Abstract
Summary: Whole-genome microarrays allow us to interrogate the entire transcriptome of a cell. Affymetrix microarrays are constructed using several probes that match to different regions of a gene and a summarization step reduces this complexity into a single value, representing the expression level of the gene or the expression level of an exon in the case of exon arrays. However, this simplification eliminates information that might be useful when focusing on specific genes of interest. To address these limitations, we present a software package for the R platform that allows detailed analysis of expression at the probe level. The package matches the probe sequences against a target gene sequence (either mRNA or DNA) and shows the expression levels of each probe along the gene. It also features functions to fit a linear regression based on several genetic models that enables study of the relationship between gene expression and genotype.
Availability and implementation: The software is implemented as a platform-independent R package available through the Bioconductor repository at http://www.bioconductor.org/. It is licensed as GPL 2.0.
Contact: lasse.folkersen@ki.se
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Expression arrays enable us to interrogate the transcription level of all known genes in a single hybridization experiment. A feature of the Affymetrix GeneChip platform is that several probes represent one gene, allowing a detailed investigation of the expression pattern along the gene sequence. However, this complexity can be detrimental when we want to focus on analyzing thousands of genes simultaneously. For that reason, traditional tools available for the analysis of expression microarrays are focused on a general approach, providing summarized values of all the probes in a probe set [e.g. RMA (Bolstad et al., 2003), MAS5 and PLIER]. Although this approach is necessary in many cases, it excludes information that can be critical for the correct interpretation of some experimental results. For example, when a non-trivial gene structure, perhaps with newly uncovered transcript variants, has rendered the established probe set structure obsolete. Individual probe sequences might also be rendered useless by updates to the reference sequence. In other cases polymorphisms in the gene sequence alter the apparent expression level of some probes. Finally, sometimes two or more probe sets map to the same gene, giving contradictory information that can be difficult to resolve using a summarizing approach. Accordingly, without further investigation, the biological relevance of these results is uncertain. Examples of these situations are provided in the Supplementary Material. To solve these complex situations, the best approach is to use all available data, and visualize the expression level and the location in the genome of individual probes. This is true for both traditional 3′ IVT arrays and for exon arrays. The information needed to perform this analysis is available (in the CEL files), but at present there is no easy way to visualize and analyze it.
Herein we present GeneRegionScan, a software package for the statistical platform R, which provides the means to extract and visualize information about individual probes in an automated fashion. The primary goal of the presented package is to facilitate the analysis of alternative splicing in the broadest sense. Since alternative splicing can be mediated by local SNPs (Kwan et al., 2008), a specific goal of the package is to assist in the analysis of the relationship between expression levels and SNP genotype. To exemplify its utility, we investigated the effect of genotype on the expression of a set of leukotriene pathway genes, which are of specific interest in cardiovascular disease.
2 DESCRIPTION
To demonstrate the package functionality, we obtained previously published data for 171 lymphoblastoid cell lines from 57 individuals from the HapMap CEU population (HapMap, 2003). Expression studies based on these data have been published (Kwan et al., 2008) using the Affymetrix Human Exon ST 1.0 arrays [available through GEO (Edgar, et al., 2002) accession number GSE9372]. In that work, Kwan and co-workers explored the association between genotypic differences and expression values for the entire genome. Leukotriene pathway genes are of interest because they are involved in the inflammatory response, which is a central part of the pathophysiology of cardiovascular disease. The relation between expression levels and a set of SNPs in the leukotriene pathway genes has recently been shown to be directly associated with ischemic stroke (Bevan et al., 2008). In this example, we combined the knowledge of genotype effects seen in the leukotriene cascade genes with the GSE9372 expression data and the HapMap genotype data for these genes, to perform an analysis with GeneRegionScan.
The ALOX5AP gene encodes for the five-lipoxygenase activating protein which, with 5-lipoxygenase, is required for leukotriene synthesis and is therefore a vital component of the inflammatory response. We investigated all the SNPs used in Bevan et al. with ALOX5AP and applied the same 0-1-2, 0-0-1 and 0-1-1 genetic models. 0-1-2 is a codominant model (three genotype groups per SNP separately) in which the heterozygote is valued as 1, and the homozygotes as 0 and 2, respectively. 0-0-1 and 0-1-1 are recessive and dominant models—essentially comparing groupings of heterozygote and homozygote samples with samples of the other homozygote type. A linear regression was fitted to the defined models to test the relation between specific genotype models and expression values. Further description of this algorithm can be found in the Supplementary Materials, as well as in the software documentation.
