Abstract
Background
The hop plant (Humulus lupulus) is a source of many secondary metabolites, with bitter acids essential in the beer brewing industry and others having potential applications for human health. This study investigated variation in DNA sequence and gene expression of valerophenone synthase (VPS), a key gene in the bitter acid biosynthesis pathway of hop.
Methods
Sequence variation was studied in 12 varieties, and expression was analysed in four of the 12 varieties in a series across the development of the hop cone.
Results
Nine single nucleotide polymorphisms (SNPs) were detected in VPS, seven of which were synonymous. The two non-synonymous polymorphisms did not appear to be related to typical bitter acid profiles of the varieties studied. However, real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis of VPS expression during hop cone development showed a clear link with the bitter acid content. The highest levels of VPS expression were observed in two triploid varieties, ‘Symphony’ and ‘Ember’, which typically have high bitter acid levels.
Conclusions
In all hop varieties studied, VPS expression was lowest in the leaves and an increase in expression was consistently observed during the early stages of cone development.
Key words: Hop, Humulus lupulus, VPS (valerophenone synthase), real-time qRT-PCR, bitter acids, alpha acid, bitterness
INTRODUCTION
Hop (Humulus lupulus) is a dioecious perennial climbing plant belonging to the family Cannabaceae (Neve, 1991). The primary commercial application of the hop plant has historically been in the beer brewing industry; however, there is potential to develop new hop products, such as phytoceuticals, due to various biological activities (anti-bacterial, anti-fungal, anti-cancer, sedative, soporific and estrogenic) of secondary metabolites unique to hop (Moir, 2000; Dietz et al., 2005; Lust et al., 2005; Possemiers et al., 2005, 2006; Vanhoecke et al., 2005; Chadwick et al., 2006; Delmulle et al., 2006; Hougee et al., 2006). The female cone of the hop plant contains many lupulin glands, which are primarily located on the adaxial surfaces of the cone bracts. Secondary metabolites (some of which impart the bitter taste and aroma to beer), such as bitter acids (alpha-acids humulone and cohumulone, and beta-acids lupulone and colupulone), prenylflavonoids (such as xanthohumol) and essential oils (such as humulene and farnesene) accumulate in the lupulin glands (Likens et al., 1978; Haunold, 1980; Verzele and de Keukeleire, 1991; Paniego et al., 1999; Okada and Ito, 2001; Matoušek et al., 2002a).
A major aim of hop breeding is to improve the content and quality of the metabolites accumulated in the lupulin glands (Okada and Ito, 2001). Hop breeding methods are time consuming, with new varieties grown to full cone maturation over two years with high growing costs in order to establish chemical profiles. While the quantitative genetics of commercial traits in hop is poorly understood (Henning et al., 1997a, b; Murakami, 1999; Henning and Townsend, 2005), the relative levels of major chemicals, including the bitter acids (alpha-acids and beta-acids) and essential oils, are reasonably constant within a variety, regardless of environment (de Cooman et al., 1998; Heyerick et al., 2002), such that chemical profiles are considered a reliable method for varietal identification (de Keukeleire et al., 1998; Murakami, 1999; Auerbach et al., 2000; Eri et al., 2000; Roberts and Lewis, 2002; Henning et al., 2004). The stability of the chemical profile within a variety indicates that biosynthesis of these compounds is under genetic control. Understanding variation in the genes of the bitter acid biosynthetic pathway, in relation to the content and quality of the accumulation of bitter acids in the lupulin glands, is essential to guide the development of marker-assisted selection techniques in hop.
The formation of bitter acid precursors, phloroisovalerophenone (PIVP) and phloroisobutyrophenone (PIBP), occurs in a reaction very similar to the first step of flavonoid synthesis (Zuurbier et al., 1998b). In the biosynthetic pathway of the hop resins, the first step of the formation of flavonoids is catalysed by chalcone synthase (CHS; Zuurbier et al., 1998a). During cone development the pattern of accumulation of bitter acid intermediates PIVP and PIBP has been shown to differ when compared with naringenin (a flavonoid precursor), indicating that the bitter acids are not produced by CHS but another enzyme, valerophenone synthase (VPS; Zuurbier et al., 1995). The VPS protein was first isolated by Paniego et al. (1999), who found the amino acid sequence to share a high degree of homology with plant chalcone synthases. A genomic DNA sequence for the gene encoding the VPS enzyme was later obtained by an investigation into genes expressed specifically in the lupulin gland (Okada and Ito, 2001). Both CHS and VPS belong to the same plant-specific family of enzymes known as type III polyketide synthases (PKSs; Austin and Noel, 2003).
