Abstract
Background and Aims
High mountain ranges of the Mediterranean Basin harbour a large number of narrowly endemic plants. In this study an investigation is made of the levels and partitioning of genetic diversity in Narcissus longispathus, a narrow endemic of south-eastern Spanish mountains characterized by a naturally fragmented distribution due to extreme specialization on a rare habitat type. By using dense sampling of populations across the species' whole geographical range, genetic structuring at different geographical scales is also examined.
Methods
Using horizontal starch-gel electrophoresis, allozyme variability was screened at 19 loci for a total of 858 individuals from 27 populations. The data were analysed by means of standard statistical approaches in order to estimate gene diversity and the genetic structure of the populations.
Key Results
Narcissus longispathus displayed high levels of genetic diversity and extensive diversification among populations. At the species level, the percentage of polymorphic loci was 68 %, with average values of 2·1, 0·11 and 0·14 for the number of alleles per locus, observed heterozygosity and expected heterozygosity, respectively. Southern and more isolated populations tended to have less genetic variability than northern and less-isolated populations. A strong spatial patterning of genetic diversity was found at the various spatial scales. Gene flow/drift equilibrium occurred over distances <4 km. Beyond that distance divergence was relatively more influenced by drift. The populations studied seem to derive from three panmictic units or ‘gene pools’, with levels of admixture being greatest in the central and south-eastern portions of the species' range.
Conclusions
In addition to documenting a case of high genetic diversity in a narrow endemic plant with naturally fragmented populations, the results emphasize the need for dense population sampling and examination of different geographical scales for understanding population genetic structure in habitat specialists restricted to ecological islands.
Key words: Allozymes, genetic diversity, geographical scale, habitat isolation, Narcissus longispathus, Mediterranean endemism, mountain range, natural fragmented distribution
INTRODUCTION
In the Betic mountain ranges of the southern Iberian Peninsula, one of the most prominent plant diversity hotspots in the Mediterranean Basin, endemic taxa often represent a substantial proportion of regional floras (Sainz Ollero and Moreno Saiz, 2002; Melendo et al., 2003; Thompson, 2005). Many of these endemics are restricted and uniquely adapted to uncommon, highly fragmented habitat types, such as patches of sandy dolomitic soils, wet overhanging cliffs, or permanently water-logged meadows associated with springs. Species restricted to these ‘ecological islands’ have naturally fragmented distributions, which may often lead to long-term genetic isolation among populations. Steep topographic and climatic gradients, characteristic of mountain habitats, may also make migration and genetic exchange among these ‘islands’ extremely difficult. These conditions, in combination with life-history features such as growth form, mating system and seed dispersal mechanism, may eventually shape the spatial distribution of genetic variation (Hamrick and Godt, 1989, 1996).
In species with fragmented distributions, population size and the level of geographical separation between populations may strongly influence the spatial structure of genetic variation (reviewed in Young et al., 1996; Leimu et al., 2006). As a result of random drift and limited gene flow, small isolated populations are expected to show lower allelic diversity and heterozygosity within populations, and higher genetic differentiation among populations, than large adjoining populations (Wright, 1951; Lande, 1988; Barrett and Kohn, 1991; Ellstrand and Elam, 1993). In addition, long-term isolated populations would be expected to be significantly differentiated with no discernible pattern of genetic structure, the overall distribution of genetic diversity should be haphazard, and species-level genetic diversity should be considerably higher than mean population values (Gonzales and Hamrick, 2005). On the other hand, for organisms like plants in which dispersal ability is restricted, theory predicts that at gene-flow-drift equilibrium a significant association between genetic and geographical distances should exist; but also, based on the stepping-stone model of population genetic structure (Kimura and Weiss, 1964), the relative influence of gene flow and drift should change as populations become more geographically separated (Hutchison and Templeton, 1999; Herrera and Bazaga, 2008).
The wild daffodil Narcissus longispathus is an example of a Mediterranean narrow endemic with naturally fragmented distribution. The species is a strict habitat specialist occupying poorly drained, deep soils around springs and permanent stream banks, and is restricted to a few contiguous mountain ranges in south-eastern Spain (Herrera et al., 1999). Throughout its small geographical range, N. longispathus occurs as discrete, small populations varying in size from dozens to a few thousand plants. Estimates of within-population genetic diversity and differentiation among N. longispathus populations reported previously by Barrett et al. (2004) support some of the expectations derived from consideration of the species' specialization on an uncommon, patchy habitat type. However, that earlier investigation considered only a few populations from a restricted area. In this study we investigate the structuring of genetic diversity in N. longispathus by sampling most known locations for the species, distributed across its whole geographical range. Specifically, the following questions are addressed. (1) How much genetic diversity does a habitat-specialist, narrow endemic like N. longispathus maintain at both the species' and local population levels? (2) How is genetic diversity spatially structured across the entire range of the species? (3) Are smaller, geographically more-isolated or peripheral populations genetically less variable and/or more differentiated than larger, less-isolated or central populations?
