Abstract
Many proteins synthesized in the cytoplasm ultimately function in non-cytoplasmic locations. In Escherichia coli, the general secretory (Sec) pathway transports the vast majority of these proteins. Two fundamental components of the Sec transport pathway are the SecYEG heterotrimeric complex that forms the channel through the cytoplasmic membrane, and SecA, the ATPase that drives the preprotein to and across the membrane. This review focuses on what is known about the oligomeric states of these core Sec components and how the oligomeric state might change during the course of the translocation of a preprotein.
Keywords: Oligomer, Protein transport, SecA, SecYEG, Translocon
All cells have mechanisms to export proteins from the cytoplasm to their final destination. Bacteria have several transport pathways including the TAT (twin arginine translocation) that transports folded proteins that often contain co-factors (for a recent review, see [1]), the YidC pathway for membrane proteins (for a recent review, see [2]), and the general secretory (Sec) pathway that translocates unfolded preproteins (for a recent review, see [3]).
The Sec pathway is found in bacteria, archaea and eukaryotic organelles such as chloroplasts and the endoplasmic reticulum. Components of the bacterial Sec translocon include two heterotrimeric membrane spanning complexes, SecYEG and SecDFYajC, and a peripherally associated SecA, although homologues for each component are not universal throughout the three kingdoms of life.
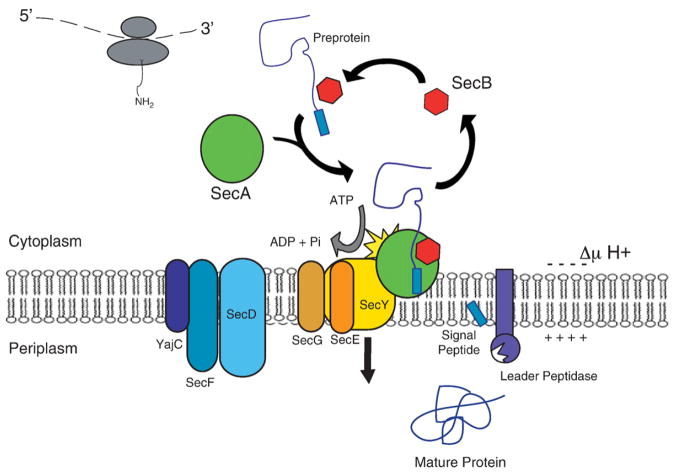
A nascent chain destined for export via the Sec pathway emerges from the ribosome in the Escherichia coli cytoplasm. This nascent chain consists of the protein to be secreted linked to a targeting signal called the signal peptide, and together known as a preprotein. It may associate directly with SecA or be targeted to SecA in complex with the cytoplasmic chaperone, SecB. SecA directs the preprotein to the membrane where the hydrolysis of ATP by SecA, along with the protonmotive force, promotes translocation across the membrane through the channel comprising three integral membrane proteins, SecY, SecE and SecG. The SecDFYajC complex may promote membrane cycling of SecA to enhance translocation through the SecYE pore [4]. The signal peptide is then cleaved from the preprotein by leader peptidase. An overview of protein export in E. coli is shown in Fig. 1. A more complete review of Sec transport components not discussed in detail here can be found in [3].
Fig. 1.
Schematic representation of the E. coli Sec transport system. Most models of Sec-dependent preprotein export share the following features. The nascent chain emerges from the ribosome and may interact with the cytoplasmic chaperone, SecB. The tetrameric SecB associates in the cytoplasm with dimeric SecA. Thus, some preproteins are delivered to SecA via SecB, while others directly interact with SecA without the participation of SecB. Upon interacting with the inner membrane-bound SecYEG, SecA may or may not dissociate to monomer. SecA hydrolyzes ATP and, with the protonmotive force (ΔμH+), drives translocation through the SecYEG pore, composed of one or more subunits. SecDFYajC may enhance translocation by regulating the membrane cycling of SecA. The signal peptide of the preprotein is cleaved on the periplasmic side of the membrane by leader peptidase, releasing the mature protein to its final location.
Components of the Sec translocase were first identified when mutations in the genes encoding SecA (prlD) and SecY (prlA) suppressed protein export defects caused by signal sequence mutations (for review, see [5]). In 1990, the translocon was reconstituted in liposomes using only purified SecYE(G), SecA and ATP, and found to be sufficient for preprotein translocation [6].
Sixteen years later, much has been learned about the components involved in the translocation of preproteins, how they function and how they interact. However, the biophysical nature of the translocon, and specifically the oligomeric states of its components, remains in question. If we are to fully understand protein export in E. coli, a physical description of the proteins involved is needed. While the recent crystal structures of bacterial SecAs [7–9] and the archaeal SecY complex [10] have provided great insight, they are static pictures of a dynamic process. This review focuses specifically on what is currently known of the oligomeric states of SecA and SecYEG.
