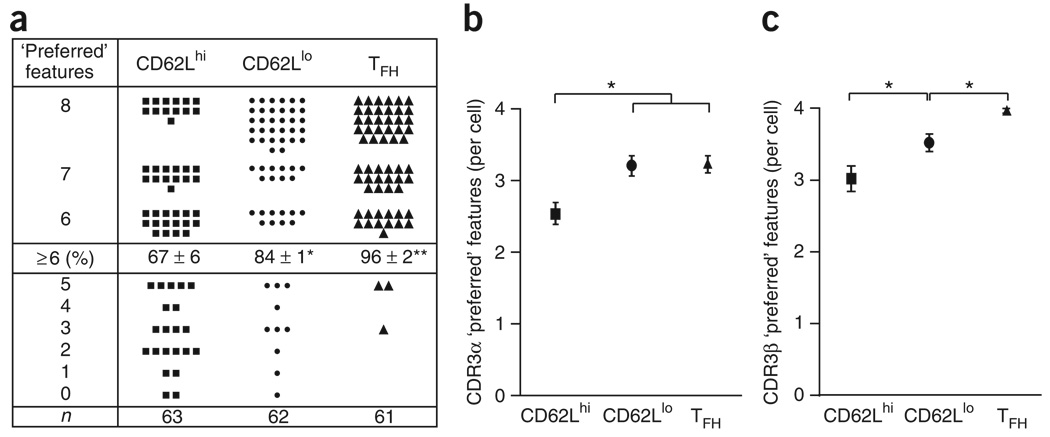
‘Resident’ TFH cells express a restricted TCR repertoire
As found in the adoptive-transfer experiments, we suspected that differences in pMHCII binding in vivo reflected differences in the expressed TCR. To test our idea more directly in the polyclonal model, we sorted single antigen-specific helper T cells from each effector helper T subset for TCR repertoire analysis. There are eight canonical features in the complementarity-determining region (CDR3) of PCC-specific TCR receptors, which for the TCRα chain consists of glutamic acid and serine at positions 93 and 95, respectively, a CDR3 length of eight amino acids and Jα16, Jα17, Jα22 or Jα34, and for TCRβ, asparagine at position 100 and alanine or glycine at position 102, a CDR3 length of nine amino acids and Jβl.2 or Jβ2.5. The presence of six or more of these features defines clonal dominance in this polyclonal TCR repertoire26,27. Most antigen-specific helper T cells in all functional subsets had six or more of these attributes (Fig. 6a). However, consistent with the pMHCII binding, there was a progressive increase in the proportion of restricted TCR use for the CD62LhiCXCR5lo ‘lymphoid’, CD62Llo CXCR5lo ‘emigrant’ and CD62LloCXCR5hi ‘resident’ effector helper T cell compartments. The differences in repertoire diversity were noticeable in the TCRα chain use of the lymphoid compartments (Fig. 6b and Supplementary Fig. 9 online) and with differences in the TCRβ chains for all three effector subsets (Fig. 6c and Supplementary Fig. 10 online). The TCR use of TFH cells resembled that of only 36% of the ‘emigrant’ helper T cells and 17% of the ‘lymphoid’ helper T cells in this response (Supplementary Fig. 11 online). Thus, the ‘resident’ TFH subset of antigen-specific effector cells expressed TCRs with stronger pMHCII binding and a much more restricted repertoire.
Figure 6. ‘Resident’ TFH cells express a more restricted TCR repertoire than do other effector helper T cells.
Single-cell repertoire analysis of individual PCC-specific effector helper T cells (Vα11+Vβ3+CD44hi) for each subset (as described in Fig. 1d), (a) Each filled symbol represents the number of the following ‘preferred’ CDR3 features known to be selected in the PCC response for a single cell: CDR3α, glutamic acid at α93, serine at α95, eight amino acids, Jα16, Jα17, Jaα22 and Jα34; CDR3β, asparagine at β100, alanine or glycine at β102, nine amino acids, Jβ1.2 and Jβ2.5. Middle row: percent cells (± s.e.m.) with six or more ‘preferred’ features, expressing a restricted TCR of the dominant clonotype (among n = number of single cells used in the analysis (bottom row); three individual mice). *, P ≤ 0.05, and **, P ≤ 0.01 (unpaired Student's t-test). (b,c) ‘Preferred’ CDR3 features for one single cell of each subset for the TCRα chain (b) or TCRβ chain (c), presented as the mean ± s.e.m. *, P ≤ 0.05 (unpaired Student's t-test). Data are representative of three experiments.