MATERIALS AND METHODS
Study species
Narcissus longispathus Pugsley (Amaryllidaceae) is a perennial herb with a narrow distribution range (Fig. 1). Aspects of its floral biology, pollination ecology and mating system have been described by Herrera (1995), Barrett et al. (2004) and Medrano et al. (2005). This large-flowered daffodil (corolla length around 50 mm) almost invariably produces a single hermaphrodite flower per inflorescence (contra Blanchard, 1990; p. 148). Flowers possess wide, greenish floral tubes and large, funnel-like yellow coronas. Flowering takes place from late February to late April, a period characterized by cool, rainy weather that frequently limits the activity of the species' main pollinator (Andrena bicolor, Andrenidae). Despite infrequent pollinator visitation, seed production is only weakly pollen-limited. Narcissus longispathus is self-compatible, but in the absence of pollinators flowers hardly ever set seed. The species has a mixed mating system, producing both outcrossed and selfed seeds, although few selfed offspring survive to maturity because of high inbreeding depression (Barrett et al., 2004). Fruit maturation and seed shedding take place in late May/early June. The seeds are thick-coated and buoyant; thus although most seeds fall directly on the ground beneath parent plants, it is possible that some seeds can occasionally be dispersed by water beyond the limits of local populations. Occasional long-distance endozoochorous seed dispersal may also occur following the ingestion of ripe fruit by large mammals. Plants often spread clonally via fision of bulbs, sometimes giving rise to dense clumps of genetically identical individuals.
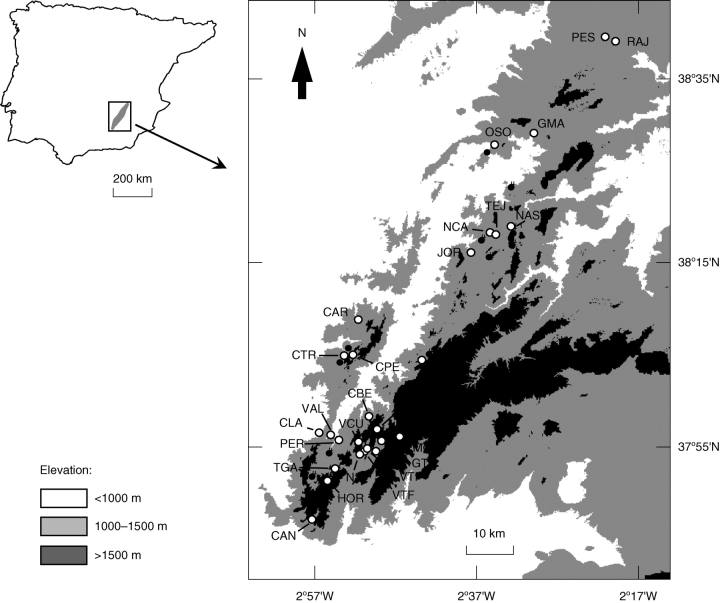
Fig. 1.
Distribution range of the narrow endemic Narcissus longispathus in the Iberian Peninsula (shaded area), and all known locations of the species (as of January 2007, main map). In the detailed map, which encompasses the mountain range running from Sierra del Pozo in the southwest to Sierra de Alcaraz in the northeast, the white dots indicate the 27 locations sampled for the present study and black dots represent the 12 unsampled locations. See Appendix for additional information on the sampled populations.
The taxonomical distinctiveness of N. longispathus has been long agreed upon by botanists, who have also generally agreed that the species' distribution range is restricted to the contiguous Sierras de Cazorla, Las Villas, Segura and Alcaraz in the south-eastern Iberian Peninsula (Webb, 1980; Blanchard, 1990; Mathew, 2002). Occasional reports from other southern Iberian mountain ranges stem from confusion with the morphologically similar Narcissus bugei (M. Medrano and C. M. Herrera, pers. obs.). Ríos et al. (1999) described three species (Narcissus segurensis, Narcissus yepesii and Narcissus alcaracensis) from a few close localities in the north-eastern range of N. longispathus, based on tenuous morphological differences. In the absence of a rigorous validation of these taxa, in this paper we have followed Webb's (1980) and Blanchard's (1990) ‘classical’ treatment, and considered all the populations sampled for our study as belonging to Narcissus longispathus. Irrespective of nomenclatural issues, however, molecular evidence demonstrates that all the populations considered here belong to the same monophyletic entity (J. Fuertes, Real Jardín Botánico de Madrid, CSIC, Spain, pers. comm.).