1. SecA—the translocation motor
The SecA gene was first described in 1982 [11] as coding for a component in secretion. The SecA gene product was identified as a large peripheral membrane protein; biochemical data in a cell free system [12] and in vitro using membrane vesicles [13] confirmed that SecA is involved in protein secretion and furthermore, that it couples ATP to protein translocation [14]. More recent studies suggest that SecA may also act as a cytoplasmic chaperone [15].
SecA may exist in monomer–dimer equilibrium [16,17] with a dissociation constant determined to be 0.25 μM–0.5 μM in a aqueous environment [17]. The cellular concentration of SecA has been estimated at about 5 μM [16] suggesting that SecA is predominantly dimeric in the cytoplasm. In fact, a number of studies indicate that SecA is an antiparallel [7,18] homodimer in the absence of detergent and lipids [18–20]. The ratio of monomer to dimer SecA populations can be altered by temperature and salt concentration [17] and by the presence of translocation ligands [16,21,22]. SecA interacts as a dimer with tetrameric SecB in the cytoplasm [23,24] and while SecB is thought to disrupt SecA’s dimer interface, it does not cause dissociation into monomers [24].
The dimerization domain of SecA was originally thought to reside in the C-terminus; when isolated from the rest of the protein, this domain was shown by size exclusion chromatography to form a dimer in solution [25,26]. However, a truncated SecA mutant without its most C-terminal 70 residues remains dimeric [27–29] although when the hydrophobicity of 6 residues along another possible dimer interface is reduced, conversion to monomer is complete [16]. More recently, the crystal structure [7] and analysis via size exclusion chromatography [24] suggested that the immediate N-terminus may also contain a dimerization domain.
While there is good agreement that SecA in the cytoplasm adopts a dimeric form, there is considerable debate as to whether SecA remains dimeric throughout translocation. Evidence that membrane-bound SecA may be dimeric was shown using chemical crosslinking and ‘membrane trapping’ [30]. A SecA amino-terminal deletion mutant that is predominantly monomeric did not integrate into the membrane and had limited ATPase activity.
Studies using translocation ATPase, crosslinking, and FRET-based assays indicate that SecA remains a dimer throughout translocation [19,29–32]. However, several lines of evidence suggest that translocation substrates including lipids [16,22], SecY [21] and signal peptides [16,33] cause SecA dissociation into monomers.
That SecA’s oligomeric state in aqueous solution is unaffected by nucleotides is suggested by studies using small angle X-ray scattering, crosslinking and native PAGE [16,21,34]. On the other hand, small angle neutron scattering suggested that the presence of either ATP or ADP promoted SecA monomer in lipids whereas SecA was dimeric in the absence of nucleotides [35]. As discussed above, the dissociation constant for the monomer–dimer equilibrium of soluble SecA is in the submicromolar range and most studies use SecA at concentrations within or near this range. Therefore the equilibrium is poised to be influenced by a number of factors including, of course, the concentration of SecA itself. Furthermore, its dissociation constant in lipids has yet to be determined. In several studies SecA dissociates to monomer in the presence of negatively-charged lipids alone [16,22]. However, the concentration of SecA used by Bu et al. [35] was slightly higher and may explain why E. coli lipids alone did not cause monomerization yet the addition of nucleotide to the lipid–SecA complex did.
While the nucleotide binding domains are essentially identical structurally, studies indicate that the B. subtilis SecA dimer [7] and monomer [9] are significantly different in the region of the proposed preprotein cross-linking domain (Fig. 2). The monomer structure is in a more open conformation similar to that observed in the presence of lipids [18,36,37] and ATP [35,38], and exposes a groove that may be the signal peptide binding site. Ligands present at the early steps of translocation appear to promote a conformational state in SecA that may represent this activated form.
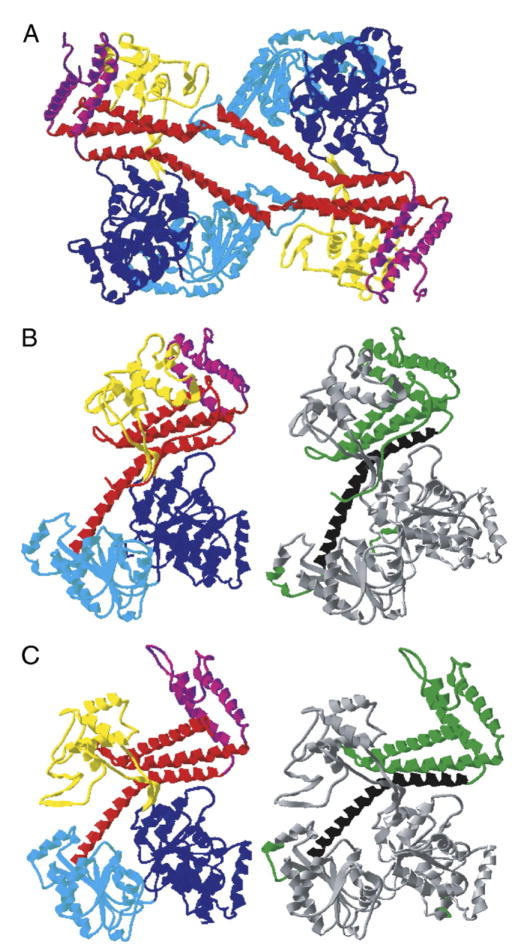
Fig. 2.
