Abstract
jClust is a user-friendly application which provides access to a set of widely used clustering and clique finding algorithms. The toolbox allows a range of filtering procedures to be applied and is combined with an advanced implementation of the Medusa interactive visualization module. These implemented algorithms are k-Means, Affinity propagation, Bron–Kerbosch, MULIC, Restricted neighborhood search cluster algorithm, Markov clustering and Spectral clustering, while the supported filtering procedures are haircut, outside–inside, best neighbors and density control operations. The combination of a simple input file format, a set of clustering and filtering algorithms linked together with the visualization tool provides a powerful tool for data analysis and information extraction.
Availability: http://jclust.embl.de/
Contact: pavlopou@embl.de; rschneid@embl.de; skossida@bioacademy.gr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
There exists a big variety of clustering algorithms, which are applicable to a wide range of problems. Most of them are available either as source code, as part of a software package like in R or Matlab packages or are available online. Beside the commercially available ones, there are a few web-based or standalone tools like NeAT (Brohee et al., 2008), Cluster 3.0 software (de Hoon et al., 2004) or Cluto (Zhao and Karypis, 2005) which provide access to some of the clustering algorithms. Nevertheless, it requires typically some effort to either implement the source code into own projects, get familiar with a specific software package or prepare the data for a specifically needed input format. A major weakness of most of the currently available tools is that they lack the interactivity and an easy visualization module to explore and navigate through the data. Here, we present the toolbox jClust, which aims to bridge the gap between analysis and visualization by integrating clustering analysis algorithms with tools able to provide these results visually. The tool provides access to a widely used set of clustering algorithms and simultaneously allows the interactive visualization of the data. It reads from a very simple input file format and produces a human readable output file. jClust comes with a user-friendly GUI that makes the functionality and the parameterization of the algorithms easy and we believe that jClust gives the users, the opportunity to analyze and visualize biological data in a fast, easy and efficient way.
2 CLUSTERING
jClust supports a variety of supervised and unsupervised clustering analysis methods. These are k-Means (MacQueen, 1967), Spectral clustering (Paccanaro et al., 2006), Affinity propagation (Frey and Dueck, 2007), Restricted neighborhood search cluster algorithms—RNSC (King et al., 2004), Markov clustering—MCL (Enright et al., 2002), MULIC (Andreopoulos et al., 2007a, b) and Bron–Kerbosch (Coen and Joep, 1973). Concerning k-Means and the Spectral clustering, the number of clusters needs to be defined by the user. The k-Means (MacQueen, 1967) algorithm requires a full, all-against-all distance matrix to run whereas this is not a requirement for the other implemented algorithms. All of the algorithms besides k-Means are suitable for sparse graphs and all of the methods are able to analyze large-scale data as long as the local computer memory permits it. The Bron–Kerbosch (Coen and Joep, 1973) algorithms is a very well-known algorithm for finding cliques in a graph, meaning that it isolates strongly connected sub-areas where every node is connected to every other node—all-against-all connections—that belongs to the same clique. All of the aforementioned clustering algorithms assign nodes to only one unique cluster whereas the Bron–Kerbosch (Coen and Joep, 1973) algorithm allows a node to belong to more than one cluster.
3 FILTERING
jCluster gives to the user the opportunity to filter noise from the predicted clusters that have been calculated by one of the previous methods. This way, in a second step, clusters can be enriched by nodes that are important or shrink by removing nodes that should not belong to the cluster. Here, we implemented the following procedures: (i) density, (ii) haircut, (iii) best neighbor and (iv) cutting edge operation. The density method applies a threshold, which filters down clusters below a certain allowed density. The haircut operation detects and excludes vertices with a low degree of connectivity from the potential cluster. In contrast to the haircut operation method, the best neighbor method tends to detect and enrich the clusters with candidate vertices that are considered as good ‘neighbors’. The cutting edge operation filters out cases of densely connected sub-areas, which are only sparsely connected to the rest of the network. A detailed explanation of how these methodologies, are mathematically defined and how they can be parameterized is given online in the Supplementary Material.
4 VISUALIZATION
We updated the Medusa (Hooper and Bork, 2005) visualization tool to graphically represent the produced clusters. Medusa can be used as an external application or can alternatively be called through the jClust application. Medusa is now more interactive and supports many layout algorithms that make the tool much more informative and the extraction of the biological knowledge easier. In contrast to the previous version, users can isolate connections of specific nodes and hyperlink them to external data sources. A predefined clustering layout algorithm is implemented to distribute nodes in an efficient way to visualize distinct clusters. According to this layout, N centers, where N is the number of clusters produced, are initially calculated on a grid distribution and then nodes that belong to the same cluster are placed circularly around these centers. This way, users can very easily see and identify distinct groups of nodes, see patterns and visually evaluate the correctness of their analysis. Through the Medusa application, users can save the final results in other formats that are readable by external visualization tools.