Sampling procedure
During the flowering seasons of 2003–2006, a total of 858 N. longispathus plants were sampled from 27 different geographical locations distributed along a latitudinal gradient spanning approximately 120 km and encompassing the entire range of the species (Fig. 1, Appendix). Nearly two-thirds of all known locations for the species were sampled. The unequal geographical distribution of sampling locations mainly reflects the decline in the frequency of the species from south to north (Fig. 1). Each sampling site corresponded to a discrete ecological population formed by a group of plants confined to a well-delimited patch of permanently damp meadow or river bank, separated from other such groups by some hundred meters or, usually, a few kilometres of unsuitable habitat that will presumably restrict gene flow. Since gene flow will be considerably greater within rather than among ecological populations, there is justification in assuming that ecological and genetic populations will be largely coincident.
Between 20–38 plants were randomly selected at each sampling location (Appendix). To avoid sampling one genetic individual more than once, the selected plants were as widely spaced as the spatial extent of the population allowed. One recently open flower was collected from each plant, kept refrigerated in a portable cooler, and subsequently stored at –80 °C until protein extraction. The total number of flowering ramets at each sampling site was estimated visually, and each population assigned to one of the following three size categories: small, <200 flowering ramets (eight populations); medium, 200–1000 flowering ramets (ten populations); and large, >1000 flowering ramets (nine populations).
Particularly among organisms whose dispersal ability is restricted, the degree of isolation of a population can be influenced not only by the average distance that separates it from all other conspecific populations, but also by the distance to the nearest population (Eckstein et al., 2006; Honnay et al., 2007). The level of geographical isolation of each sampled population was thus estimated using two different distance measures: (1) the straight-line distance from each population to the nearest sampled population (range: 0·1–13·0 km; Appendix); and (2) the average straight-line distance from each population to the other 26 sampled populations (range: 25·7–77·2 km; Appendix). These two distance measurements were uncorrelated in the sample of populations studied (r = 0·16, n = 27, P = 0·41). Other measures descriptive of the level of isolation or marginality of each population were also estimated (e.g. distance to nearest known population, distance to the centroid of the species range), but were closely correlated with the other two measures and were therefore not included in the analyses.
Allozyme electrophoresis
Starch-gel electrophoresis was used to determine allozyme diversity. Homogenization, electrophoresis and enzyme assay procedures followed the general methods of Soltis et al. (1983) and Wendel and Weeden (1989). In the laboratory, small pieces of chilled flower tissue were ground with four drops of 0·02 m Na2HPO4 extraction buffer (pH = 7·4) containing 1 mg mL–1 DL-dithiothreitol. The crude extract was immediately absorbed onto chromatography paper wicks (Whatman 3MM or 17MM), and kept at –80 °C until electrophoresis was performed on 11–12 % starch gels. Based on previous work (Barrett et al., 2004), eleven enzyme systems were resolved using two electrophoretic buffer systems. A continuous histidine-citrate buffer (pH = 6·5) was used to resolve aconitase (ACO, E.C. 4·2·1·3), acid phosphatase (ACP, E.C. 3·1·3·2), isocitrate dehydrogenase (IDH, E.C. 1·1·1·42), glucose-6-phosphate isomerase (PGI, E.C. 5·3·1·9), malate dehydrogenase (MDH, E.C. 1·1·1·37) and phosphoglucomutase (PGM, E.C. 5·4·2·2). A discontinuous lithium-borate buffer (pH = 8·3) was used to resolve alcohol dehydrogenase (ADH, E.C. 1·1·1·1), aspartate amino transaminase (AAT, E.C. 2·6·1·1), diaphorase (DIA, E.C. 1·6·4·3), formate dehydrogenase (FDH, E.C. 1·2·1·2) and leucyl aminopeptidase (LAP, E.C. 3·4·11·1). Although they appeared highly variable, other enzyme systems such as 6-phosphogluconate dehydrogenase (6PGD, E.C., 1·1·1·44) and shikimate dehydrogenase (SKDH, E.C., 1·1·1·25) could not be consistently resolved and were excluded from the analyses. A total of 19 loci with consistent, clearly interpretable banding patterns were scored: MDH, five loci; AAT, ACO, PGI and LAP, two loci each; and all other enzyme systems, a single scorable locus. For each enzyme, gene loci and alleles were inferred and labelled following numerical and alphabetical sequence, respectively.