Despite a high degree of homology between the VPS and CHS genes and protein products, there are differences that result in a slight, but significant, change in substrate specificity (Paniego et al., 1999; Matoušek et al., 2002b; Novak et al., 2006). Chalcone synthase enzymes catalyse a claisen condensation reaction to form naringenin-chalcone from three molecules of malonyl-CoA and one molecule of p-coumaroyl-CoA (Austin and Noel, 2003). However, VPS preferentially uses a molecule of isovaleryl-CoA (or isobutyryl-CoA) in place of the p-coumaroyl-CoA molecule to catalyse the formation of PIVP (or PIBP) in a very similar reaction (Paniego et al., 1999; Austin and Noel, 2003). Many CHS homologues have been isolated in hop and analyses have been undertaken to compare them with VPS at a genetic and enzymatic level. Okada et al. (2004) studied the enzymatic activities for five of the CHS homologues and showed that the formation of naringenin-chalcone (a flavonoid precursor) is catalysed by CHS_H1 and at a much lower rate by VPS. The synthesis of PIVP was shown to be catalysed by VPS, and at a much lower rate by CHS_H1 (Okada et al., 2004). The synthesis of PIVP or naringenin-chalcone was not catalysed by CHS 2, CHS 3 or CHS 4. Several researchers have conducted Southern analyses with various VPS and CHS probes in hop with varying results. Matoušek et al. (2002a) detected at least five CHS-like genes related to CHS_H1 and at least two VPS-like genes with variation in gene copy number observed according to the specific genotype. Novak et al. (2003) also undertook Southern analyses using CHS_H1, VPS, CHS 2 and CHS 4 probes. In that study no cross-hybridization was observed between CHS_H1 and VPS. The differences between CHS 2, CHS 4 and VPS were less clear, with some cross-hybridization occurring (Novak et al., 2003). Both CHS 3 and CHS 4 were shown to contain an intron identical to that found in VPS, making them more VPS-like than CHS-like (Novak et al., 2003). Further, Novak et al. (2003) detected several as yet unidentified members of the VPS homologue gene family. Both Matoušek et al. (2002a) and Novak et al. (2003) found that the number of bands relating to these unidentified VPS-like genes differed between varieties. However, the number of genes detected is dependent on the stringency of the Southern blot and the probe used. Okada and Ito (2001) detected two VPS genes using a high-stringency analysis and several more with a low-stringency analysis using hop VPS cDNA as a probe. When the VPS promoter region was used as a probe, Okada et al. (2003) reported a single clearly identifiable VPS gene product. Novak et al. (2006) investigated the enzymatic properties of four chalcone synthase homologues, including VPS from hop by heterologous expression in Escherichia coli. They also determined that there were high levels of expression of chalcone synthase-like genes in maturing hop cones of cultivars with high bitter acid content by northern and western blotting using probes specific for VPS, CHS_H1, CHS 4 and polyspecific serum raised against recombinant protein CHS 4, respectively.
This study investigated sequence variation (single nucleotide polymorphisms, SNPs) in VPS in twelve varieties, and investigated changes in the expression of VPS during cone development in four varieties. SNPs are the most common type of sequence variation between gene alleles and can be used as tools in genetic mapping, estimating population diversity and correlating genotype with phenotype. Of specific relevance to polyploid hop breeding is the fact that allelic discrimination through SNP detection has been demonstrated in polyploid potato, permitting distinction not only between homo- and heterozygosity, but also among different heterozygous states (Rickert et al., 2002, 2005). Expression levels were determined using real-time quantitative PCR (qRT-PCR) and ‘normalized’ using hop polyubiquitin as a control gene. Polyubiquitin is a small, highly conserved protein that has been found in the cells of all eukaryotes so far analysed (Kawalleck et al., 1993). The gene encoding the polyubiquitin protein is often used as a control or ‘housekeeping’ gene to normalize real-time qRT-PCR results due to its constitutive expression in plant tissues at all developmental stages (Sun et al., 1997; Brunner et al., 2004). A comparison of ten common housekeeping genes for real-time qRT-PCR in a Populus hybrid revealed polyubiquitin to display the highest stability index (least variation) when comparing different tissue samples (Brunner et al., 2004), indicating that it is currently the most suitable candidate for use as an internal control in real-time qRT-PCR studies. Few gene expression analyses using real-time qRT-PCR have been carried out in hop. Genbank searches revealed that no actin-like or polyubiquitin-like genes exhibiting constitutive expression have been isolated from the hop genome. For example, Matoušek et al. (2002a) used 7SL RNA as a reference sample for real-time qRT-PCR analyses. However, this has been shown in a previous study (Matoušek et al., 1999) to exhibit varying transcription rates, making it unsuitable as an internal control gene. The present study is the first to isolate polyubiquitin (UBI) in hop for use as an internal control for normalizing results in real-time qRT-PCR.
MATERIALS AND METHODS
Plant material
Plant material from twelve hop (Humulus lupulus L.) varieties was sampled from the breeding garden maintained by Hop Products Australia at the Bushy Park Estates, Tasmania, Australia (42°42′33′′S, 146°53′54′′E). Three of these varieties are closely related. The diploid variety ‘J78’ is an open-pollinated (OP) progeny of the diploid variety ‘Pride of Ringwood’, and the variety ‘Ember’ is an OP progeny of tetraploid ‘Pride of Ringwood’. The variety ‘Victoria’ is an OP progeny of tetraploid ‘J78’. From four varieties (‘Saaz ×’, ‘J78’, ‘Symphony’ and ‘Ember’; where ‘Saaz ×’ refers to an open-pollinated progeny of diploid ‘Saaz’ grown in Tasmania), four samples were freshly collected and snap-frozen in liquid nitrogen on site over a period of 5 weeks as follows: (1) young leaf, and cones at (2) early, (3) mid, and (4) late stages of development. Samples were stored at –80 °C until nucleic acid extraction. The early cone stage was characterized by the onset of cone development (from flowers); all early-stage cones were <1 cm in width. Mid-development cones were of a size intermediate between the early stage and the most developed cones for each variety. Cones at the mid-development stage were soft in texture and varied from 1–3 cm in width. The late cone development stage was defined by the maximum size that cones would reach prior to drying out for each variety. All cones in the late category were showing the first signs of cone maturation as determined by texture (the cones were still soft, but with some bracts papery).
DNA sequencing
DNA extraction
DNA was extracted from young leaves of the twelve hop varieties for use in sequencing VPS. Several leaves from each variety were ground to a fine powder under liquid nitrogen using a mortar and pestle. DNA was extracted from approximately 100 mg of ground leaf tissue using a Qiagen DNeasy Plant Minikit (Qiagen, Hilden, Germany). The yield of genomic DNA was quantified by electrophoresis through a 2 % agarose gel dissolved in Tris-borate-ethylenediaminetetraacetic acid (TBE) stained with ethidium bromide and examined under ultraviolet light.