5 FUNCTIONALITY
The input file is very simple. It only requires a list of weighted connections where the weight determines the importance of the connection. These files could contain, for example, protein–protein interaction data resulting from experiments or other data sources like protein–chemical interactions coming from the Stitch database (Kuhn et al., 2008) or experimentally calculated sets like yeast protein–protein datasets (Gavin et al., 2006). jClust provides a Java interface, which allows parameterization for any of the available algorithms and shows the final and intermediate results in the GUI jtext areas, which are simultaneously saved as text files. These files also keep the track about the information regarding the distinct clusters, the nodes that belong to them and the connections between the member and nodes of each cluster.
6 CONCLUDING REMARKS
We believe that the jClust toolbox provides a simple but yet powerful tool for researchers in the life science field as it integrates a very strong collection of lately implemented clustering algorithms with an easy to use visualization tool. jClust can be used to address various questions like classifying similar literature abstracts, identifying protein families according to their sequence or domain similarity or predicting protein complexes from protein–protein interaction data. The usefulness of the tool was already shown in a biological case study recently published (Moschopoulos et al., 2008). There, we show how the combination of clustering (in that case a RNSC and MCL) and filtering algorithms can be applied to protein–protein interaction data to predict protein complexes (see Figure 1). The newer version of the Medusa visualization application provides an enriched functionality and interactivity, which makes exploration of data and navigation easier. Further information about the algorithms, the filters, their parameters, some typical application examples and real biological datasets are offered online in the Supplementary Material section.
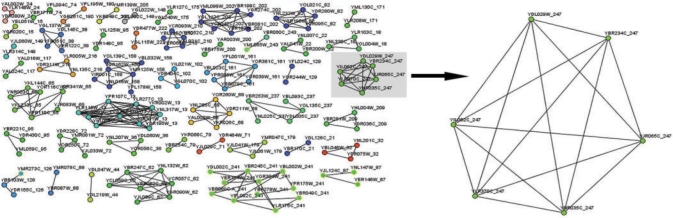
Fig. 1.
This figure shows some protein complexes that were predicted after applying Spectral clustering algorithm and filtering the results with parameters density=0.7 and haircut=3 in a yeast protein–protein dataset (Gavin et al., 2006). The budding yeast Arp2/3 complex shown on the right part of the figure was successfully predicted as it is mentioned in the literature (Winter et al., 1999).
Conflict of Interest: none declared.
REFERENCES
- Andreopoulos B, et al. Clustering by common friends finds locally significant proteins mediating modules. Bioinformatics. 2007a;23:1124–1131. doi: 10.1093/bioinformatics/btm064. [DOI] [PubMed] [Google Scholar]
- Andreopoulos B, et al. Finding molecular complexes through multiple layer clustering of protein interaction networks. Int. J. Bioinform. Res. Appl. 2007b;3:65–85. doi: 10.1504/IJBRA.2007.011835. [DOI] [PubMed] [Google Scholar]
- Brohee S, et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–W451. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, et al. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Enright AJ, et al. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- Coen B, Joep K. Communications of the ACM. Vol. 16. New York, USA: ACM Press; 1973. Algorithm 457: finding all cliques of an undirected graph. [Google Scholar]
- King AD, et al. Protein complex prediction via cost-based clustering. Bioinformatics. 2004;20:3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- Kuhn M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen JB. 5th Berkeley Symposium on Mathematical Statistics and Probability. Berkeley: University of California Press; 1967. Kmeans some methods for classification and analysis of multivariate observations; pp. 281–297. [Google Scholar]
- Moschopoulos CN, et al. 8th IEEE International Conference on Bioinformatics and Bioengineering. Athens, Greece: 2008. An enhanced Markov clustering method for detecting protein complexes. [Google Scholar]
- Paccanaro A, et al. Spectral clustering of protein sequences. Nucleic Acids Res. 2006;34:1571–1580. doi: 10.1093/nar/gkj515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DC, et al. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl Acad. Sci. USA. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Karypis G. Data clustering in life sciences. Mol. Biotechnol. 2005;31:55–80. doi: 10.1385/MB:31:1:055. [DOI] [PubMed] [Google Scholar]