Statistical analyses
Estimates of genetic diversity at the species' (combining all the populations into a single sample) and population levels were obtained using the following standard genetic parameters: percentage of polymorphic loci (P99, 0·99 criterion), average number of alleles per locus (A), observed heterozygosity (Ho), and gene diversity or Nei's unbiased expected heterozygosity under Hardy–Weinberg equilibrium (He = 1 – Σfi2). These parameters were calculated using GDA 1·1 (Lewis and Zaykin, 2001). In addition, a multilocus fixation index (FIS), a measure of inbreeding within populations, was calculated for each population using FSTAT, version 2·9·3 (Goudet, 2001). The significance of the FIS values calculated over all loci for each population was tested by randomization tests (randomizing alleles among individuals). Total genetic diversity was partitioned into within- and among-population components using Nei's (1973, 1987) genetic diversity statistics. For each polymorphic locus, total gene diversity (HT) was partitioned into diversity within populations (HS) and diversity among populations (DST), as HT = HS + DST. A measure of genetic differentiation among populations relative to the total genetic diversity (GST) was calculated at each polymorphic locus (GST = DST/HT) and then averaged over all loci. Theoretically, GST ranges from zero (all genetic variation maintained within populations) to 1 (all genetic variation maintained among populations).
The expectation of reduced genetic diversity in small, marginal or isolated populations was tested by regressing each measurement of within-population genetic variation (dependent variables: P99, A, Ho and He) on the distance to the nearest population, the average distance to all other populations, and population size (coded as an ordinal variable) using multiple regression. To account for possible clinal patterns, latitude was also included in all these analyses as a continuous independent variable.
Spatial structuring of genetic variation was investigated by combining two different approaches, based on genetic parameters of pre-defined populations (sampling locations) and on individual multilocus genotypes, respectively. For the first approach, pairwise FST values between all pairs of populations were estimated according to Weir and Cockerham (1984), and statistical significance of differentiation between populations was tested with a permutation test using FSTAT. The relative importance of gene flow and drift in determining genetic structure, as well as departures from equilibrium, were tested by plotting population pairwise FST values against geographical distances (Hutchison and Templeton, 1999; see also Slatkin, 1993; Rousset, 1997). Untransformed FST and distance values were used in the analyses to allow direct comparison of our results with Hutchison and Templeton's (1999) theoretical scenarios. The strength and significance of the relationships were evaluated using regression analysis and a Mantel test (10 000 permutations) as implemented in FSTAT. The results suggested that the shape of the relationship between FST and geographical distances varied with distance. This possibility was examined by means of piecewise regression, a form of regression that allows multiple linear models to be fitted to the data for different ranges of the independent variable (x). The ‘breakpoint’, or the value of x where the slope of the regression changes, was estimated using Ryan and Porth's (2007) method, which is based on simultaneously solving independent regressions by means of non-linear least-squares fitting.
Two methods based on individual multilocus genotypes and not requiring a pre-defined delimitation of genetic populations were used to study spatial genetic structuring at the whole-species range. The assignment of individuals into genetic groups was examined with the program STRUCTURE version 2·2 (Pritchard et al., 2000; Falush et al., 2007). This program probabilistically assigns genotyped individuals into genetic groups in order to minimize departures from Hardy–Weinberg equilibrium and linkage equilibrium. The number of genetic groups (I) was explored by performing five replicates of each simulation from K = 1 to K = 28, with a burn-in of 50 000, and MCMC of 100 000, assuming admixture and correlated allele frequencies as recommended by Pritchard et al. (2000). We applied Evanno et al.'s (2005) modal ΔK parameter as the choice criterion to detect the true number of genetic groups in the set of n = 858 individuals assayed. In addition to estimating the number of genetic groups, STRUCTURE estimates Q, the proportion of membership of each individual into a given genetic cluster. When genetic variation is hierarchically organized, the algorithm underlying STRUCTURE detects only the uppermost level of population structure (Evanno et al., 2005), thus the results of this analysis can provide a view of genetic structuring at the higher spatial scale corresponding to the whole distribution of the species. In order to investigate spatial structuring of genetic variation at smaller spatial scales, we used spatial autocorrelation analysis of multi-allele, multilocus genetic similarity of individual plants, using the Smouse and Peakall (1999) method as implemented in GenAlEx version 6 (Peakall and Smouse, 2006). This method differs from classical spatial autocorrelation analysis in that it employs a multivariate approach to simultaneously assess the spatial signal generated by multiple loci and alleles. The starting point for the analysis, in which the n = 858 individuals were treated as a single population, was a pair of 858 × 858 matrices of geographical and genetic distances between individuals. Geographical distances between individuals from the same populations were set to zero. The number and width of distance classes were chosen so as to obtain a greater spatial resolution at the shortest distances without compromising the number of data points per interval. This resulted in the choice of 15 distance classes that increased in width with increasing distance. Standard errors of estimates of autocorrelation coefficients were obtained by bootstrapping.