Primer design and PCR
Hop VPS primers 5′VPS-START and 3′VPS-END for PCR and 5′VPS-S2 and 3′VPS-S1 for sequencing, were designed by eye to the only hop VPS gDNA sequence available in Genbank (Genbank accession number AB047593 from variety ‘9418R’; Okada and Ito, 2001; Table 1). The gDNA sequence AB047593 is 3910 bp in length and includes the promoter and 5′ untranslated region of VPS. This study sequenced the coding region of VPS and, as such, the start of the coding region at base 2642 of AB047593 was designated here as base 1. The forward PCR primer 5′VPS-START was positioned at base 3–22 (bases 2644–2663 of AB047593) and the reverse PCR primer 3′VPS-END at base 1250–1269 (bases 3891–3910 of AB047593). This 1269-bp sequence spans the complete coding region, which includes a first exon of 186 bp, a single intron of 84 bp and a second exon of 999 bp. Primer properties were determined using Oligo Analyzer 1·0·2 software (Teemu Kuulasmaa©, Kuopio, Finland). Polymerase chain reaction was conducted for each of the twelve varieties (Table 2) using approx. 100 ng of gDNA in a 100-μL reaction containing a final concentration of 0·25 mm dNTPs, 3 mm MgCl2 and 1 mm primers. The thermal-cycling program was a first cycle of 2 min at 94 °C followed by 35 amplification cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 52 °C and extension of 1 min at 72 °C, with a final cycle of 5 min at 72 °C. PCR products were visualized by electrophoresis on a 2 % agarose gel stained with ethidium bromide, purified using a QIAquick PCR purification kit (Qiagen) and quantified using a spectrophotometer (BioRad Laboratories, Hercules, CA, USA).
Table 1.
Primers used for PCR sequencing and real-time qRT-PCR, amplicon length and optimal annealing temperatures
| Gene | Primer | Sequence (5′ → 3′) | Amplicon length (bp) | Optimum Ta (°C) |
|---|---|---|---|---|
| VPS | 5′VPS-START | GGCGTCCGTAACTGTAGAGC | 1269 | 52 |
| 3′VPS-END | TTAGACGTTTGTGGGCACGC | |||
| 5′VPS-S2 | GTTGAAGTTCCCAAGCTTGG | 582 | 60 | |
| 3′VPS-S2 | AATCCCAATCGGAGTGAAGG | |||
| 5′VPS-RTF1 | CCAACGAATGTGTGAAAAATCCAC | 119 | 60 | |
| 3′VPS-RTR1 | CTTGGCGTGTGTTCAGAGATGG | |||
| UBI | UBI-F | ATGCAGATYTTTGTGAAGAC | 55 | |
| UBI-R | ACCACCACGRAGACGGAG | |||
| Plasmid | pGEMT-F | GCCCGACGTCGCATGCTCC | 50 | |
| pGEMT-R | GAGCTCTCCCATATGGTCG |
Table 2.
The varieties studied, their typical alpha-acid levels, the start, end, and length of DNA sequences obtained from the twelve samples relative to ‘9418R’ (where base 1 of the sequences from the varieties in this study refers to base 2642 of AB047593), the nucleotide bases at which polymorphisms were observed, and the amino acids encoded by non-synonymous polymorphisms observed. ‘Saaz ×’ refers to a progeny of ‘Saaz’, and ‘Hallertau mf’ refers to ‘Hallertau mittelfrueh’. Triploid varieties are indicated by bold, all other varieties are diploid
| Nucleotide Base |
Amino acid residue |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variety | Genbank accession | α-acid | Start | End | 351 | 723 | 762 | 779† | 780† | 874‡ | 909 | 1005 | 1084 | 232† | 264‡ |
| ‘9418R’ | AB047593 | – | 1 | 1269 | T | G | T | C | C | G | A | C | C | Ala | Ala |
| ‘Saaz ×’* | EU685798 | Low | 45 | 1238 | T/T | A/A | G/G | A/C | G/G | G/G | A/A | C/C | C/C | Ala/Glu | Ala |
| ‘Cascade’ | EU685789 | Low | 34 | 1238 | T/T | A/G | T/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘Cluster’ | EU685791 | Low | 44 | 1230 | T/T | A/A | G/G | A/A | G/G | G/G | A/A | C/C | C/C | Glu | Ala |
| ‘Golding’ | EU685792 | Low | 44 | 1238 | T/T | A/G | G/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘Hallertau mf’ | EU685790 | Low | 44 | 1239 | T/T | A/G | G/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘Eastern Gold’ | EU685793 | High | 27 | 1238 | T/T | A/G | T/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘Nugget’ | EU685794 | High | 44 | 1238 | T/T | A/A | G/G | A/A | G/G | G/G | A/A | C/C | C/C | Glu | Ala |
| ‘Opal’ | EU685795 | High | 45 | 1230 | T/T | A/G | G/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘J78’* | EU685797 | High | 165 | 1269 | T/T | A/G | T/T | C/C | G/C | G/G | A/A | C/C | C/C | Ala | Ala |
| ‘Symphony’* | EU685799 | High | 177 | 1238 | C/T | A/A | G/T | A/C | G/G | G/T | A/C | G/C | C/T | Ala/Glu | Ala/Ser |
| ‘Ember’* | EU685800 | High | 45 | 1238 | T/T | A/G | G/T | A/C | G/C | G/G | A/A | C/C | C/C | Ala/Glu/Asp | Ala |
| ‘Victoria’ | EU685796 | High | 45 | 1238 | T/T | A/G | G/T | A/C | G/C | G/G | A/A | C/C | C/C | Ala/Glu/Asp | Ala |
* Indicates varieties in which the gene expression of VPS was studied.
† and ‡ indicate non-synonymous nucleotide polymorphisms, and the corresponding amino acid residue of the VPS protein encoded. Ala = alanine; Glu = glutamic acid; Asp = aspartic acid; and Ser = serine.