Vaccine adjuvants affect TFH development in vivo
To further address the mechanism underlying functional ‘programming’ in vivo, we sought ways to alter the responder clonal composition in the polyclonal vaccine model. Published studies have shown that decreasing the antigen dose over a two-log range does not affect clonal composition in this model28,29. Using a dose of protein that was tenfold higher or one-tenth as high, we found no effect on clonal expansion (Supplementary Fig. 12a online), overall pMHCII tetramer binding (Supplementary Fig. 12b) or the proportion or number of antigen-specific TFH cells (Supplementary Fig. 12c). With these very different doses of antigen stimuli, we also detected similar numbers of antigen-specific helper T cells from all three subsets (Supplementary Fig. 12d), which indicated there was no effect on functional differentiation.
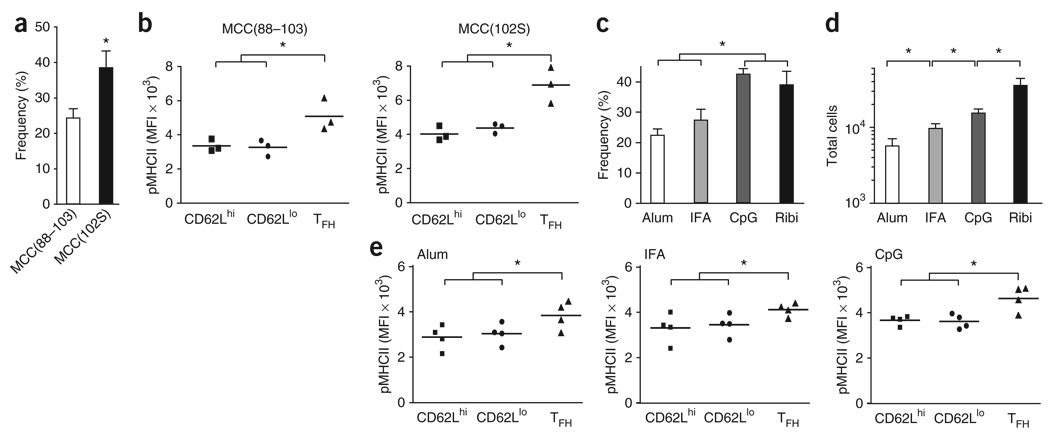
Next we used the agonist MCC peptide consisting of amino acids 88-103 (MCC(88-103)) and the altered peptide MCC(102S) as immunogens to vaccinate B10.BR mice. Vaccination with MCC(102S) induced substantial clonal expansion but less than that obtained with the MCC agonist peptide (Supplementary Fig. 13a online). However, the helper T cells selected into this response had an overall greater pMHCII binding intensity than that of the total responding population obtained by vaccination with the agonist MCC peptide (Supplementary Fig. 13b). These trends suggested the imposition of a higher selection threshold at the initiation of the immune response. In these conditions of more stringent clonal selection, there were significantly larger proportions of antigen-specific TFH cells with stronger pMHCII binding with MCC(102S) than with the agonist MCC peptide (Fig. 7a,b).
Figure 7. ‘Resident’ TFH cells ‘preferentially’ develop after priming with adjuvants that promote highaffinity antigen-specific helper T cells.