RESULTS
Genetic diversity
The 19 loci scored gave a total of 40 alleles in the 27 populations of N. longispathus studied. Five loci (26·3 %) were monomorphic in all populations (Pgi-1, Aco-1, Mdh-2, Mdh-3 and Mdh-4), and the remaining 14 (73·7 %) were polymorphic in more than one population except Idh-1, which showed variation only in CBE. Polymorphic loci had two to three alleles, with the exception of Pgi-2 that had four alleles. Four private alleles (i.e. alleles restricted exclusively to one population) were found: Idh-1a to CBE, Lap-1c to NAS, Pgi-2b to CTR and Pgm-1c to GMA. A complete table of the allele frequencies is available online as Supplementary Information, and estimates of overall genetic diversity at the species level are shown in the Appendix. The proportion of loci polymorphic according to the 0·99 criterion (P99) was 68·4 %, and the mean number of alleles per locus (A) was 2·1. Observed heterozygosity for the species as a whole (Ho = 0·110) was slightly lower than expected heterozygosity (He = 0·139), which suggests a slight heterozygote deficiency.
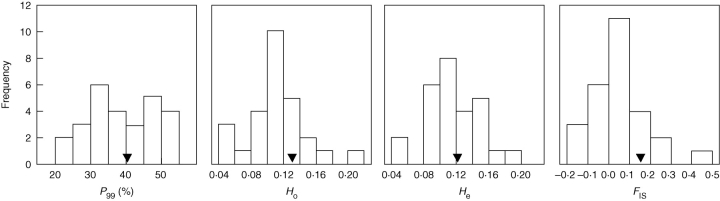
Population-level estimates of genetic diversity varied widely among populations (Fig. 2). Values of P99 ranged between 21·1–52·6 %, Ho between 0·049–0·202, and He between 0·046–0·184. The mean number of alleles per locus (A) was less variable among populations (range = 1·21–1·58). The mean inbreeding coefficient (FIS) for the set of 27 populations was 0·051 (see Appendix), which suggests a negligible level of inbreeding for the species as a whole. Broad among-population variation becomes apparent, however, when population-level estimates are considered, with FIS values ranging between –0·135 and 0·468 (Fig. 2, Appendix).
Fig. 2.
Frequency distributions of population-level estimates of genetic diversity (P99, Ho, He) and inbreeding coefficient (FIS) for the set of populations of Narcissus longispathus sampled (n = 27). In each graph the means are indicated by black triangles. See Appendix for population-level data and additional information on the populations sampled.
Nei's genetic diversity statistics are shown in Table 1 separately for all polymorphic loci. Average total heterozygosity (HT) and intra-population genetic diversity (HS) were 0·140 and 0·117, respectively. The coefficient of genic differentiation among populations (GST) ranged between 0·013 (Idh-1) and 0·314 (Aat-2), with an overall mean of 0·163. These results indicate that approx. 16 % of total genetic variation in N. longispathus was due to variation among populations, and that most genetic variability (approx. 84 %) occurred within populations.
Table 1.
Gene diversity statistics for the 14 polymorphic loci considered in this study that were identified in N. longispathus
| Nei's genetic diversity indices |
||||
|---|---|---|---|---|
| Locus | HT | HS | DST | GST |
| Pgm-1 | 0·038 | 0·034 | 0·004 | 0·111 |
| Idh-1 | 0·002 | 0·002 | 0·000 | 0·013 |
| Pgi-2 | 0·388 | 0·307 | 0·081 | 0·208 |
| Acph-1 | 0·101 | 0·088 | 0·013 | 0·128 |
| Dia-1 | 0·429 | 0·324 | 0·105 | 0·244 |
| Lap-1 | 0·030 | 0·029 | 0·001 | 0·042 |
| Lap-2 | 0·459 | 0·404 | 0·054 | 0·118 |
| Aat-2 | 0·100 | 0·068 | 0·031 | 0·314 |
| Aat-3 | 0·082 | 0·062 | 0·020 | 0·245 |
| Mdh-1 | 0·075 | 0·057 | 0·018 | 0·235 |
| Mdh-5 | 0·464 | 0·437 | 0·027 | 0·059 |
| Adh-1 | 0·026 | 0·023 | 0·003 | 0·099 |
| Fdh-1 | 0·343 | 0·273 | 0·069 | 0·202 |
| Aco-2 | 0·116 | 0·111 | 0·006 | 0·049 |
| Overall | 0·140 | 0·117 | 0·023 | 0·163 |
HT = total genetic diversity for the species; HS = mean within-population genetic diversity; DST = mean genetic diversity among populations; GST = proportion of total genetic diversity among populations.