Structure of SecA. (A) Dimeric form of Mycobacterium tuberculosis SecA (PDB ID: 1NL3; [4]). The domains are colored as follows: nucleotide binding fold I, dark blue; nucleotide binding fold II, light blue; preprotein crosslinking domain, yellow; helical scaffold domain, red; helical wing domain, purple. An elliptical pore of l0 Å × 35 Å is evident at the center of the dimer which could align with the cavity formed by SecYEG to provide a channel for preprotein movement. (B) Closed form of an individual subunit of Bacillus subtilis SecA (PDB ID: 1M6N; [3]). On the left, domains are colored as in (A). On the right, the dimer interface based on the crystal structure (residues 572–618; [3]) is indicated in black and other domains that may impact dimerization, suggested by a variety of studies (residues 1–10 [3,17], 558–841[21], 609–771 [20]), are indicated in green. (C) Open form of monomeric Bacillus subtilis SecA (PDB ID: 1TF5; [5]). The left and right structures are as in (B). In this open conformation, a deep groove is apparent between the preprotein crosslinking domain and the helical scaffold and helical wing domains. It has been proposed that this may accommodate the translocating preprotein.
Remarkably, two very similar SecA mutants with small (up to 10 residues) N-terminal deletions and 70 residue C-terminal deletions displayed different oligomeric states. Karamanou et al. [29] showed that the mutant SecA remained dimeric and functioned like wild type in binding to SecY, ATPase activity and translocation of precursor. In contrast, Or et al. [39] describe a monomeric mutant SecA that functioned reasonably well in binding to SecY (2-fold lower than wild-type SecA) with ATPase activity similar to wild type, but functioned poorly in preprotein translocation (16% of wild type). Each study’s authors argued that they had identified SecA’s functional oligomeric state, illustrating the continuing controversy.
2. SecYEG intra-complex associations
SecY, SecE and SecG exist in a 1:1:1 stoichiometry within the SecYEG heterotrimer [40]. A variety of methods, including cysteine-scanning mutagenesis and disulfide cross-linking as well as genetic and crystallographic analyses, show that SecY and SecE have many points of contact including periplasmic loops [41–43], cytoplasmic loops [10,44,45] and transmembrane segments [10,43,46–50]. Furthermore, the association between TM7 of SecY and TM3 of SecE does not change upon preprotein insertion [49]. Cytoplasmic loops [51,52] and transmembrane segments [53] of SecY also crosslink to SecG.
Interactions have been detected, via cysteine crosslinking, between TMs of neighboring SecEs in cells overexpressing SecYEG [48] although another study, using near wild-type levels of proteins, showed no crosslinking upon treatment with formaldehyde [40]. As discussed above for SecA, and again reflected here, the levels of protein present will likely influence the observed oligomeric state; thus results must be interpreted with care. Another possibility is that the crosslinking agent used in the latter study induced monomerization [54]. Additional evidence, including crosslinking of cytoplasmic loops of neighboring SecYs [51], and formation of stable, functional homodimers of SecG [55], suggests (at least) dimeric SecYEG complexes exist although individual YE subunits do not exchange among neighbors during translocation [56,57].
Translocation did not go to completion in the presence of crosslinked SecY–SecE [49,50] or SecE–SecE [48] yet function was maintained when the N- and C-termini of neighboring SecYs were linked [51]. It seems reasonable that conformational flexibility during translocation is required, whether by rearrangements of transmembrane segments or by oligomer dissociation; if this flexibility is prevented by crosslinking, a translocation defect would likely be the result.
3. SecYEG—the protein conducting channel
The SecYEG heterotrimer forms the basic unit of the integral membrane pore through which translocating preproteins travel. The recent crystal structure of the Methanococcus jannaschii SecYEβ (homologous to E. coli SecYEG; Fig. 3) suggests that the monomer could act as the functional unit [10]. The crystal structure reveals the presence of a plug in the closed state of the channel [10], and crosslinking studies provide evidence that the functional translocon requires dimeric SecYEG for displacement of the plug from, and transport of a preprotein through, a monomeric channel [57]. Similarly, dimers of E. coli SecYEG were observed in complex with a ribosome and nascent chain by cryo-electron microscopy [58]. These dimers demonstrated a front-to-front orientation, i.e., each protomer resembles a clam and in the dimer, the mouths open facing each other. In other words, if the lateral gates of each protomer are open, the pores become connected. (In back-to-back orientation, the lateral gates would open to lipids). Despite this orientation, it appears that the pore of each protomer remains a separate translocation channel; the two pores do not open to create a single larger channel. The authors suggest that during post-translational translocation, one channel actively translocates the preprotein while the other remains in the closed state with plug in place.