(a) Frequency of TFH cells (CD62LloCXCR5hi) among PCC-specific helper T cells (CD8−CD11b−B220−PI−CD4+Vα11+Vβ3+CD44hi) from lymph nodes on day 7 after priming with MCC(88–103) or MCC(102S) in Ribi (mean ± s.e.m.; n = 3 mice). *, P ≤ 0.05 (unpaired Student's t-test). (b) Staining of pMHCII tetramers for each PCC-specific effector helper T cell subset (as in Fig. 5b) from the mice in a. *, P ≤ 0.05 (one-tail paired Student's t-test). (c,d) Frequency of TFH cells among PCC-specific helper T cells (c) or total PCC-specific TFH cells (d) from lymph nodes on day 7 after priming with PCC in alum, IFA, CpG or Ribi (mean and s.e.m.; n = 4 mice). *, P ≤ 0.05 (unpaired Student's t-test). (e) Staining of pMHCII tetramers for each PCC-specific effector helper T cell subset (as in Fig. 5b) from lymph nodes on day 7 after priming with PCC in alum, IFA or CpG. *, P ≤ 0.05 (one-tail paired Student's t-test). Data are representative of at least three experiments.
Different vaccine adjuvants alter the selection threshold and clonal composition of antigen-responsive helper T cells29. Aluminum hydroxide (alum) and incomplete Freund's adjuvant (IFA) allowed clonotypes of lower affinity to emerge and persist, whereas the oligodinucleotide CpG adjuvant promoted a high-affinity clonal composition similar to that obtained with the monophosphoryl lipid A-based adjuvant Ribi (Supplementary Fig. 14a,b online). Here, we noted a significantly smaller proportion of antigen-specific TFH cells (Fig. 7c) and fewer total antigen-specific TFH cells (Fig. 7d) when we used alum or IFA than when we used CpG or Ribi. The TFH composition of the responding population correlated with the different effects of these adjuvants on clonal expansion in vivo. With each adjuvant, the antigen-specific TFH cell subset expressed TCRs with the strongest pMHCII binding relative to that of the other two antigen-specific effector compartments (Fig. 7e). These data further support the idea of a causal relationship between the strength of TCR signals delivered during clonal selection and the functional program subsequently adopted by the antigen-specific effector TFH cell compartment in vivo (Supplementary Fig. 15 online).
DISCUSSION
The capacity for lymphoid recirculation, regional ‘placement’ in lymphoid tissue and migratory activity can be used to categorize cells of the antigen-responsive effector helper T cell compartment. The CD62Lhi ‘lymphoid’ effector helper T cells retain the ability to reenter lymph nodes but remain CCR7hi with negligible exit from lymph nodes during the first week of priming. Hence, these cytokine-producing CD44hiICOShi effector cells may provide additional cognate help for antigen-primed B cells already in the extrafollicular regions2. These specialized effectors could also maintain35 or reestablish contact with pMHCII-expressing dendritic cells for their own survival and development36. Alternatively, this effector helper T subset may be needed to ‘license’ dendritic cells for help to CD8 T cells in cross-presentation functions or perhaps an extrafollicular function for helper T cells to promote isotype switching without somatic diversification37. As there are CD62Lhi memory helper T cells with this cell surface phenotype38, it is plausible that this ‘lymphoid’ effector program may also give rise to a recirculating lymphoid ‘central memory’ helper T cell compartment.
CXCR5hi effector helper T cells had all lost CCR7 expression, ensuring their ‘placement’ in the follicular B cell zones of lymphoid tissues7,8. These cells had lost CD62L expression, which indicated that if they left the lymph node, reentry would be a rare event. The results obtained with AAL-R treatment32,33 strongly supported the idea of the ‘resident’ status of these follicular helper T cells14. In contrast, the expression of ICOS, PD-1, IL-4, IL-10 and IL-21 associated with TFH function1,3,4 was not exclusive to the CXCR5hi effector TFH compartment. ICOS expression and the regulation of its amount in vivo11 are important for effective B cell immunity and are required for TFH maintenance in vivo. Furthermore, ICOS ligand10,15 expressed by B cells is required for TFH development and effective B cell immunity in vivo. Similarly, IL-21 is critical to TFH development and can act in an autocrine way in the generation of TFH cells and can costimulate initial TCR-triggering events16. The absence of IL-21 or its receptor on helper T cells leads to much lower B cell responses in vivo16. Our findings presented here are consistent with an early function for ICOS and IL-21 in effector helper T cell function that requires chemokine receptor ‘reprogramming’ for the delivery of cognate effector TFH functions to antigen-primed B cells.