Sequencing
Purified VPS PCR products for each of the twelve varieties were sequenced using Beckman CEQ2000 dye terminator cycle-sequencing technology (CEQ DTCS Quick Start Kit, Beckman Coulter, Fullerton, CA, USA) in a Beckman CEQ8000 according to the manufacturer's instructions. Sequencing reactions were performed using the original PCR primers 5′VPS-START and 3′VPS-END, and internal primers 5′VPS-S2 and 3′VPS-S1 (Table 1). Forward and reverse sequences for each variety were aligned using Sequencher software (version 4·1·2, Gene Codes Corporation MI, USA), the automated base calls checked visually by inspection of the electropherograms, a consensus sequence generated for each variety, and aligned to the reference VPS sequence AB047593. Where polymorphisms were detected, the genotype for each variety was recorded, except in the case of the triploid varieties (‘Symphony’, ‘Ember’ and ‘Victoria’), where it was possible to identify the bases present at heterozygous sites, but not possible to determine the allelic frequencies. Nucleotide sequences were translated and the predicted amino acid sequences aligned using ClustalX software (Thompson et al., 1997) to determine if SNPs altered the amino acid sequence encoded.
RNA extraction and cDNA synthesis
For each of the four varieties (‘Saaz ×’, ‘J78’, ‘Symphony’ and ‘Ember’) in which the expression of VPS was studied (Table 2), duplicate leaf, early, mid- and late-stage cones were ground to a fine powder under liquid nitrogen using a sterile mortar and pestle. RNA was extracted from approximately 100 mg of frozen ground tissue using the Qiagen RNeasy Plant Minikit and RLT extraction buffer with 50 µL Plant RNA Isolation Aid (Ambion, Austin, TX, USA) added. RNA was DNase treated (Ambion DNA-free kit), the quality checked by electrophoresis through a 2 % agarose gel stained with ethidium bromide and high-quality RNA quantified using a spectrophotometer. Synthesis of cDNA was from 2 µg of RNA in 60-μL reaction volume using the Superscript III first-strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) and random hexamers, and from 1 µg of RNA in 30-μL reaction volume without the reverse transcriptase enzyme (negative control denoted RT-) for each sample.
Gene expression with real-time qRT-PCR
Real-time qRT-PCR VPS primers, 5′VPS-RTF1 and 3′VPS-RTR1, were designed to regions where no polymophisms were detected in the Genbank sequence AB047593 and the new VPS sequence obtained for the twelve varieties in this study (Table 2). Primers were checked for cross-reactivity to hop CHS Genbank sequences: CHS 2 mRNA (AB061021), CHS 2 gDNA (AB061020), CHS_H1 gDNA (AJ304877), CHS 3 gDNA (AB061022), CHS 4 gDNA (AJ430353), novel CHS 1 (BD182131), novel CHS 2 gDNA (BD182132), novel CHS 3 (BD182133) and novel CHS 4 gDNA (BD182134) to ensure specificity to VPS. The forward primer 5′VPS-RTF1 was positioned across the boundary of exon 1 and the single intron to prevent amplification of background gDNA. The amplicon size from cDNA was 119 bp and the position of the intron was known from comparing gDNA (AB047593) to the corresponding mRNA sequence (AB015430; Okada and Ito, 2001).
Polyubiquitin (UBI) was used as an internal control gene to normalize real-time qRT-PCR data. Degenerate UBI primers (Albrecht et al., 1998) were used to amplify hop UBI from gDNA and cDNA using a PCR program of: one cycle of 3 min at 94 °C, 35 cycles of 30 s at 94 °C, 45 s at 55 °C and 1 min at 72 °C, with a final cycle of 5 min for 72 °C. The same cycling parameters were used for the amplification of VPS using 5′VPS-RTF1 and 3′VPS-RTR1, with an annealing temperature of 60 °C (Table 1), with Promega GoTaq Green and cDNA as a template. For use as standards in real-time qRT-PCR, purified PCR products for VPS and UBI were cloned into the pGEM-T-Easy Plasmid Vector System II (Promega, Madison, WI, USA). Plasmid DNA was purified using the Wizard Plus SV Minipreps DNA Purification System (Promega) and at least five clones sequenced in both directions using the plasmid promoter primers SP6 5′-ATTTAGGTGACACTATAGAA-3′ and T7 5′-AATACGACTCACTATAGGG-3′. Forward and reverse sequences were aligned, the automated base calls checked visually by inspection of the electropherograms, and a BLAST search of Genbank performed in order to confirm specific amplification of VPS and UBI. A single plasmid clone for 119-bp VPS and 238-bp UBI was selected and seven 10-fold serial dilutions from 1 ng μL–1 to 1 × 10–7 ng μL–1 (standard curves) were used in all real-time qRT-PCR assays to determine relative gene expression levels between assays.
Duplicate VPS and UBI real-time qRT-PCR reactions were performed for each of two samples per variety. Reactions were performed on RT+ (cDNA) and RT– (no RT) using 2 µL cDNA or RT– (diluted 1 : 5), 0·5 µm primers and 1× iQ SYBR Green Supermix (BIO-RAD) in 10-μL reaction volumes, with two negative controls per assay on a Rotorgene2000 (Corbett Research, Sydney, Australia) using the following program: 95 °C for 3 min, 50 cycles of 95 °C for 5 s and 60 °C (VPS) and 55 °C (UBI) for 40 s. A melt-curve to check for a single product was determined by ramping from 72 °C to 95 °C, acquiring every 1 °C, waiting 45 s after the first step and then waiting 5 s after each step thereafter. Expression of VPS (Ct values) was calculated relative to the VPS standard curve as an average of duplicate reactions for each sample. That result was then normalized against the expression of UBI (also in relation to a UBI standard curve). All assays (including standard curves) had an R2 ≥ 0·99 and efficiency above 80 %.
