Abstract
The advent of high-throughput DNA sequencers has increased the pace of collecting enormous amounts of genomic information, yielding billions of nucleotides on a weekly basis. This advance represents an improvement of two orders of magnitude over traditional Sanger sequencers in terms of the number of nucleotides per unit time, allowing even small groups of researchers to obtain huge volumes of genomic data over fairly short period. Consequently, a pressing need exists for the development of personalized genome browsers for analyzing these immense amounts of locally stored data. The UTGB (University of Tokyo Genome Browser) Toolkit is designed to meet three major requirements for personalization of genome browsers: easy installation of the system with minimum efforts, browsing locally stored data and rapid interactive design of web interfaces tailored to individual needs. The UTGB Toolkit is licensed under an open source license.
Availability: The software is freely available at http://utgenome.org/.
Contact: moris@cb.k.u-tokyo.ac.jp
1 INTRODUCTION
Browsing genomic information has been central to life science analysis since large-scale genomes became available for many species including mammals, invertebrates, plants and insects. To support the task of analyzing genomic information, two categories of genome databases are available. The first category includes generic genome database species maintained by large organizations, such as Ensembl (Hubbard et al., 2009), UCSC (Kuhn et al., 2009) and NCBI (Wheeler et al., 2007). The other category includes species-specific genome databases, such as SGD (Hong et al., 2007), FlyBase (Wilson et al., 2008) and Wormbase (Rogers et al., 2007), which utilize general-purpose genome browsers, such as GBrowse (Stein et al., 2002). Both types of genome browser represent centralized resources because of the high cost of building and maintaining web database servers. Due to the limited number of experts available to maintain the servers, updates of these centralized web servers are likely to be slow, which is tolerated because the amount of genomic data output by Sanger sequencers is relatively small, yielding on the order of ∼10 million nt per week.
Today, however, the rate at which genome information is produced has outperformed the pace of centralized web browser maintenance. Indeed, the recent advent of high-throughput DNA sequencers (e.g. Solexa/Illumina, SOLiD/ABI and 454/Roche) allows even small groups of people to collect huge amount of genomic data in a fairly short period, billions of nucleotides per week, requiring the data analysis to be done as quickly as possible. In particular, generating tracks for investigating personalized data becomes a crucial step in conducting specific analysis. The UCSC Genome Browser, for example, offers the function of managing custom tracks that upload and display personal data in local disks to the UCSC Genome Browser together with well-annotated existing tracks. These functions in traditional genome browsers are useful but suffer from three major drawbacks. First, the users have to upload their novel and confidential data to the server, though they want to keep these datasets in their local files before publication. Second, although anonymization of the data is partially supported by UCSC, uploading large volumes of data (e.g. 1 GB of Solexa reads data) to the remote web server still needs enourmous amount of time. In order to avoid the costs of data uploads, installing a private version of these genome browsers is desirable; however, the task is extremely hard for non-programming biologists because it demands expertise in programming. Third, personalization of web interface is limited and time consuming, though personalizing web interface through a variety of rearrangements is an important step toward finding new ideas. For example, Affymetrix developed the Integrated Genome Browser (IGB) (Affymetrix, http://www.affymetrix.com/partners_programs/programs/developer/tools/affytools.affx.) for integrating various types of biological data provided by DAS (Distributed Annotation System) (Dowell et al., 2001) servers.