The transcription factor T-bet is required for the differentiation of T helper type 1 cells39. In our study here, all subsets had equivalent expression of T-bet mRNA that would account for the presence of IFN-γ in all effector compartments. However, the cognate delivery of IFN-γ to different target cells would induce separate immune functions in vivo, such as immunoglobulin G2a isotype switching in antigen-primed B cells12,39. Such a pattern would suggest the presence of T helper type 1–like cells in the broad class of effector TFH cells. The presence of IL-4 in the antigen-specific TFH compartment supports the same idea, with evidence of the presence of T helper type 2 cells as well in the TFH compartment. Published data indicate that TFH cells are able to produce IL-17 (ref. 40). Such trends suggest that effector TFH cells have heterogeneous patterns of production of cytokines all known to be involved in the regulation of B cell immunity.
The larger amounts of transcriptional repressor Blimp-1 only in the ‘lymphoid’ effector compartment suggested different programs of development. Blimp-1 controls the plasma cell program of B cells41,42 and is involved in regulatory helper T cells43 and T helper type 1 differentiation44. Bcl-6 is also involved in antigen-driven B cell development and is required for germinal center formation45. Notably, Bcl-6 is a direct target for Blimp-1 repression, and the mutual repression of these two factors is important for maintaining the separate B cell fates44. In the CD4+ compartment, Blimp-1-deficient helper T cells have higher Bcl-6 expression46. Thus, Blimp-1 and Bcl-6 may regulate mutually exclusive ‘lymphoid’ effector helper T cell and ‘resident’ effector TFH cell developmental programs in the response to vaccination with protein.
TCR recognition of pMHCII complexes on antigen-presenting cells defines the first main checkpoint in the development of antigen-specific helper T cells in vivo2. Most models favor the idea that TCR affinity21,22,29,47 or the strength and duration48,49 of the binding of pMHCII to antigen-specific helper T cells controls these initial recognition events. Unlike CD8+ cells, the helper T cell compartment requires continued35 or multiple36 contacts with antigen-presenting cells for maximum clonal expansion in vivo. These sequential cognate interactions provide opportunities for the antigen-specific helper T cells to deliver effector functions and receive new developmental programming signals at later phases of the immune response. The costimulatory context of pMHCII expression and the cytokines produced by the antigen-presenting cell and by the responding helper T cells can affect the differentiation of helper T cells. IL-21 is important in the costimulation of antigen-specific TFH differentiation15,16. Hence, soluble and cell-associated microenvironmental factors also influence the differentiation of antigen-specific effector helper T cells during the acute phase of an immune response.
TFH cells represent a class of effector helper T cells responsible for the antigen-specific regulation of B cell immunity1,3,4. Published studies have further identified a function for IL-6 and STAT3 in the IL-21-dependent genetic program that initiates TFH development15. Notably, these studies have also demonstrated that expression of the ICOS ligand on B cells is needed to promote this specialized effector TFH compartment in vivo. Other studies have also indicated that ICOS expression on helper T cells is needed for the production of IL-4 and IL-21 but not for CXCR5 expression37. The costimulation of TCR signals has been identified as an important factor in IL-21 production, thereby promoting effector TFH development in vivo16. Our analyses here have added involvement of the strength of TCR signal itself in the early programming events involved in the development of antigen-specific TFH cells. It remains unclear whether the initial selection events between naive helper T cells and antigen-primed dendritic cells impose the critical selection events that affect the functional program. Alternatively, it may be the secondary encounter with pMHCII-expressing antigen-primed B cells that is critical for the ‘imprinting’ of effector TFH function, as suggested before17.