RESULTS
Sequence variation
Only clean, unambiguous sequences were used for analysis. Polymorphic and heterozygous sites identified within the 12 varieties studied are recorded in Table 2. The length of clean VPS sequence obtained ranged from 1062 bp in the variety ‘Symphony’ to 1212 bp in the variety ‘Eastern Gold’. The start of the VPS sequence was consistently ambiguous in the varieties ‘J78’ and ‘Symphony’. Comparative analysis of the VPS sequence in all 12 varieties revealed nine polymorphic loci (Table 2). ‘Symphony’ was the only variety with polymorphisms at bases 874, 909, 1005 and 1084. Translation of the VPS sequence revealed that three of the polymorphisms (at bases 779, 780 and 874) were non-synonymous. The polymorphisms at bases 779 and 780 occurred within the same codon, resulting in a change in the amino acid encoded, with either alanine (GCN, AB047593; ‘Saaz ×’, ‘Cascade’, ‘Golding’, ‘Hallertau mittelfrueh’, ‘Eastern Gold’, ‘Opal’, ‘J78’,), glutamic acid (GAG; ‘Saaz ×’, ‘Cluster’, ‘Nugget’, ‘Symphony’, ‘Ember’ and ‘Victoria’) or aspartic acid (GAC; ‘Ember’ and ‘Victoria’) at residue 232 of the VPS protein (Table 2). ‘Ember’ and ‘Victoria’ were heterozygous at bases 779 and 780, resulting in three possible amino acids at residue 232 of the VPS protein. All varieties were homozygous for the G allele at nucleotide base 874, except for ‘Symphony’. This polymorphism resulted in a non-synonymous change in the amino acid encoded, with either alanine (GCC) or serine (TCC) at residue 264 of the VPS protein (Table 2).
Expression of VPS
The degenerate UBI primers of Albrecht et al. (1998) amplified three fragments of 238, 480 and 700 bp in hop. The shortest fragment (238 bp) showed a high degree of identity to other polyubiquitin sequences from plants in Genbank [for example, 86 % with polyubiquitin 1, AF527441, from Phaseolus vulgaris, and 84 % with polyubiquitin (UBQ10), NM_178970, from Arabidopsis thaliana]. The cloned 238-bp fragment (UBI238; Genbank accession number EU700059) was successfully used as an internal control to normalize VPS expression measured by real-time qRT-PCR.
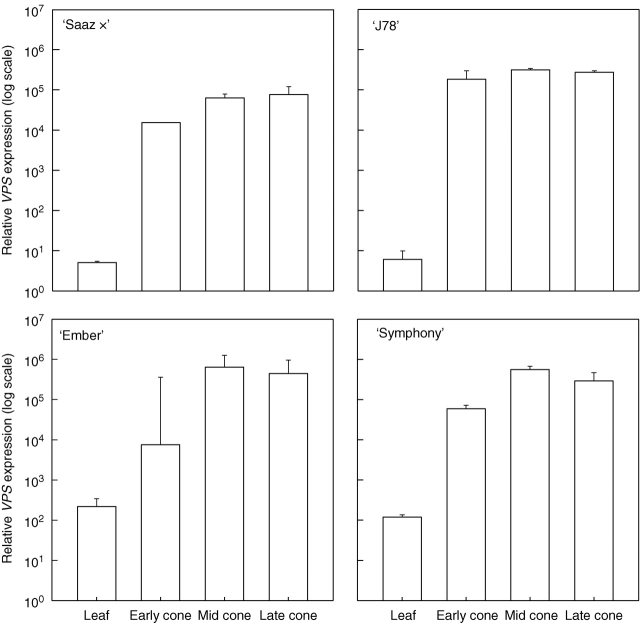
Real-time qRT-PCR analyses revealed that VPS was expressed at low levels in leaves of all four varieties tested (Fig. 1). All varieties exhibited higher levels of VPS expression in cones, with a large increase in the expression of VPS occurring early in cone development. Expression of VPS appeared to reach a plateau at relatively high levels later in cone development in all four varieties (Fig. 1). The lowest maximum expression of VPS was observed in ‘Saaz ×’, the only low alpha-acid variety studied. The maximum expression of VPS in ‘Saaz ×’ (∼1 × 105; Fig. 1) was approximately one tenth of the maximum expression in ‘J78’, ‘Symphony’ or ‘Ember’ (∼1 × 106; Fig. 1), all of which are high alpha-acid varieties.
Fig. 1.
Relative VPS gene expression during cone development in four varieties of hop.
DISCUSSION
Sequence variation
Prior to this study the only other published VPS sequences available were the genomic DNA sequence with Genbank accession number AB047593 and the corresponding messenger RNA with Genbank accession number AB015430 (Okada et al., 2001). This study examined DNA sequence variation in the VPS gene between hop varieties exhibiting a broad range of alpha-acid levels. Nine polymorphic sites were documented. The majority (six of the nine) of polymorphisms observed in the VPS gDNA sequences were found to be silent, encoding the same amino acid in the twelve hop varieties studied. The other polymorphisms (at bases 779, 780 and 874; Table 2) were non-synonymous, encoding changes in the amino acids at residues 232 and 264 of the VPS protein. Further, the amino acids encoded exhibit chemical properties differing from those of the amino acid encoded in the reference sequence (AB047593). Alanine (encoded at residue 232 in all varieties but ‘Cluster’ and ‘Nugget’) is an inactive hydrophobic amino acid. Both these varieties are homozygous for nucleotides encoding glutamic acid, while other varieties (‘Saaz ×’, ‘Symphony’ and ‘Ember’) are heterozygous. In the triploid varieties ‘Ember’ and ‘Victoria’, a third amino acid, aspartic acid, is potentially encoded at residue 232. Both glutamic acid and aspartic acid are acidic, and therefore may change the behaviour of the protein when compared to alanine. The polymorphism observed in ‘Symphony’ at base number 874 encodes the amino acid serine, as opposed to alanine found in all other varieties. These two residues were not documented as sites that influence the substrate specificity of VPS in a comprehensive study by Austin and Noel (2003).