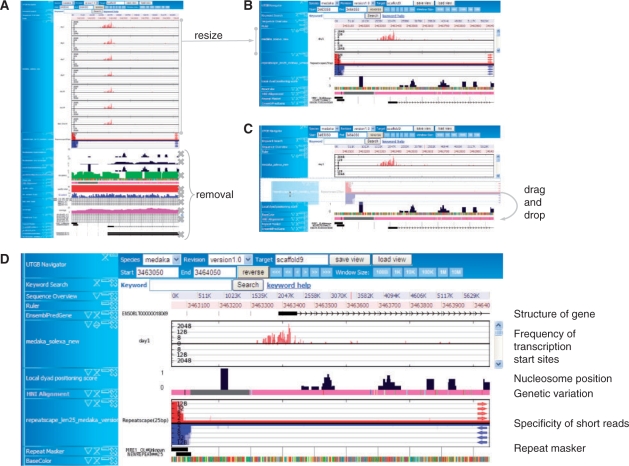
We developed the (University of Tokyo Genome Browser) UTGB Toolkit to provide solutions to these three major requirements: browsing locally stored data, ease of system installation with minimum effort and custom web interface design. Installing the UTGB Toolkit locally on one's own computer is quite easy for inexperienced users because the system can be installed with a few steps, and also avoids uploading confidential data to the remote server. Furthermore, the UTGB Toolkit packages ready-to-use functions, such as a stand-alone web server, HTML rendering functionality, database engines, into one component so that these functions can be made available at private sites immediately after the installation. Figure 1 shows how the UTGB Toolkit facilitates personalization of the web interface in the study of epigenomics. Studying epigenomics involves searching for meaningful combinations from a variety of genome-wide data resources such as DNA methylation, nucleosome positions, transcription start sites, gene expression and evolutionary conservation (Kasahara et al., 2007; Sasaki et al., 2009), which can now be observed via high-throughput sequencers. Our toolkit makes it quite easy to develop a genome browser with drag-and-drop functions for rearranging, juxtaposing and resizing relevant tracks side-by-side interactively to highlight how epigenetic controls are responsible for gene expression and evolution.
Fig. 1.
Rapid creation of tailored interface. (A) A number of tracks in a UTGB genome browser. A long track with a large amount of information can be resized to a shorter track interactively using the scroll bar, which allows the user to browse the content of the original long track. It is also quite easy to eliminate tracks irrelevant to a particular analysis. (B) The resulting tracks can be reordered to facilitate the further analysis. (C) Tracks can be rearranged using the interactive drag-and-drop interface. (D) A genome browser tailored to the analysis of nucleosome positioning surrounding transcriptional start sites and its effect on genetic variation. The track for specificity of short reads is useful in assessing the uniqueness of short-read alignments on the genome. It takes <1 min to perform all the steps.
2 METHODS
2.1 Genome browser interface
To enhance the portability of the system, the UTGB Toolkit is implemented in Java, a portable programming language, such that its machine codes run on top of the Java Virtual Machine. Thus, the UTGB Toolkit is executable on most commonly available platforms, including Mac OS X, Windows, Linux, Solaris and FreeBSD. In addition, UTGB Toolkit is designed such that the browser interface runs on most common web browsers, such as IE, Safari, Firefox and Opera, although it is still necessary to settle discrepancies between these web browsers. To achieve this goal, the interface is compiled into JavaScript code via the Google Web Toolkit (GWT) compiler, which is capable of subsuming the differences between JavaScript engines in the individual web browsers.
2.2 Portable web server for quickly browsing local resources
UTGB Toolkit contains a portable web server so that the genome browser can be launched from the user's personal computer. To avoid manual installation of the web server program (e.g. Apache), the UTGB Toolkit has an embedded Tomcat web server engine. The Tomcat server in the UTGB Toolkit works as a stand-alone web server that is not resident on the system. UTGB Shell launches an instance of Tomcat with the ‘utgb server’ command, and then deploys the genome-browser program on the local Tomcat server.
The portable web server is useful not only for avoiding the installation process, but also for browsing locally stored data resources without losing data privacy. With the web server running on the user machine, no need exists to upload confidential data, such as personal genomic data, to a remote database center. Although anonymization of the data is supported in the UCSC genome browser, the cost of uploading large volumes of data is still prohibitive. As a solution to this problem, the user can utilize the UTGB running on the local machine to simultaneously display local tracks, whose data are kept on the local hard disk, and publicly available tracks.
2.3 Ensuring portability via the embedded database engine
The database management system (DBMS) is an essential component of the genome browser used to provide genomic data for drawing tracks. However, its installation and setup are quite complicated tasks even for database experts. To avoid problems in setting up database engines, we embedded the SQLite (http://www.sqlite.org/) database engine into the UTGB Toolkit. Connections to other DBMS, such as MySQL, PostgreSQL, are also supported in the UTGB Toolkit through JDBC (Java Database Connection) (http://java.sun.com/products/jdbc/). However, unlike these DBMS that use several files to store the database contents, SQLite is portable in that it uses a single file with a universal format, which can work across several operating systems.