Fig. 3.
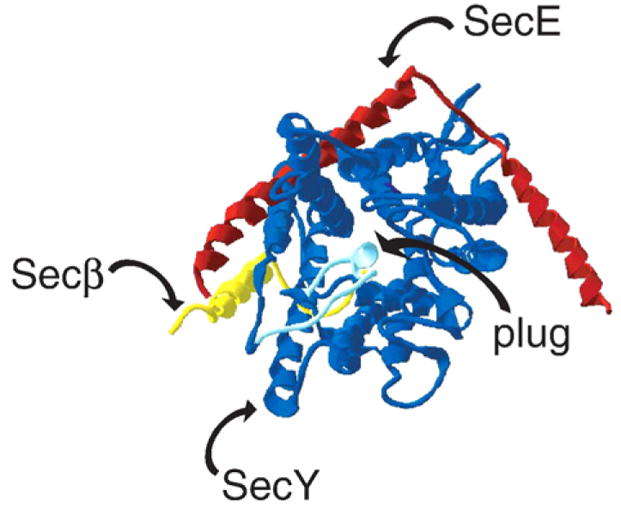
Structure of an archaeal SecYEβ (PDB ID: 1RHZ; [6]). SecY is depicted in blue (with the plug, formed by a portion of TM2, in light blue), SecE in red, and Secβ (homologous to E. coli SecG) in yellow. Signal peptide binding could induce plug displacement, exposing a channel for the translocating polypeptide. Alternatively, binding of SecA or another SecYEβ/SecYEG complex may play a role in opening the cavity and producing the translocation-competent structure. To date, information regarding possible oligomerization domains is speculative.
FRET analysis of the SecYEG of Thermus thermophilus indicates that 2 or more complexes are closely associated in the membrane [59]. Dimers of E. coli SecYEG were observed using cryo-EM in phospholipid bilayers [60]; a 16 × 25 Å cavity formed at the dimer interface was of similar dimensions to other translocation pores and was closed on the periplasmic side by highly tilted helices. On the other hand, analytical ultracentrifugation of E. coli SecYEG in detergent solution [61] detected the presence of monomers and tetramers. Freeze-fracture electron microscopy recently showed that monomeric, dimeric and tetrameric SecYEG complexes exist in the membrane and the ratio of oligomeric states changes in the presence of translocation ligands [62]. The analysis of the state of the preprotein conducting channel is made more difficult by the fact that detergent and protein concentrations, as well as the presence of SecG in overexpressed SecYE complexes, effect oligomeric associations [54], yet these parameters are not always well controlled.
The crystal structure of the monomeric archaeal complex [10] aligns well with a single subunit of the dimeric E. coli SecYEG [60] suggesting that oligomerization does not significantly change the conformation of the heterotrimer. Crystal structure data have, thus far, come from the channel in its “resting” state, i.e. not in complex with other transport components. However, studies of the channel that include translocation intermediates indicate monomer [40], dimer [54,58,61] or tetramer [63] SecYEG are present during transport. Negative staining electron microscopy of Bacillus subtilis SecYE suggests a complex of three protomers in the presence or absence of SecA [64]. Sec61p, the eukaryotic homologue of SecYEG, has been reported to form tetramers in the absence and presence of other translocation-associated proteins [65,66]. At this stage, it is evident that SecYEG are detected in multimeric forms though it is not clear to what extent the method of analysis biases the observations. For example, it is conceivable that in some analyses of SecYEG complexes, sufficiently high concentrations could generate FRET among neighboring molecules that do not form intimate non-covalent contacts. Complicating the picture is the possibility that the oligomeric states of the structural and functional units are not equivalent. Perhaps the translocon functions as a monomer but the recruitment of additional translocons to protein export sites during active periods of transport puts multiple subunits in close association. The functional oligomeric state of the translocon is discussed in more detail below.
4. The SecA–SecYEG complex or What happens during translocation?
SecA interacts with the SecYEG channel at the cytoplasmic membrane during preprotein transport. Immunostaining has shown that SecA and SecY co-localize in B. subtilis membranes [67], and SecA and SecY crosslink to each other in the absence of nucleotide and preprotein (a.k.a. the resting state; [68]). SecA and SecY crosslink concurrently to a trapped preprotein [69] and, during translocation crosslinks between SecA and SecG in the membrane increase [70]. The C-terminus of SecA binds SecY [71,72] at its two most C-terminal cytoplasmic domains [47,73–76]. The studies mentioned here, although hardly an exhaustive presentation of the work, clearly show that SecA and SecYEG interact during preprotein transport. But what is the oligomeric status of the complex?