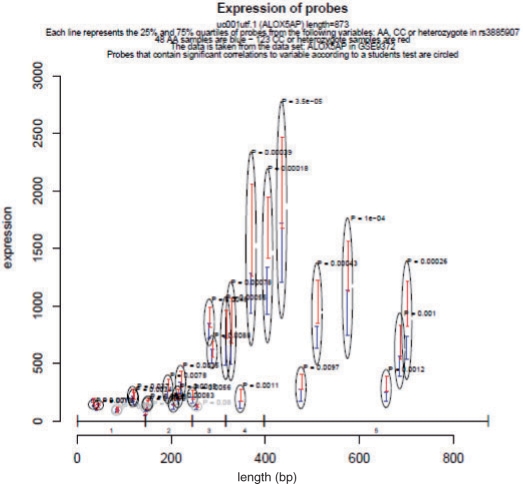
The most interesting result was SNP rs3885907, also referred to as FL10 by Bevan et al., which evidenced a highly significant expression change when comparing the risk allele AA samples with the heterozygote and the non-risk allele CC samples (Fig. 1). In addition, this SNP was also found to confer a 1.473-fold increased risk of ischemic stroke (Bevan et al., 2008). Figure 1 shows that: (i) possessing two copies of the risk allele A results in a decreased intensity for all probes across the entire ALOX5AP gene, and(ii) this effect is not mediated through alternative splicing, since all probes matching to the mRNA show the same trend. These results suggest that the SNP rs3885907 or a linked SNP confers a mechanism for controlling the expression level of the ALOX5AP transcript, which could have implications in the development of cardiovascular disease.
Fig. 1.
Effect of the rs3885907 SNP on the expression level of ALOX5AP. Visualization of the expression levels of all probes with sequences mapping to current ALOX5AP sequence. The data have been stratified by risk allele (AA) or non-risk allele (CC and heterozygote) of rs3885907. The y-axis indicates intensity values of each probe. The x-axis shows the length of the gene ALOX5AP, measured in base pair. Vertical bars show the span of the 25% and 75% quartiles of samples that are either AA (blue) or CC/heterozygote (red). Gray and black circles highlight probes, which have a significant relation to the genotype of the SNP. The significance of this relation has been calculated using a linear additive model as implemented in R. The exon structure of ALOX5AP is shown along the x-axis. It has been created using the exonStructure function based on sequence data from the UCSC genome browser. Expression data were taken from 171 samples in the Gene Expression Omnibus (GEO) dataset GSE9372. Genotype data were downloaded from the HapMap project.
3 CONCLUSION
Herein, we present a software package that enables fine-grained probe-level analysis on a gene-by-gene level. Individual probes are matched against the gene sequence, and the probe intensities are plotted. The per-probe approach gives a more versatile tool to investigate different transcript variants and discover all information available about differences across the length of each transcript. Package utility was demonstrated by analyzing data from lymphoblastoid cells, which evidenced an association between a SNP that is associated with a higher risk of myocardial infarction and the expression level of ALOX5AP.
ACKNOWLEDGEMENTS
Disclaimer: The report reflects only the author's views and the European Commission is not liable for any use that may be made of the information therein.
Funding: Swedish Research Council (grant #20854); Japanese Society for the Promotion of Science post-doctoral fellowship (to D.D.); Centre for Allergy Research fellowship (to C.E.W.); The Swedish Heart-Lung Foundation (to A.G.); European Commission FP6 (LSHM-CT-2004-005033).
Conflict of Interest: none declared.
REFERENCES
- Bevan S, et al. Genetic variation in members of the leukotriene biosynthesis pathway confer an increased risk of ischemic stroke: a replication study in two independent populations. Stroke. 2008;39:1109–1114. doi: 10.1161/STROKEAHA.107.491969. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Edgar R, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HapMap. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]