Multiple-regressions testing for the effects on genetic diversity estimates of population size, latitude, distance to the nearest population and mean distance to the rest of populations provided contrasting results depending on the parameter used as the dependent variable. Statistically significant regressions were found for the proportion of polymorphic loci (P99; R2 = 0·345) and mean number of alleles per loci (A; R2 = 0·272), but not for observed (Ho) or expected (He) heterozygosity (Table 2). In the former two cases, only geographical latitude and distance to the nearest population had significant effects on both P99 (b = 0·747 and –0·498, respectively) and A (b = 0·666 and –0·369, respectively; Table 1). As denoted by the signs of the respective regression coefficients, the greater the latitude of a population and the smaller its distance to the nearest population, the greater its genetic diversity as measured with these two parameters. Northern populations and those with closer neighbours thus tended to have greater polymorphism and allelic richness than southern or more isolated populations.
Table 2.
Results of multiple regression analyses relating four within-population genetic diversity estimates to population size (treated as an ordinal variable), geographical latitude, distance to the nearest population (Dist1), and mean distance to the rest of the populations (Dist2), for the set of 27 Narcissus longispathus populations studied
| Standardized partial regression coefficients |
Significance of regression |
||||||
|---|---|---|---|---|---|---|---|
| Dependent variable | Population size | Latitude | Dist1 | Dist2 | R2 | F4,22 | P-value |
| P99 | 0·019 | 0·747** | –0·498*** | –0·260 | 0·345 | 4·42 | 0·009 |
| A | 0·096 | 0·666* | –0·369* | –0·138 | 0·272 | 3·43 | 0·025 |
| Ho | 0·059 | 0·247 | –0·247 | 0·174 | 0·047 | 1·32 | 0·29 |
| He | –0·036 | 0·393 | –0·335 | 0·128 | 0·185 | 2·47 | 0·074 |
*0·05 < P < 0·07; **P < 0·05; ***P < 0·01. Significant coefficients are shown in bold type.
Geographical structuring of genetic variation
The analysis of the relationships between populations using STRUCTURE showed a distinct modal maximum of ΔK at K = 3 genetic groups. Membership assignments to the three inferred genetic groups exhibited a distinct geographical pattern. The highest proportions of membership to groups 1, 2 and 3 occurred at populations located at the north-eastern, north-western and south-western extremes of the species distribution, respectively (Fig. 3). With only minor exceptions, populations located at the central and south-eastern portions of the range were consistently characterized by high admixture levels and intermediate mean group memberships (Fig. 3).
Fig. 3.
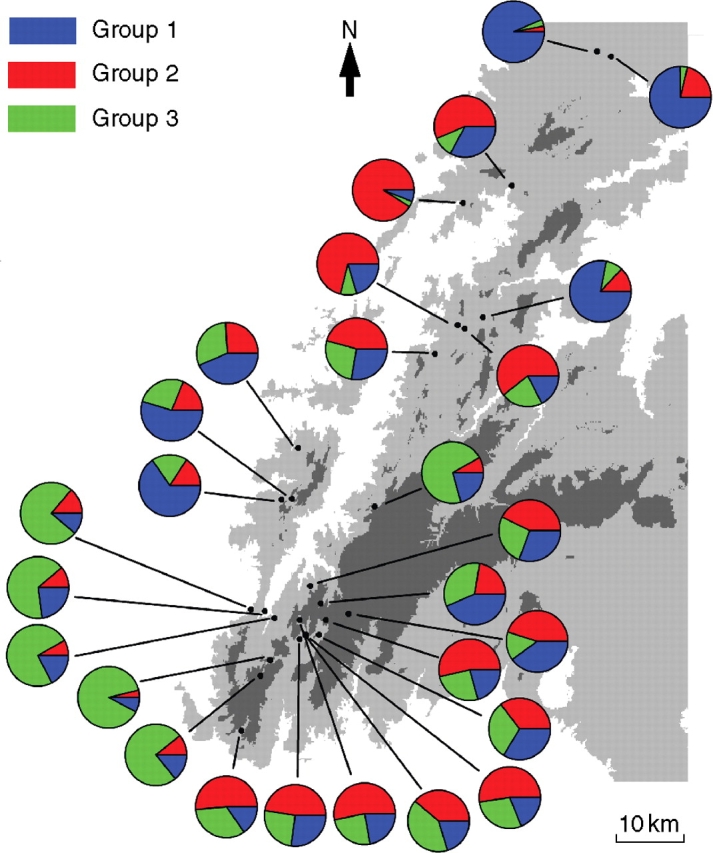
Population structure of Narcissus longispathus inferred using the model-based clustering method implemented in STRUCTURE. The individual pie charts indicate the mean proportion of membership of each local population for the inferred number of K = 3 genetic groups.