A monomeric mutant of SecA binds strongly to SecYEG but translocation activity is low [39], whereas a similar but dimeric mutant SecA binds and translocates like wild type [29]. Studies on SecYEG in the presence of SecA and a translocation intermediate preprotein showed SecYEG in the monomeric state using crosslinking [40], whereas native gel electrophoresis appeared to indicate dimeric SecYEG [54]. However, it was later determined that since SecA and SecYEG co-migrate in native gel electrophoresis, the gel likely showed monomeric SecYEG in association with monomeric SecA rather than dimeric SecYEG [21]. When SecYEG dimers were created, either by covalent linkage [21] or by antibody stabilization [77], they associated with SecA monomers and dimers. However, during translocation SecA monomer remained associated with a SecYEG protomer and the preprotein [21]. Recently, a preprotein was shown by disulfide crosslinking to come in close contact to residues on the interior of SecY but not to those on the exterior [78]; the authors propose this as evidence that a single SecYEG forms the translocation channel but do not rule out the possibility that several heterotrimers in open front-to-front conformations together could form the channel through the membrane.
Other studies propose that during translocation the functional complex contains SecA dimers and SecYEG tetramers. Manting et al. [63] inferred, from mass analysis using scanning transmission electron microscopy, that purified SecYEG was mostly dimeric but the presence of SecA and preprotein induced about half to form tetrameric associations. Quantitative western blotting showed that the ratio of SecA:SecYEG was 1:2, consistent with dimeric SecA in complex with tetrameric SecYEG. In another study, a SecA dimer trapped in translocation by a non-hydrolyzable ATP analogue or by the presence of preprotein associated with large SecYEG oligomers proposed to be tetramers [76].
5. Insights from other bacterial transporters
The SecA ATPase component of the Sec translocon is homologous to the DEAD box helicases, and the recent crystal structure of B. subtilis SecA highlights the structural similarities [7]. These helicases are generally composed of two parallel α/β domains and interdomain flexibility allows for substantial conformation changes [79]. The E. coli DEAD box helicases, UvrB [80] and Rep [81], appear to function as dimers. Alternatively, E. coli RecG [82] and B. stearothermophilus PcrA [83] are helicases that function as monomers. The functional oligomeric state of the helicase determines the method for DNA unwinding.
Like the Sec transport system, ABC (ATP-binding cassette) transporters utilize the energy of ATP hydrolysis to transport substrates across a membrane [84]. Functionally, the ATP-catalytic component of SecA might be compared with that of the nucleotide binding domains of these transporters. MalK functions as a dimer in complex with membrane components, MalF and MalG, to transport maltose [85] but has been shown to be monomeric in solution [86,87]. Similarly, Rad50 [88] and BtuD [89] are dimeric and associate with dimeric partners to form heterotetramers. The periplasmic histidine permease, HisP [90], and the membrane bound lipid A flippase, MsbA [91], are both also functional dimers. One model suggests that during one transport cycle both protomers of dimeric ATPases of the ABC transporters bind and hydrolyze ATP [84]. In fact, a hetero-dimeric SecA in which one protomer was inactivated for ATP hydrolysis did not support preprotein translocation [19].
Transmembrane transporters, SecYEβ included, have structural mechanisms for minimizing water and ion transport [92] and promoting the transport of hydrophobic moieties across a membrane through a proteinaceous channel. These structures may include an hourglass shape, and/or a hydrophobic pore or ring.
The recent structure of SecYEβ suggests that it functions as a monomer with an hourglass-shaped pore, formed by pseudo-twofold symmetry of the 10 α-helices of the SecY subunit. The pore constricts from 20–25 Å down to 5–8Å [10] and is plugged on its periplasmic side by a portion of helix 2. The authors propose that the plug moves away from the pore concomitant with shifts in the helices to allow translocation of a preprotein. E. coli lactose permease [93] and glycerol-3-phosphate transporter [94], each composed of 12 α-helices showing pseudo-symmetry of six helix bundles with pore diameters similar to SecYEβ, are thought to function as monomers.
The narrow opening of SecYEβ in the middle of the membrane is lined with a ring of hydrophobic residues. Such a hydrophobic constriction has been seen in an E. coli mechanosensitive channel, McsS, which narrows to 8–11 Å and is closed by a hydrophobic seal [95]; its transmembrane region is composed of a homoheptamer of three α-helices. Similarly, the 12 αhelices of the Na+/H+ antiporter, NhaA, form a funnel lined with hydrophobic residues [96]. Interestingly, NhaA is thought to function as a dimer, its role being to regulate activity while the monomer forms the translocation pathway [97]. This is very much in line with a recent study suggesting that dimerization of SecYEG is necessary for the plug movement that allows preprotein translocation through a single subunit [57].