On the basis of this small sample, no polymorphisms appear to consistently co-occur in either high or low alpha-acid varieties. Linkage mapping and quantitative trait loci analysis, using the SNPs identified in the present study would be required to establish a quantitative relationship between sequence variation observed in VPS and known hop acid levels. Further comparative sequence analyses of the upstream promoter region the VPS may reveal DNA sequence variation more closely linked with the accumulation of bitter acids. The importance of the variation at residues 232 and 264 cannot be ascertained without further studies testing the different efficiencies and specificities of the translation products of each VPS sequence.
Copy number of the VPS gene
Our inability to obtain clear sequence data at the start of VPS in varieties ‘J78’ and ‘Symphony’ may be due to the presence in the genome of more than one VPS homologue. The high level of heterozygosity observed (Table 2) may imply that multiple copies of VPS are present. It was not possible to discern whether the heterozygosity observed in ‘Symphony’ (at seven out of nine polymorphic sites) was due to allelic variation or the presence of a VPS homologue. However, the purity of the PCR product from which VPS was sequenced suggests that a single VPS sequence was amplified and any variation was due to allelic copies, or homologues of identical size.
The variation observed in the VPS coding sequence relating to residues 232 and 264 of the protein may not be translated into protein if the polymorphisms occur in redundant homologues of VPS, or, if translated, the protein formed may be non-functional. However, it is important to note that if VPS was present in the hop genome at more than one locus and the homologues are being expressed, then real-time qRT-PCR analyses would only detect these copies if sequences were identical at the primer locations (Table 1). The real-time qRT-PCR primers for VPS were checked against all CHS-like sequences available from hop to ensure specificity to VPS. PCR using these primers was shown to amplify only one clean fragment of the expected size and sequence. This suggests that regardless of whether more VPS homologues exist, the real-time qRT-PCR analyses quantified the relative expression levels of only one VPS copy, identical to that studied by Okada et al. (2004).
VPS expression
This study was the first to use real-time qRT-PCR to quantify VPS expression in the leaf and during the development of hop cones. The results of the real-time qRT-PCR analyses revealed the same overall trend for VPS expression within both the high and low alpha-acid varieties. Extremely low levels of expression of VPS were found in the leaves of all of the four varieties analysed (Fig. 1). Similarly, other studies have found that hop acids are present at extremely low levels in leaves (de Keukeleire et al., 2003). The expression of VPS was found to be considerably higher in all cone samples of all four varieties, and the expression of VPS appeared to reach a plateau at relatively high levels in the mid and late cones sampled (Fig. 1). A histochemical assay using the VPS promoter by Okada et al. (2003) revealed that the centre of the lupulin gland was the site at which VPS was expressed, and that mature glands did not stain as strongly as young glands. Okada et al. (2003) suggested that VPS expression levels were higher at an earlier cone stage. The late stage cones sampled in the present study were ripe, but not yet senescing. It is possible that the older lupulin glands sampled by Okada et al. (2003) were obtained from cones at a later stage of development than those sampled in this study. However, Okada et al. (2003) also suggested that oily metabolites in the lupulin gland were possibly interfering with the histochemical assay performed.
Studies monitoring the levels of alpha-acids and beta-acids during development from female inflorescences to cones have revealed a gradual increase in these compounds, with rates of hop-acid accumulation differing between varieties (Zuurbier et al., 1995; de Keukeleire et al., 2003; Novak et al., 2006). If VPS were responsible for the levels of the alpha-acids and beta-acids, then a characteristic rate of accumulation would be apparent in the rate of increase in VPS expression throughout cone development in a particular variety. Figure 1 shows that the rate of increase of VPS gene expression did appear to differ between varieties. Varieties with high alpha-acid levels (‘J78’, ‘Symphony’ and ‘Ember’) showed the highest outright levels of VPS expression and the highest rates of increase in VPS expression between leaf and hop cone. It is also noteworthy that the expression of VPS in leaves and the maximum expression in cones were higher in the triploid varieties (‘Symphony’ and ‘Ember’; Fig. 1). While this study incorporated only four varieties, the pattern of VPS expression observed between the triploid and diploid varieties may be suggestive of an effect of ploidy on the base level of expression of VPS, possibly through changes in the regulation of gene expression.
Concluding remarks
No significant correlation was observed between the high alpha-acid varieties and a particular VPS gene sequence or polymorphism. The quantification of VPS expression in leaves and cones at different stages of development and between varieties revealed a clear link between VPS expression and typical alpha-acid levels for each variety. The two varieties with the highest typical levels of bitter acids, the triploids ‘Symphony’ and ‘Ember’, also displayed the highest relative VPS expression levels. The general trend for VPS expression that was observed in all varieties was low levels of VPS expression in leaves and an increase in expression with the development of the hop cone from early to mid and late stages.
The bitter acid pathway requires further investigation in order to characterize the steps and genes involved. Importantly, this study has shown that increases in the expression of VPS occur early in cone development, and as such further studies of changes in expression of VPS during cone development in hop should be based on a more refined sampling strategy, such as harvesting cones at precise times post-flowering and fertilization, and determination of the chemical composition of the lupulin glands of the individual cones investigated. The target gene approach taken in this study has allowed direct assessment of the expression of the VPS gene in four varieties. Sequencing this gene in further samples has shown a degree of polymorphism in this key enzyme of the bitter acid biosynthesis pathway. Future comparative studies of DNA sequence variation in VPS should use a larger sample size, from full sib material in linkage mapping and quantitative trait loci studies to test for quantitative relationships between the SNPs identified in the present study and the chemical profiles of the genotypes under investigation. If bitter acid accumulation is controlled by VPS, and polymorphisms in the coding region of VPS do not influence bitter acid accumulation, then variation in the DNA sequence of the upstream regulatory regions of VPS may be more closely linked to bitter acid biosynthesis. Understanding the regulation of bitter acid biosynthesis would assist in the development of an understanding about how environmental variation affects the production of bitter acids, leading to improved crop management, and could also lead to the development of marker-assisted selection techniques for breeding plants with improved production of these economically important plant secondary metabolites.