To make the DBMS available in any OS environment, we developed an SQLite JDBC connection library, which packs natively compiled SQLite binaries (SQLite is written in C) for operating systems such as Windows, Mac OS and Linux. To support other operating systems for which the SQLite binary is not available, our SQLite JDBC library also contains a pure Java SQLite database engine, which works in any environment that supports Java. Therefore, even if the computer has no DBMS, running the UTGB browser with database support is possible. In addition, with the embedded SQLite database engine, we can easily port the genome browser to other OS environments; e.g. we can make a clone of the genome browser simply by copying the browser code and database files. This portability of the genome browser is a novel feature made possible by the UTGB Toolkit.
2.4 Server-side programming support
The UTGB Toolkit is designed to accommodate various requirements of data visualization, as visualization for genomic data tends to be different for each scientific study. Standard graphical representations provided by existing database centers do not always fulfill user needs. The UTGB Toolkit allows developers to use their own data visualization programs, e.g. CGI-based graphic image generators, HTML content renderer. Although many variations in data visualization exist, a common implementation pattern is used in writing web-based graphic generation programs. For example, a typical pattern of server-side graphic drawing is as follows: the web server receives a user request from the browser, and then issues a query to the database. The results of the query are translated to class objects (e.g. gene objects), and finally, image data for visualizing these gene objects are returned to the browser. This pattern contains three major processes: web request handling, database connection and database object mapping. Here, we describe libraries for supporting implementation of these common tasks, and how the UTGB Toolkit eases server-side programming.
2.4.1 Web Action
The web request handler in the UTGB Toolkit is called a web action, which is a Java class for receiving HTTP requests. Web actions in the UTGB Toolkit enable developers to rapidly begin coding web interfaces; the ‘utgb action’ command in UTGB Shell generates a web action instantly. Each web action is directly mapped to a web server URL. For example, a web action named ‘sequence’ corresponds to the URL, http://(server_base_url)/sequence. Parameter values attached to the URL (e.g. ‘sequence?name=chr1&start=100’) are passed to the web action, and these values are automatically assigned to corresponding variables in the web action class. Data types of the request parameters, such as integer and string, are detected automatically from the class definitions of web action class, and parameter values in the URL, which are merely a string type, are automatically converted into appropriate data types. Individual web actions correspond to the genome browser web API. With the web action mechanism, it is possible to avoid writing repetitive code for request handling and data type conversion.
2.4.2 Database connection
The second core component is database connection support. In general, web database development with Java requires a database server installation and complex configuration files. Inclusion of the embedded SQLite database engine removes the requirement for database installation, and connections to SQLite databases or other DBMS are immediately available within the web action codes. UTGB Toolkit supports both local SQLite database files and remote databases connected through JDBC. These databases can be used by specifying their system types and database location (file names or URLs) in the config/track-config.xml file.
2.4.3 Object database mapping
Relational database engines serve table-formatted data, and an impedance mismatch occurs between table data and their memory representations in computer programs. To use the database data in a program, it is necessary to convert the table data into a more usable format, such as array or class objects in main memory. The UTGB Toolkit provides the BeanUtil library, which supports translation of table data into class objects. Similar to the web action handling, table format data are converted to appropriate class objects by investigating the Java class definitions. Our matching algorithm translates the table data to a set of class objects by comparing column names in the table data and parameter names in the class definition. The matching algorithm automatically converts the data types between table and class objects using the reflection mechanism in the Java language, which provides the information of parameter names and types in the class definitions. Thus, no manual mapping configuration is required to bind table data to class objects.
2.5 Importing biological data
The UTGB Toolkit supports visualiztion of commonly used biological data formats, such as BED, DAS (Dowell et al., 2001), etc. To create a track for displaying biological features mapped onto the genome, the user has to specify the data files to load (see the documentation at http://utgenome.org/toolkit/ for details). The UTGB parses these biological data files in a stream manner and generates graphics that display the features in the region specified in the genome browser. Access to DAS data is also supported in the UTGB Toolkit for utilizing existing biological data resources in conjunction with the user's own tracks. We are continuing the effort of improving the UTGB Toolkit to support other biological data formats, such as AGP, GFF, PSL, AXT (alignment data), etc.