Vaccine adjuvants affect clonal-selection mechanisms, changing the distribution of the ‘preferred’ TCR affinity for responding clonotypes and altering the clonal composition of the effector helper T cell compartment. Our studies here indicate that the initial strength of TCR binding also regulates the function of antigen-specific effector helper T cells. TCR with stronger pMHCII binding ‘preferentially’ drive antigen-specific precursors into the ‘resident’ effector TFH compartment with the ability to control B cell immunity. Thus, manipulation of the clonal selection in helper T cells and regulation of the balance of effector helper T cell differentiation will provide a powerful new strategy for the future design of protein vaccines.
METHODS
Mice
B10.BR, B10.BR–Thy−1.1+ congenie and TCR-transgenic mice (2B4 TCRαβ, 5C.C7 TCRαβ, 5C.C7 TCRβ) were maintained in pathogen-free conditions at The Scripps Research Institute. Experiments were approved by the Institutional Animal Care and Use Committee and The Scripps Research Institute.
Immunization, adoptive transfer and AAL-R treatment
Mice were immunized subcutaneously with 400 µg whole PCC (Sigma), 60 µg MCC(88-103) (ANERADLIAYLKQATK; Anaspec) or 60 µg MCC(102S) (ANERADLIAYLK QASK; Anaspec) in Ribi (lab formulation based on ref. 29), IFA (Sigma), alum (Sigma) or CpG (ODN1826; Coley). For adoptive transfer, sorted naive Thy-1.2+ PCC-specific helper T splenocytes from 5C.C7 TCRαβ and 2B4 TCRαβ mice (1 × 104 of each) or total splenocytes from 5CC7β mice (2 × 106) were transferred intravenously into B10.BR-Thy-1.1 mice. Recipient mice were then immunized as described above. For AAL-R treatment, mice received intraperitoneal administration (1 mg AAL-R per kg body weight) daily from the day before PCC immunization to day 6 after immunization.
Flow cytometry
For population analysis, draining lymph nodes (inguinal and periaortic) were removed from immunized mice and were prepared and pelleted, then were resuspended in PBS with 5% (vol/vol) FCS and stained for flow cytometry at a density of 2.0 × 108 cells per ml. For tetramer staining, cells were first stained for 2 h at 20 °C with the optimal concentration of pMHCII tetramer (240 nM, prepared as described28). Where required, cytochalasin D was used at a final concentration of 50 µM (Sigma). Then, cells were incubated for 45 min at 4 °C with predetermined optimal concentrations of the following fluorophore-labeled (or biotin-labeled) monoclonal antibodies: fluorescein isothiocyanate-conjugated anti-Vα11 (RR8.1) or anti-Thy-1.2 (53-2.1), phycoerythrin-conjugated anti-CD4 (L3T4), allophycocyanin-conjugated anti-CD44 (Pgpl) or anti-Vβ3 (KJ25), indodicarbocyanine-phycoerythrin-conjugated anti-B220 (6B2) or anti-CD8 (53-6.7), and biotin-conjugated anti-CXCR5 (2G8) or anti-CD28 (37.51; all from BD Pharmingen); indodicarbocyanine-phycoerythrin– conjugated anti-Thy-1.1 (HIS51), indotricarbocyanine-phycoerythrin-conjugated anti-CD62L (Mell4), indotricarbocyanine-allophycocyanin-conjugated or allophycocyanin-Alexa Fluor 750-conjugated anti-CD44 (IM7), and biotin-conjugated anti-CCR7 (4B12), anti-CD69 (H1.2F3), anti-PD-1 (J43), anti-ICOS (7E.17G9) or anti-OX40 (OX86; all from eBioscience); and indodicarbocyanine-phycoerythrin-conjugated anti-CD11b (M1/70.15) or anti-CD4 (L3T4), and Pacific blue-conjugated anti-B220 (6B2; all from Biolegend). Streptavidin-phycoerythrin (BD Pharmingen) or streptavidin-indotricarbocyanine-allophycocyanin (BD Pharmingen) was used as a second-step visualization reagent. Cells were washed, were resuspended in propidium iodide (2 µg/ml in 5% (vol/vol) FCS in PBS) for exclusion of dead cells and were analyzed with a FACSVantage SE (BD Biosciences) with CellQuest or FACSDiva software (BD Biosciences). Data were analyzed with FlowJo software (Tree Star). Profiles are presented as 5% probability contours with outliers.