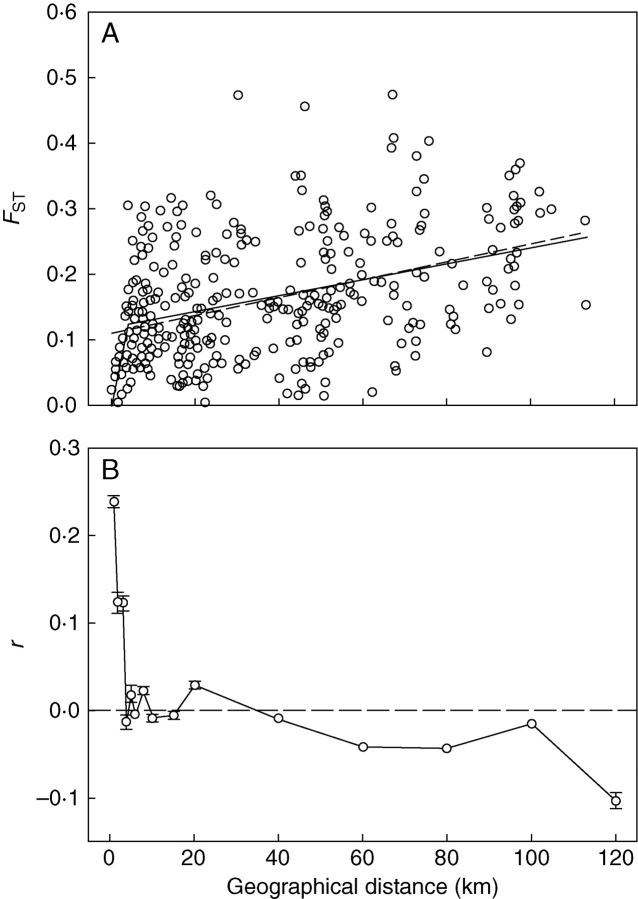
Genetic distance among populations, as measured by pairwise FST, correlated positively with geographical distance across the geographical range of the species (r = 0·421, P < 0·0001, Mantel's test with 10 000 permutations; Fig. 4A), with approx. 18 % of the variation in genetic differentiation between populations being explained by geographical separation. The scatter of points around the regression increased with geographical distance, as revealed by the significant positive correlation between residuals and geographical distance (r = 0·907, P < 0·0001, Mantel's test with 10 000 permutations). The graph also suggests that the relationship between genetic and geographical distance is distance-dependent. This possibility was confirmed by fitting a piecewise, non-linear regression to the data (Fig. 4A), which reveals an abrupt discontinuity in the slope of the relationship between genetic and geographical distance taking place at 3·9 km (95 % confidence interval estimated by bootstraping = 3·3–6·1 km). Up to that distance, there is a close and steep genetic–geographical distance relationship, which becomes looser and considerably more shallow beyond that distance. Although no formal test is available for comparing the goodness of fit of conventional and non-linear piecewise regressions, the model squared error of the piecewise regression (0·00699) is one order of magnitude smaller than the corresponding value for the conventional linear regression (0·08443), which reveals a considerably better fit of the relationship between genetic and geographical distance.
Fig. 4.
(A) Relationship between pairwise genetic (FST) and geographical distances for the 27 Narcissus longispathus populations included in this study. Shown are the conventional linear regression (dashed line) and the non-linear piecewise regression (solid line) fitted to the data. (B) Correlogram showing the multilocus, multiallelic genetic correlation (r) between individual Narcissus longispathus plants across the full range of distances occurring over the distribution area of the species (error bars are ± 2 s.e., as estimated by bootstrapping). Unequal distance classes were used to improve resolution at shorter distances (upper limits of distance classes: 1, 2, 3, 4, 5, 6, 8, 10, 15, 20, 40, 60, 80, 100, 120 km).
The spatial autocorrelation analysis provides additional evidence of strong small-scale patterning of genetic variation across the distribution range of N. longispathus (Fig. 4B). Genetic correlations between individuals are high and significantly greater than zero at the three shorter, narrowest distance classes (0–1, 1–2 and 2–3 km), declining steadily down to zero at the 3–4 km distance class. Beyond that distance, autocorrelations are either zero or negative. The first x-intercept, which provides an estimate of the extent of non-random (positive) genetic structure, occurs at 3·91 km; thus remarkably close to the 3·93 km break-point obtained from the piecewise regression of pairwise genetic and geographical distances between populations.
DISCUSSION
Genetic diversity
This study confirms for the whole distribution range of the species earlier results showing that, despite its small geographical range, N. longispathus is characterized by high genetic diversity at both the species' and local population levels (Barrett et al., 2004). The results also indicate that most of the total genetic variation was present within populations, a pattern that has often been described for mountain plants (Wesche et al., 2006, and references therein). Contrary to expectations, there were no significant effects of either population size or degree of marginality in relation to the distribution area of the species on any of the within-population genetic diversity measures considered. Genetic diversity, as measured by allelic richness and percent polymorphism, was inversely related to the distance to the nearest population and positively related to geographical latitude. We discuss below possible interpretations of these results.