6. Conclusions
Determining the biologically relevant oligomeric state of a membrane-associated protein is complicated, and inherent problems exist in many of the experimental strategies used to study this issue. Protein concentration influences the dissociation of complexes and the concentration at which an analysis is done may not be physiological. In addition, nonequilibrium methods are often used so that the oligomeric state of the component may change during the analysis. Oligomers may also be underestimated by crosslinking because of inaccessible target sites. Equally confounding, an overestimate of oligomers may result from non-specific crosslinking, or components may be present in sufficiently high density in the membrane such that near neighbors cannot be distinguished from proteins in complex. The oligomeric state of a protein may also be influenced by the environment in which it is studied. For example, membrane mimetics such as detergent differ significantly from the native lipid environment, and the crystals required for X-ray structure analysis are analyzed in a non-native environment. Electron microscopy is a useful technique for studying macromolecular complexes. However, negative staining electron microscopy uses heavy metal salts and drying of the specimen that may distort its structure; on the other hand, cryo-EM maintains the molecule in an aqueous environment. Both techniques provide only moderate resolution, but in combination with crystal structure information can provide much greater detail for multi-unit complexes [98]. Therefore, no one strategy is likely to unambiguously delineate the oligomeric state of the Sec components, and several complementary approaches must be used.
The process of protein translocation is a dynamic one with proteins associating and dissociating throughout. This may also be true of the oligomeric state of the protein components of the pathway. Oligomerization may act to regulate function, perhaps by opening or closing the translocation channel. Or, a multi-meric form of the protein might be important for maintaining structure while the monomer is the functional unit. A variation of this model involves multimers whose subunits function cooperatively to propel successive rounds of preprotein transit. This may involve individual subunits in the oligomer alternating to bind and release progressively more C-terminal segments of the preprotein. Studies to examine the oligomeric states of SecA and SecYEG are ongoing and will ultimately define the functional Sec translocon.
Acknowledgments
We thank Aaron D’Antona for creating Fig. 1. This work was supported in part by National Institutes of Health Grant GM37639 (to D.A.K.).
References
- 1.Sargent F, Berks BC, Palmer T. Pathfinders and trailblazers: a prokaryotic targeting system for transport of folded proteins. FEMS Microbiol Lett. 2006;254:198–207. doi: 10.1111/j.1574-6968.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- 2.Luirink J, von Heijne G, Houben E, de Gier JW. Biogenesis of Inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- 3.de Keyzer J, van der Does C, Driessen AJM. The bacterial translocase: a dynamic protein channel complex. Cell Mol Life Sci. 2003;60:2034–2052. doi: 10.1007/s00018-003-3006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson SA, Hall MN, Silhavy TJ. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- 6.Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 7.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 8.Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci U S A. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci U S A. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 11.Oliver D, Beckwith J. Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J Bacteriol. 1982;150:686–691. doi: 10.1128/jb.150.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabelli RJ, Chen L, Tai PC, Oliver DB. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver D. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 15.Eser M, Ehrmann M. Sec-dependent quality control of intracellular protein localization. Proc Natl Acad Sci. 2003;100:13231–13234. doi: 10.1073/pnas.2234410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodbury RL, Hardy SJS, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding H, Hunt JF, Mukerji I, Oliver D. Bacillus subtilis SecA ATPase exists as an antiparallel dimer in solution. Biochemistry. 2003;42:8729–8738. doi: 10.1021/bi0342057. [DOI] [PubMed] [Google Scholar]
- 19.Driessen AJM. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 20.Doyle SM, Braswell EH, Teschke CM. SecA folds via a dimeric intermediate. Biochemistry. 2000;39:11667–11676. doi: 10.1021/bi000299y. [DOI] [PubMed] [Google Scholar]
- 21.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem. 2003;278:3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 23.Fekkes P, van der Does C, Driessen AJM. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJS. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol. 2005;348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 25.Hirano M, Matsuyama S, Tokuda H. The carboxyl-terminal region is essential for SecA dimerization. Biochem Biophys Res Commun. 1996;229:90–95. doi: 10.1006/bbrc.1996.1762. [DOI] [PubMed] [Google Scholar]
- 26.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsuyama S, Kimura E, Mizushima S. Complementation of two overlapping fragments of SecA, a protein translocation ATPase of Escherichia coli, allows ATP binding to its amino-terminal region. J Biol Chem. 1990;265:8760–8765. [PubMed] [Google Scholar]