3 RESULTS
3.1 UTGB framework for browsing various data sources
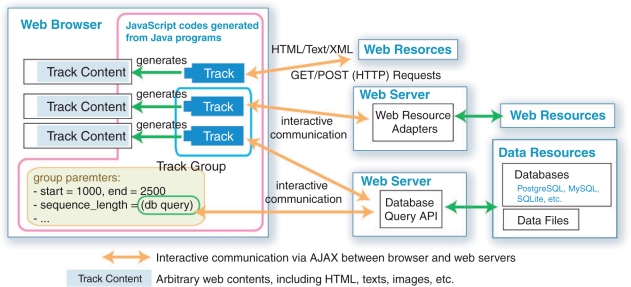
A notable feature of the UTGB Toolkit is its framework design for integrating multiple web resources (Figure 2). The interface of the UTGB browser consists of tracks, which display individual data resources. To manipulate a set of tracks at the same time, we provide the notion of the track group, which holds variables that are shared between multiple tracks. For example, the user can relocate the window positions of several tracks by changing their track group state, e.g. by clicking the scroll button. The genome browsers generated by using the UTGB Toolkit can display both public and private data sources on a local machine simultaneously.
Fig. 2.
Illustration of the UTGB framework. The UTGB framework has a two-sided design, client- (browser) and server-side code. For the client side, the UTGB Toolkit generates a web browser interface that consists of a set of tracks. Individual tracks communicate with the web servers, and produce track contents from the received data in the form of, for example, graphics or table data. Track groups, which manage a set of tracks, hold common parameters shared among tracks, such as window location on the genome sequence. On the server side, arbitrary web data sources (e.g. HTML, text, XML, database query results, etc.) can be used to generate track contents using mini-browser (iframe) tracks or web resource adapters. Advanced users can implement tracks in Java, which are compiled into JavaScript code, to provide a more sophisticated user interface.
3.2 Fast and flexible genome browser interface revision
The interface of the UTGB genome browser displays a list of tracks, each of which can be dragged to a different location, allowing the user to customize the browser interface online. The track interface of the UTGB Toolkit is implemented using JavaScript technology, which can dynamically rewrite HTML components in the browser window, so track relocation can be performed without accessing HTML pages on the server. This feature greatly reduces user frustration when employing traditional genome browsers that reload the entire pages after the track relocation. Reloading of track contents is independent for each track in the browser. Therefore, even if the response time of the server providing a track contents is slow, other track contents served by other servers can be displayed immediately after the arrival of the data. Therefore, the users can continue to relocate sequence positions on the UTGB genome browser seamlessly, eliminating the latency caused by the slowest server to display tracks.
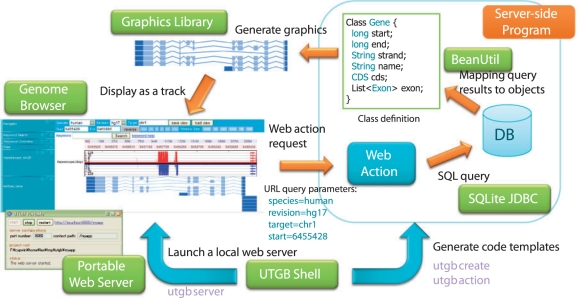
3.3 UTGB shell for quick genome browser development
To facilitate rapid genome browser development, we developed the UTGB Shell, a command-line user interface of the UTGB Toolkit. Figure 3 shows the overview of the UTGB Toolkit and what can be done with the UTGB Shell. The UTGB Shell has several user commands that support development of a personalized genome browser. For example, the ‘utgb create’ command creates a new genome browser in a few seconds, and the ‘utgb server’ command launches a web server on the local machine, which enables immediate use of the genome browser. For developers, track programs that use, for example, database searches, data visualization, can be implemented with minimal programming effort because ‘utgb create’ command generates a genome browser code that already has features such as database connection support, a rich user interface and standard track implementations.
Fig. 3.