ACKNOWLEDGEMENTS
Funding from Hop Products Australia and Horticulture Australia Limited supported this research. The authors would like to acknowledge the work of Grey Leggett, the plant breeder for Hop Products Australia from 1988 to 2006.
LITERATURE CITED
- Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant Journal. 1998;15:605–614. doi: 10.1046/j.1365-313x.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- Auerbach RH, Dost K, Davidson G. Characterization of varietal differences in essential oil components of hops (Humulus lupulus) by SFC-FTIR spectroscopy. Journal of the Association of Analytical Communities International. 2000;83:621–626. [PubMed] [Google Scholar]
- Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Natural Product Reports. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology. 2004;4:14. doi: 10.1186/1471-2229-4-14. doi:10·1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LR, Pauli GF, Farnsworth NR. The pharmacognosy of Humulus lupulus L. (hops) with an emphasis on estrogenic properties. Phytomedicine. 2006;13:119–131. doi: 10.1016/j.phymed.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cooman L, Everaert E, de Keukeleire D. Quantitative analysis of hop acids, essential oils and flavonoids as a clue to the identification of hop varieties. Phytochemical Analysis. 1998;9:145–150. [Google Scholar]
- Delmulle L, Bellahcene A, Dhooge W, Comhaire F, Roelens F, Huvaere K, et al. Anti-proliferative properties of prenylated flavonoids from hops (Humulus lupulus L.) in human prostate cancer cell lines. Phytomedicine. 2006;13:732–734. doi: 10.1016/j.phymed.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Dietz BM, Kang YH, Liu GW, Eggler AL, Yao P, Chadwick LR, et al. Xanthohumol isolated from Humulus lupulus inhibits menadione-induced DNA damage through induction of quinone reductase. Chemical Research in Toxicology. 2005;18:1296–1305. doi: 10.1021/tx050058x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eri S, Khoo BK, Lech J, Hartman TG. Direct thermal desorption-gas chromatography and gas chromatography-mass spectrometry profiling of hop (Humulus lupulus L.) essential oils in support of varietal characterization. Journal of Agricultural and Food Chemistry. 2000;48:1140–1149. doi: 10.1021/jf9911850. [DOI] [PubMed] [Google Scholar]
- Haunold A. In: Hybridization of crop plants. Fehr WR, Hadley HH, editors. Madison, WI: American Society of Agronomy; 1980. Hop. [Google Scholar]
- Henning JA, Townsend MS. Field-based estimates of heritability and genetic correlations in hop. Crop Science. 2005;45:1469–1475. [Google Scholar]
- Henning J, Haunold A, Nickerson G. Genetic parameter estimates for five traits in male hop accessions: a preliminary study. Journal of the American Society of Brewing Chemists. 1997;a 55:157–160. [Google Scholar]
- Henning J, Haunold A, Nickerson G, Gampert U. Estimates of heritability and genetic correlation for five traits in female hop accessions. Journal of the American Society of Brewing Chemists. 1997;b 55:161–165. [Google Scholar]
- Henning JA, Steiner JJ, Hummer KE. Genetic diversity among world hop accessions grown in the USA. Crop Science. 2004;44:411–417. [Google Scholar]
- Heyerick A, De Keukeleire D, Van Peteghem C, De Saeger S. Modulation of the phytoestrogenicity of beer by monoterpene alcohols present in various hop oil fractions. Journal of the Institute of Brewing. 2002;108:94–101. [Google Scholar]
- Hougee S, Faber J, Sanders A, van den Berg WB, Garssen J, Smit HF, Hoijer MA. Selective inhibition of COX-2 by a standardized CO2 extract of Humulus lupulus in vitro and its activity in a mouse model of zymosan-induced arthritis. Planta Medica. 2006;72:228–233. doi: 10.1055/s-2005-916212. [DOI] [PubMed] [Google Scholar]
- Kawalleck P, Somssich I, Feldbrugge M, Hahlbrock K, Weisshaar B. Polyubiquitin gene expression and structural properties of the ubi4-2 gene in Petroselinum crispum. Plant Molecular Biology. 1993;21:673–684. doi: 10.1007/BF00014550. [DOI] [PubMed] [Google Scholar]
- de Keukeleire D, David F, Haghebaert K, Sandra P. Automated reporting on the quality of hops and hop products. Journal of the Institute of Brewing. 1998;104:75–82. [Google Scholar]
- de Keukeleire J, Ooms G, Heyerick A, Roldan-Ruiz I, Van Bockstaele E, De Keukeleire D. Formation and accumulation of α-acids, β-acids, desmethylxanthohumol, and xanthohumol during flowering of hops (Humulus lupulus L.) Journal of Agricultural and Food Chemistry. 2003;51:4436–4441. doi: 10.1021/jf034263z. [DOI] [PubMed] [Google Scholar]
- Likens ST, Nickerson GB, Haunold A, Zimmermann CE. Relationship between alpha-acids, beta-acids, and lupulin content of hops. Crop Science. 1978;18:380–386. [Google Scholar]
- Lust S, Vanhoecke B, Janssens A, Philippe J, Bracke M, Offner F. Xanthohumol kills B-chronic lymphocytic leukemia cells by an apoptotic mechanism. Molecular Nutrition & Food Research. 2005;49:844–850. doi: 10.1002/mnfr.200500045. [DOI] [PubMed] [Google Scholar]
- Matoušek J, Junker V, Vrba L, Schubert J, Patzak J, Steger G. Molecular characterization and genome organization of 7SL RNA genes from hop (Humulus lupulus L.) Gene. 1999;239:173–183. doi: 10.1016/s0378-1119(99)00352-2. [DOI] [PubMed] [Google Scholar]