- 28.Shinkai A, Mei LH, Tokuda H, Mizushima S. The conformation of SecA, as revealed by its protease sensitivity, is altered upon interaction with ATP, presecretory proteins, everted membrane vesicles and phospholipids. J Biol Chem. 1991;266:5827–5833. [PubMed] [Google Scholar]
- 29.Karamanou S, Sianidis G, Gouridis G, Pozidis C, Papanikolau Y, Papanikou E, Economou A. Escherichia coli SecA truncated at its termini is functional and dimeric. FEBS Lett. 2005;579:1267–1271. doi: 10.1016/j.febslet.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Jilaveanu LB, Zito CR, Oliver D. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci U S A. 2005;102:7511–7516. doi: 10.1073/pnas.0502774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Keyzer J, van der Sluis EO, Spelbrink REJ, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJM. Covalently dimerized SecA is functional in protein translocation. J Biol Chem. 2005;280:35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- 32.Jilaveanu LB, Oliver D. SecA dimer cross-linked at its subunit interface is functional for protein translocation. J Bacteriol. 2006;188:335–338. doi: 10.1128/JB.188.1.335-338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musial-Siwek M, Rusch SL, Kendall DA. Probing the affinity of SecA for signal peptide in different environments. Biochemistry. 2005;44:13987–13996. doi: 10.1021/bi050882k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shilton B, Svergun DI, Volkov VV, Koch MH, Cusack S, Economou A. Escherichia coli SecA shape and dimensions. FEBS Lett. 1988;436:277–282. doi: 10.1016/s0014-5793(98)01141-7. [DOI] [PubMed] [Google Scholar]
- 35.Bu Z, Wang L, Kendall DA. Nucleotide binding induces changes in the oligomeric state and conformation of SecA in a lipid environment: a small-angle neutron-scattering study. J Mol Biol. 2003;332:23–30. doi: 10.1016/s0022-2836(03)00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulbrandt ND, London E, Oliver DB. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 37.Ding H, Mukerji I, Oliver D. Lipid and signal peptide-induced conformational changes within the C-domain of Escherichia coli SecA protein. Biochemistry. 2001;40:1835–1843. doi: 10.1021/bi002058w. [DOI] [PubMed] [Google Scholar]
- 38.den Blaauwen T, Fekkes P, de Wit JG, Kuiper W, Driessen AJM. Domain interactions of the peripheral preprotein translocase subunit SecA. Biochemistry. 1996;35:11194–12004. doi: 10.1021/bi9605088. [DOI] [PubMed] [Google Scholar]
- 39.Or E, Boyd D, Gon S, Beckwith J, Rapaport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 40.Yahr TL, Wickner WT. Evaluating the oligomeric state of SecYEG in preprotein translocase. EMBO J. 2000;19:4393–4401. doi: 10.1093/emboj/19.16.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flower AM, Osborne RS, Silhavy TJ. The allele-specific synthetic lethality of prlA–prlG double mutants predicts interactive domains of SecY and SecE. EMBO J. 1995;14:884–893. doi: 10.1002/j.1460-2075.1995.tb07070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris CR, Silhavy TJ. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba T, Taura TT, Shimoike T, Akiyama T, Yoshihisa K. A cytoplasmic domain is important for the formation of a SecY–SecE translocator complex. Proc Natl Acad Sci U S A. 1994;91:4539–4543. doi: 10.1073/pnas.91.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pohlschröder M, Murphy C, Beckwith J. In vivo analyses of interactions between SecE and SecY, core components of the Escherichia coli protein translocation machinery. J Biol Chem. 1996;271:19908–19914. doi: 10.1074/jbc.271.33.19908. [DOI] [PubMed] [Google Scholar]
- 46.Manting EH, van der Does C, Driessen AJM. In vivo crosslinking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J Bacteriol. 1997;179:5699–5704. doi: 10.1128/jb.179.18.5699-5704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taura T, Yoshihisa T, Ito K. Protein translocation functions of Escherichia coli SecY: in vitro characterization of cold-sensitive SecY mutants. Biochimie. 1997;79:517–521. doi: 10.1016/s0300-9084(97)82744-7. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann A, Manting EH, Veenendaal AKJ, Driessen AJM, van der Does C. Cystein-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry. 1999;38:9115–9125. doi: 10.1021/bi990539d. [DOI] [PubMed] [Google Scholar]
- 49.Veenendaal AKJ, van der Does C, Driessen AJM. Mapping the sites of interaction between SecY and SecE by cystein scanning mutagenesis. J Biol Chem. 2001;276:32559–32566. doi: 10.1074/jbc.M103912200. [DOI] [PubMed] [Google Scholar]
- 50.Veenendaal AKJ, van der Does C, Driessen AJM. The core of the bacterial translocase harbors a tilted transmembrane segment 3 of SecE. J Biol Chem. 2002;277:36640–36645. doi: 10.1074/jbc.M205713200. [DOI] [PubMed] [Google Scholar]
- 51.Van der Sluis EO, Nouwen N, Driessen AJM. SecY–SecY and SecY– SecG contacts revealed by site-specific crosslinking. FEBS Lett. 2002;527:159–165. doi: 10.1016/s0014-5793(02)03202-7. [DOI] [PubMed] [Google Scholar]
- 52.Satoh Y, Matsumoto G, Mori H, Ito K. Nearest neighbor analysis of the SecYEG complex. I. Identification of a SecY–SecG interface. Biochemistry. 2003;42:7434–7441. doi: 10.1021/bi034331a. [DOI] [PubMed] [Google Scholar]
- 53.Mori H, Shimokawa N, Satoh Y, Ito K. Mutational analysis of transmembrane regions 3 and 4 of SecY, a central component of protein translocase. J Bacteriol. 2004;186:3960–3969. doi: 10.1128/JB.186.12.3960-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagamori S, Nishiyama K, Tokuda H. Two SecG molecules present in a single protein translocation machinery are functional even after cross-linking. J Biochem (Tokyo) 2000;128:129–137. doi: 10.1093/oxfordjournals.jbchem.a022723. [DOI] [PubMed] [Google Scholar]