Overview of the UTGB Toolkit. The UTGB Toolkit supports development of personalized genome browsers in various ways. The UTGB Shell generates code templates for handling web requests from the genome browser interface (web action), database access support through SQLite JDBC and mapping support from SQL query results to the specified class objects (BeanUtil). Developers can generate track graphics by using the library included in the UTGB Toolkit or their own programs. The generated graphics (or arbitrary HTML contents) can be displayed as track contents in the genome browser interface. To browse both of the locally and remotely stored data, these steps can be performed in a local user machine by launching a local web server from the UTGB Shell.
3.4 Stand-alone and web mode
Genome browsers generated by the UTGB Toolkit work in two modes, stand-alone and web modes. In the stand-alone mode, the genome browser runs as a user program on the local computer, so a privileged account is not required to run the genome browser program. This stand-alone mode is useful in testing the behavior of the browser before publishing. The web mode is for publishing the genome browser on a server machine. The browser code is sent to the Tomcat engine, which is a standard web server program for running web applications written in Java. With ‘utgb deploy’ command in the UTGB Shell, we can immediately deploy the genome browser on the Tomcat server. Even if the server machine already is running another web server, such as Apache, these server programs can coexist by bypassing HTTP requests received by the Apache server to the Tomcat engine through the proxy module, and users can use the genome browser contents as if they were served by the Apache web server. Detailed information regarding such settings is available from http://utgenome.org/.
3.5 Personalization of genome browser
Maintenance of biological databases consists of data conversions to accommodate site-specific data formats, and data submission to appropriate sites. In general, however, this process is not suited to visualizing the huge amounts of data that are now common in the era of large-scale genome analysis. In addition, site-specific data formats and their graphical representations may not be ready or extendable to publish a variety of research results. The UTGB Toolkit tackles these problems by providing a web browser interface to display tracks hosted by multiple web servers in a single window. A set of standard tracks supporting visualization of data resources is already available. The framework design of the UTGB Toolkit provides users with flexibility to browse their own data using both preinstalled and their own visualization programs. To enhance the user experience, we also incorporated the modern web application technologies, such as the Google Web Toolkit (GWT), AJAX and client-side graphic drawing into our toolkit. These technologies make several features of UTGB, such as drag-and-drops, flexible resizing and smooth scrolling, available to both users and web application developers.
3.6 Efficiency of client-side resource integration
The Ensembl's Genome Browser (Hubbard et al., 2009) is composed of a single set of image data integrating images of several tracks using server-side programs. However, this architecture suffers from a major drawback: when the user clicks a browser button to move the location to display, the genome browser must redraw the whole contents already shown on the page. This type of implementation, which we call server-side integration, is problematic as the server program has to perform similar database queries and repeatedly draw graphics even for slight window relocation. Without a caching mechanism, this type of implementation cannot work efficiently. The major reason why the Ensembl Genome Browser merges images is that the HTTP protocol is stateless; i.e. the browser cannot remember the presented HTML data when the user clicks a link and moves to the next page. To display the next page, the browser must retrieve the entire contents again from the server as stateless web browsers do not allow partial updating of the genome tracks.
With recent advances in browser technology, it has become possible to change the HTML contents dynamically after loading HTML and image data using the JavaScript language. Several other technologies are available for drawing graphical contents in the browser, such as Flash, Adobe Air and Microsoft Silverlight. However, these extensions of the web browser sometimes do not work; e.g. if the browser does not have the required plug-ins to run these extensions or if plug-in installation is not allowed for non-privileged users. Therefore, we chose JavaScript which is commonly supported in most modern web browsers, including Internet Explorer (IE), Firefox, Safari and Opera. These browsers already have an embedded JavaScript engine, and therefore no additional installation process is required. Scripts written in JavaScript language can update the displayed browser content in situ, and enable the web browsers to remember the state of the web page. Therefore, modern web browsers already have the capability to partially modify displayed content while retaining necessary information in memory on the browser (client) side. The UTGB Toolkit fully utilizes this browser (client)-side memory to preserve track contents, which enables the genome browser to support drag-and-drop and resizing of tracks without reloading.