Single-cell repertoire analysis
Single cells with the appropriate surface phenotype were sorted for repertoire analysis with a FACSVantage SE and CloneCyt software (BD Biosciences). Synthesis of cDNA and amplification of the TCR Vα11 and Vβ3 regions were done as described26. The purified PCR products were sequenced directly with a Vα11-specific or Vβ3-specific primer and the BigDye Terminator cycle sequencing kit (Applied Biosystems). Sequences were analyzed on an ABI 373A DNA sequencer (Applied Biosystems).
Quantification of Jβ-gene-segment use
Single-cell cDNA was amplified with primers specific for the Vβ3 region and constant region, then 4% of the PCR product was used for Jβ-specific amplification with nested primers specific for the Vβ3 region and antisense primers specific for the Jβl.2 or Jβ2.5 gene segment as described29.
Quantitative PCR
Naive and PCC-specific helper T cells (2 × 103) were sorted by flow cytometry and were suspended in lysis buffer (Qiagen). RNA was purified and cDNA was generated as described14. Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and a RotorGene RG-3000 (Corbett Research) or StepOne Real-Time PCR system (Applied Biosystems) were used for PCR of 10% of the cDNA (primers, Supplementary Table 1 online). The results of quantitative PCR were analyzed with Rotor-Gene 6.0 software (Corbett Research) or StepOne software v2.0 (Applied Biosystems).
In vitro stimulation
Sorted naive transgenic helper T splenocytes were cultured for 3 d in RPMI medium supplemented with 10% (vol/vol) FCS, 1 mM sodium pyruvate, nonessential amino acids, 50 µM 2-mercaptoethanol (Gibco, Invitrogen) in the presence of 5 × 103 CD11c+ APCs loaded with MCC(88-103) or MCC(102S) at various concentrations.
Statistical analysis
Mean values, standard error of the mean and Student's t-test (paired and unpaired) were calculated with GraphPad Prism (GraphPad Software).
Supplementary Material
Note: Supplementary information is available on the Nature Immunology website.
ACKNOWLEDGMENTS
Supported by the National Institutes of Health (AI047231, AI040215 and AI059475 to M.G.M.-W.). This is manuscript 19762 from The Scripps Research Institute.
Footnotes
Accession codes
UCSD-Nature Signaling Gateway (http://www.signaling-gate way.org): A001262, A000369, A001258, and A003268.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Fazilleau N, McHeyzer-Williams LJ, McHeyzer-Williams MG. Local development of effector and memory T helper cells. Curr. Opin. Immunol. 2007;19:259–267. doi: 10.1016/j.coi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 2.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu. Rev. Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 3.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu. Rev. Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 4.Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 5.Breitfeld D, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim CH, et al. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 1999;190:1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell DJ, Kim CH, Butcher EC. Separable effector T cell populations specialized for B cell help or tissue inflammation. Nat. Immunol. 2001;2:876–881. doi: 10.1038/ni0901-876. [DOI] [PubMed] [Google Scholar]
- 9.Quezada SA, Jarvinen LZ, Lind EF, Noelle RJ. CD40/CD154 Interactions at the interface of tolerance and immunity. Annu. Rev. Immunol. 2004;22:307–328. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- 10.Mak TW, et al. Costimulation through the inducible costimulator ligand is essential for both T helper and B cell functions in T cell-dependent B cell responses. Nat. Immunol. 2003;4:765–772. doi: 10.1038/ni947. [DOI] [PubMed] [Google Scholar]
- 11.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 12.Snapper CM, Paul WE. Interferon-γ and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 13.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 14.Fazilleau N, et al. Lymphoid reservoirs of antigen-specific memory T helper cells. Nat. Immunol. 2007;8:753–761. doi: 10.1038/ni1472. [DOI] [PubMed] [Google Scholar]
- 15.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interlukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelzang A, et al. A fundamental role for iterleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal center formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nat. Immunol. 2003;4:355–360. doi: 10.1038/ni908. [DOI] [PubMed] [Google Scholar]