- 56.Joly JC, Leonard MR, Wickner WT. Subunit dynamics in Escherichia coli preprotein translocase. Proc Natl Acad Sci U S A. 1994;91:4703–4707. doi: 10.1073/pnas.91.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tam PCK, Maillard AP, Chan KKY, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, III, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mori H, Tsukazaki T, Masui R, Kuramitsu S, Yokoyama S, Johnson AE, Kimura Y, Akiyama Y, Ito K. Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J Biol Chem. 2003;278:14257–14264. doi: 10.1074/jbc.M300230200. [DOI] [PubMed] [Google Scholar]
- 60.Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;413:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 61.Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kuhlbrandt W. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 2001;20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheuring J, Braun N, Nothdurft L, Stumpf M, Veenendaal AKJ, Kol S, van der Does C, Driessen AJM, Weinkauf S. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 63.Manting EH, van der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer TH, Ménétret JF, Breitling R, Miller KR, Akey CW, Rapoport TA. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 65.Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 66.Ménétret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 67.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Müller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 68.Manting EH, Kaufmann A, van der Does C, Driessen AJ. A single amino acid substitution in SecY stabilizes the interaction with SecA. J Biol Chem. 1999;274:23868–23874. doi: 10.1074/jbc.274.34.23868. [DOI] [PubMed] [Google Scholar]
- 69.Joly JC, Wickner W. The SecA and SecY subunits of translocase are the nearest neighbors of the translocating preprotein, shielding it from phospholipids. EMBO J. 1993;12:255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagamori S, Nishiyama K, Tokuda H. Membrane topology inversion of SecG detected by labeling with a membrane impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J Biochem (Tokyo) 2002;132:629–634. doi: 10.1093/oxfordjournals.jbchem.a003266. [DOI] [PubMed] [Google Scholar]
- 71.Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 72.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 73.Shimoike T, Akiyama Y, Baba T, Taura T, Ito K. SecY variants that interfere with Escherichia coli protein export in the presence of normal SecY. Mol Microbiol. 1992;6:1205–1210. doi: 10.1111/j.1365-2958.1992.tb01559.x. [DOI] [PubMed] [Google Scholar]
- 74.Matsumoto G, Yoshihisa T, Ito K. SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. EMBO J. 1997;16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mori H, Ito K. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci U S A. 2001;98:5128–5133. doi: 10.1073/pnas.081617398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiba K, Mori H, Ito K. Roles of the C-terminal end of SecY in protein translocation and viability of Escherichia coli. J Bacteriol. 2002;184:2243–2250. doi: 10.1128/JB.184.8.2243-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schägger H, Haase W, Duong F, Collinson I. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol. 2004;340:513–524. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 78.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 80.Verhoeven EE, Wyman C, Moolenaar GF, Goosen N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng W, Hsieh J, Brendza KM, Lohman TM. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 82.McGlynn P, Mahdi AA, Lloyd RG. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 2000;28:2324–2332. doi: 10.1093/nar/28.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 84.van der Does C, Tampé R. How do ABC transporters drive transport? Biol Chem. 2004;385:927–933. doi: 10.1515/BC.2004.121. [DOI] [PubMed] [Google Scholar]
- 85.Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider E, Wilken S, Schmid RJ. Nucleotide-induced conformational changes of MalK, a bacterial ATP binding cassette transporter protein. J Biol Chem. 1994;269:20456–20461. [PubMed] [Google Scholar]
- 87.Greller G, Horlacher R, DiRuggiero J, Boos WJ. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem. 1999;274:20259–20264. doi: 10.1074/jbc.274.29.20259. [DOI] [PubMed] [Google Scholar]
- 88.Hopfner KP, Karcher A, Crais L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 89.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 90.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GFL, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 91.Chang G, Roth CB. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 92.Beckstein O, Sansom MS. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol. 2004;1:42–52. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 93.Abramsom J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 94.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 95.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 96.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 97.Veenhoff LM, Heuberger EHML, Poolman B. Quaternary structure and function of transport proteins. Trend Biochem Sci. 2002;27:242–249. doi: 10.1016/s0968-0004(02)02077-7. [DOI] [PubMed] [Google Scholar]
- 98.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]