The utilization of client-side memory has attracted a great deal of attention not only for the genome browsers, but also for developing general web applications. In September 2008, Google launched a new web browser, Google Chrome. This browser has its own implementation of the JavaScript engine called V8 with major performance improvements in the JavaScript garbage collection mechanism, which efficiently reuses browser-side memory. The open-source web browser Firefox 3 is also continuing to develop the JavaScript engine, and has achieved faster performance than the previous version, Firefox 2. This trend of improving the JavaScript engine lends marked support for client-side resource integration.
3.7 Server-side and client-side graphic drawing
With regard to graphical visualization of genomic data, the UTGB Toolkit provides several ready-to-use graphical representations, such as drawing genes and graph representations. In the UTGB Toolkit, these visualization supports are available in server-side code, and we are continuing to develop other types of data visualization. The UTGB Toolkit also supports browser-side graphic drawing with the canvas tag, which will be a standard of browser-side graphic drawing in HTML 5, the next version of HTML. Several web browsers, including Google Chrome, Firefox, Safari and Opera, support drawing graphics with the canvas tag, with IE being the only major browser not providing support for this tag (as of 2008). However, IE can draw graphics using Vector Markup Language (VML), which has similar functionality to canvas for drawing graphics. A team at Google has developed GWT Canvas, a graphic drawing library, which hides differences between VML in IE and the canvas tag, so developers can draw graphics in both IE and other browsers that support this tag. The UTGB Toolkit uses GWT Canvas to draw custom user tracks.
The capability of drawing graphics in the browser has an impact on genome browser design because it simplifies the role of the server; the server machine has no need to generate graphics, but rather its role is to serve data objects that represent genomic information such as genes and CDS. The client (browser) receives these data objects and draws their graphical representation on behalf of the server. This two-sided genome browser design greatly reduces the workload on the server machine and simplifies server-side programming; the server needs only to perform database queries and publish the query results.
3.8 Availability of the UTGB Toolkit
The UTGB Toolkit is freely available from the UTGB Project Page (http://utgenome.org/). All source code is managed using the source revision control system Subversion, so the latest code is made available immediately. All of our source code is licensed under the Apache License version 2.0, which is an open-source license allowing free use and distribution for both personal and commercial purposes. The Apache License is applied on a file basis; i.e. modified source files must be licensed under the Apache License Version 2.0, while users are free to apply any license they wish to source code generated by the UTGB Toolkit and user-developed source code that simply uses the UTGB Toolkit.
4 DISCUSSION
In general, database integration takes various forms: a portal, compound and fully integrated. A portal collects links to internal or external data sources (e.g. PubMed and ENCODEdb); search engines, such as Google and Yahoo, also belong to this category. The second type, compound, displays several database contents at the same time in the browser window. The interface of the UTGB can support this type of data visualization. The IGB (Affymetrix, http://www.affymetrix.com/partners_programs/programs/developer/tools/affytools.affx.) is also a compoond browser and supports the integration of genome data resources provided by DAS protocol using the Java-based GUI. Another example is iGoogle (http://www.google.co.jp/ig), a web service consisting of user-customizable gadgets, sub windows that can display, for example, news or a calendar. The third type comprises fully integrated databases that merge several databases into a single DBMS to support queries across these databases; this type of integration is common in centralized database centers such as NCBI and Ensembl.
These various types of database integration each have their own benefits: portals can be used as directories to data resources, compound browsers can collect resources provided by several organizations and full integration is necessary to process queries that cannot be evaluated without integration of databases. For example, a query listing all SNPs surrounding user-selected genes requires both SNP and gene locus databases. Without integration of these databases, the user must click the browser buttons numerous times to navigate through unintegrated database browsers. Our UTGB framework is not restricted to a certain pattern of database integration, and all of three of the above types can be used as back-end databases. However, to achieve faster query performance and maintenance of the integrated databases, further implementation effort are required, for example, integrated query support, query optimization and user-friendly database management interfaces.
Funding: Japan Science and Technology Agency (JST).
Conflict of Interest: none declared.
REFERENCES
- Dowell R, et al. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2007;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2009. Nucleic Acids Res. 2009;37(Suppl.1):D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, et al. The UCSC genome browser database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A, et al. Wormbase 2007. Nucleic Acids Res. 2007;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DLL, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, et al. Flybase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]