- 19.lezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T ells. Immunlty. 1998;8:89–95. doi: 10.1016/s1074-7613(00)80461-6. [DOI] [PubMed] [Google Scholar]
- 20.Fasso M, et al. T cell receptor (TCR)-mediated repertoire selection and loss of TCR Vb diversity during the initiation of a CD4+ T cell response in vivo. J. Exp. Med. 2000;192:1719–1730. doi: 10.1084/jem.192.12.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malherbe L, et al. Selective activation and expansion of high-affinity CD4+ T cells in resistant mice upon infection with Leishmania major. Immunity. 2000;13:771–782. doi: 10.1016/s1074-7613(00)00075-3. [DOI] [PubMed] [Google Scholar]
- 22.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 23.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blander JM, Sant'Angelo DB, Bottomly K, Janeway CA., Jr Alteration at a single amino acid residue in the T cell receptor α chain complementarity determining region 2 changes the differentiation of naive CD4 T cells in response to antigen from T helper cell type 1 (Th1) to Th2. J. Exp. Med. 2000;191:2065–2074. doi: 10.1084/jem.191.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Constant S, Pfeiffer C, Woodard A, Pasquallni T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J. Exp. Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHeyzer-Williams LJ, Panus JF, Mlkszta JA, McHeyzer-Williams MG. Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs. J. Exp. Med. 1999;189:1823–1838. doi: 10.1084/jem.189.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 28.Malherbe L, Hausl C, Teyton L, McHeyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunlty. 2004;21:669–679. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Malherbe L, Mark L, Fazilleau N, McHeyzer-Williams LJ, McHeyzer-Williams MG. Vaccine adjuvants alter TCR-based selection thresholds. Immunity. 2008;28:698–709. doi: 10.1016/j.immuni.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panus JF, McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific T helper cell function: differential cytokine expression in primary and memory responses. J. Exp. Med. 2000;192:1301–1316. doi: 10.1084/jem.192.9.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bikah G, Pogue-Caley RR, McHeyzer-Williams LJ, McHeyzer-Williams MG. Regulating T helper cell immunity through antigen responsiveness and calcium entry. Nat. Immunol. 2000;1:402–412. doi: 10.1038/80841. [DOI] [PubMed] [Google Scholar]
- 32.Mandala S, et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 33.Wei SH, et al. Sphingosine 1-phosphate type 1 receptor agonism inhibits transendothelial migration of medullary T cells to lymphatic sinuses. Nat. Immunol. 2005;6:1228–1235. doi: 10.1038/ni1269. [DOI] [PubMed] [Google Scholar]
- 34.Haynes NM, et al. Role of CXCR5 and CCR7 In follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated sub-population. J. Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 35.Obst R, van Santen HM, Mathis D, Benoist C. Antigen persistence is required throughout the expansion phase of a CD4+ T cell response. J. Exp. Med. 2005;201:1555–1565. doi: 10.1084/jem.20042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celli S, Garcia Z, Bousso P. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J. Exp. Med. 2005;202:1271–1278. doi: 10.1084/jem.20051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odegard JM, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 39.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 40.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and Th-17 cells. Nat. Immunol. 2008;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 42.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 43.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 44.Martins G, Calame K. Regulation and functions of Blimp-1 in Tand B lymphocytes. Annu. Rev. Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 45.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 46.Cimmino L, et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 47.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–545. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celli S, Lemaitre F, Bousso P. Real-time manipulation of T cell-dendritic cell interactions in vivo reveals the importance of prolonged contacts for CD4+ T cell activation. Immunity. 2007;27:625–634. doi: 10.1016/j.immuni.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Henrickson SE, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol. 2008;9:282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Immunology website.