- Matoušek J, Novak P, Briza J, Patzak J, Niedermeierova H. Cloning and characterisation of chs-specific DNA and cDNA sequences from hop (Humulus lupulus L.) Plant Science. 2002;a 162:1007–1018. [Google Scholar]
- Matoušek J, Novak P, Patzak J, Briza J, Krofta K. Analysis of true chalcone synthase from Humulus lupulus L. and biotechnology aspects of medicinal hops. Rostlinna Vyroba. 2002;b 48:7–14. [Google Scholar]
- Moir M. Hops – a millennium review. Journal of the American Society of Brewing Chemists. 2000;58:131–146. [Google Scholar]
- Murakami A. Inheritance of major chemical components in hops. Journal of the Institute of Brewing. 1999;105:107–111. [Google Scholar]
- Neve RA. Hops. London: Chapman and Hall; 1991. [Google Scholar]
- Novak P, Matousek J, Briza J. Valerophenone synthase-like chalcone synthase homologues in Humulus lupulus. Biologia Plantarum. 2003;46:375–381. [Google Scholar]
- Novak P, Krofta K, Matousek J. Chalcone synthase homologues from Humulus lupulus: some enzymatic properties and expression. Biologia Plantarum. 2006;50:48–54. [Google Scholar]
- Okada Y, Ito K. Cloning and analysis of valerophenone synthase gene expressed specifically in lupulin gland of hop (Humulus lupulus L.) Bioscience Biotechnology and Biochemistry. 2001;65:150–155. doi: 10.1271/bbb.65.150. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamazaki Y, Suh DY, Sankawa U, Ito K. Bifunctional activities of valerophenone synthase in hop (Humulus lupulus L.) Journal of the American Society of Brewing Chemists. 2001;59:163–166. [Google Scholar]
- Okada Y, Saeki K, Inaba A, Suda N, Kaneko T, Ito K. Construction of gene expression system in hop (Humulus lupulus) lupulin gland using valerophenone synthase promoter. Journal of Plant Physiology. 2003;160:1101–1108. doi: 10.1078/0176-1617-01116. [DOI] [PubMed] [Google Scholar]
- Okada Y, Sano Y, Kaneko T, Abe I, Noguchi H, Ito K. Enzymatic reactions by five chalcone synthase homologs from hop (Humulus lupulus L) Bioscience Biotechnology and Biochemistry. 2004;68:1142–1145. doi: 10.1271/bbb.68.1142. [DOI] [PubMed] [Google Scholar]
- Paniego NB, Zuurbier KWM, Fung SY, van der Heijden R, Scheffer JJC, Verpoorte R. Phlorisovalerophenone synthase, a novel polyketide synthase from hop (Humulus lupulus L.) cones. European Journal of Biochemistry. 1999;262:612–616. doi: 10.1046/j.1432-1327.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Possemiers S, Heyerick A, Robbens V, De Keukeleire D, Verstraete W. Activation of proestrogens from hops (Humulus lupulus L.) by intestinal microbiota; conversion of isoxanthohumol into 8-prenylnaringenin. Journal of Agricultural and Food Chemistry. 2005;53:6281–6288. doi: 10.1021/jf0509714. [DOI] [PubMed] [Google Scholar]
- Possemiers S, Bolca S, Grootaert C, Heyerick A, Decroos K, Dhooge W, et al. The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. Journal of Nutrition. 2006;136:1862–1867. doi: 10.1093/jn/136.7.1862. [DOI] [PubMed] [Google Scholar]
- Rickert AM, Premstaller A, Gebhardt C, Oefner PJ. Genotyping of SNPs in a polyploid genome by pyrosequencing(TM) Biotechniques. 2002;32:592–603. doi: 10.2144/02323rr01. [DOI] [PubMed] [Google Scholar]
- Rickert AM, Ballvora A, Matzner U, Klemm M, Gebhardt C. Quantitative genotyping of single-nucleotide polymorphisms by allele-specific oligonucleotide hybridization on DNA microarrays. Biotechnology and Applied Biochemistry. 2005;42:93–96. doi: 10.1042/BA20040177. [DOI] [PubMed] [Google Scholar]
- Roberts MT, Lewis AC. Rapid characterization of hop essential oils using gas chromatography – time of flight mass spectrometry. Journal of the American Society of Brewing Chemists. 2002;60:116–121. [Google Scholar]
- Sun C-W, Griffen S, Callis J. A model for the evolution of polyubiquitin genes from the study of Arabidopsis thaliana ecotypes. Plant Molecular Biology. 1997;34:745–758. doi: 10.1023/a:1005848828368. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoecke B, Derycke L, Van Marck V, Depypere H, De Keukeleire D, Bracke M. Antiinvasive effect of xanthohumol, a prenylated chalcone present in hops (Humulus lupulus L.) and beer. International Journal of Cancer. 2005;117:889–895. doi: 10.1002/ijc.21249. [DOI] [PubMed] [Google Scholar]
- Verzele M, de Keukeleire D. Chemistry and analysis of hop and beer bitter acids. Vol. 27. Amsterdam: Elsevier; 1991. Developments in food science. [Google Scholar]
- Zuurbier KWM, Fung SY, Scheffer JJC, Verpoorte R. Formation of aromatic intermediates in the biosynthesis of bitter acids in Humulus lupulus. Phytochemistry. 1995;38:77–82. [Google Scholar]
- Zuurbier KWM, Leser J, Berger T, Hofte AJP, Schroder G, Verpoorte R, Schroder J. 4-hydroxy-2-pyrone formation by chalcone and stilbene synthase with nonphysiological substrates. Phytochemistry. 1998;a 49:1945–1951. doi: 10.1016/s0031-9422(98)00346-x. [DOI] [PubMed] [Google Scholar]
- Zuurbier KWM, Yung SY, Scheffer JJC, Verpoorte R. In-vitro prenylation of aromatic intermediates in the biosynthesis of bitter acids in Humulus lupulus. Phytochemistry. 1998;b 49:2315–2322. [Google Scholar]