In order to determine if rare species exhibit lower diversity than common ones, congeneric comparisons where phylogenetic effects can be controlled are recommended (e.g. Karron, 1987; Gitzendanner and Soltis, 2000). This kind of comparison is not possible for N. longispathus because genetic diversity studies of congeneric species with broad distributions using allozymes are not available. Comparisons are possible, however, with other plant species sharing similar geographical ranges and life histories. Values for P, A and He are higher for N. longispathus than for other endemic (P99 = 40·0 and 26·3; A = 1·80 and 1·39; He = 0·096 and 0·063, respectively, for species and population levels) or narrowly distributed plant taxa (P99 = 45·1 and 30·6; A = 1·83 and 1·45; He = 0·137 and 0·105, respectively, for species and population levels; Hamrick and Godt, 1989, 1996). Although the generalized assumption of low levels of genetic variability in rare species (Stebbins, 1980) is confirmed for many taxa by most compilations of plant allozyme data (Hamrick and Godt, 1989, 1996; Gitzendanner and Soltis, 2000), this generalization does not always hold (see for example Neel and Ellstrand, 2001; Kang et al., 2005; Ellis et al., 2006, and references therein), and the Mediterranean flora provides many exceptions. In the Iberian Peninsula, examples of high levels of genetic diversity in ecologically specialized species with narrow distributions include Antirrhinum microphyllum (Torres et al., 2003), Rosmarinus tomentosus (Martín and Hernández Bermejo, 2000), Petrocoptis montsicciana and P. pardoi (López-Pujol et al., 2001), Echinospartum algibicum (Aparicio et al., 2002) and Delphinium pentagynum subsp. formenteranum (López-Pujol et al., 2003). These examples support Rabinowitz's (1981) suggestion that the genetic consequences of the different forms of rarity existing in nature may also be very diverse.
Although N. longispathus has a narrow geographical range and high habitat specificity it has a relatively large number of populations, most of which consist of a relatively large number of individuals, and one possible explanation for the high levels of genetic variation often exhibited by rare species is that they consist of relatively large populations (Ellstrand and Elam, 1993). Two findings of our study support this hypothesis. On the one hand, the high proportion of polymorphic loci and mean number of alleles per locus occurring within populations suggest that these have not experienced severe or long-lasting population bottlenecks causing loss of genetic diversity. On the other hand, the predominantly low levels of inbreeding likewise suggest large historical populations and/or a predominantly outcrossing mating system (Barrett et al., 2004), any of which could also contribute to maintain the high levels of genetic variation observed. Other ecological and demographical characteristics of the species, such as high habitat stability, low population turnover (Barrett et al., 2004) or extended persistence of individual genotypes through clonal reproduction are all likely to favour the maintenance of high levels of genetic variation.
Although numerous studies have reported a positive relationship between population size and within-population genetic diversity (van Rossum et al., 2004; Prentice et al., 2006; Honnay et al., 2006, and references therein), failures to detect such relationships are not uncommon (Leimu and Mutikainen, 2005; Honnay et al., 2007). In long-lived perennials such as N. longispathus, present-day population sizes may not be predictive of the genetic composition of populations (Gonzales and Hamrick, 2005). Fluctuations between years in the number of flowering plants are frequent in N. longispathus populations, and thus a possible explanation for the lack of a population size effect is that our instantaneous estimates of population size do not adequately reflect the long-term average for effective population size (Young et al., 1993; Hurtrez-Bousses, 1996). Likewise, although in our analysis the effect of population size on genetic diversity was controlled for isolation level, it can not be ruled out that population characteristics not considered here (e.g. density of individuals, level of clonality) or interactions at the landscape level (e.g. connectivity among populations, altitudinal or topographical barriers) may counteract the population size effect.
As indicated by the significant positive slopes of P99 and A on latitude, genetic diversity increased steadily towards the northern distribution edge of N. longispathus. This result seems contrary to most empirical evidence to date, as genetic diversity has generally been found to decline towards the periphery of species' ranges (see for example Durka, 1999; Griffin and Barrett, 2004; Arnaud-Haond et al., 2006; and references therein). It has been suggested that increased genetic diversity in peripheral populations could occur if distinct genotypes are favoured compared to core populations because of suboptimal or more stressful conditions that presumably occur on distribution edges (Safriel et al., 1994; Lesica and Allendorf, 1995), but alternative explanations can also apply (e.g. demographic processes leading to variable effective population sizes). The first scenario could apply in N. longispathus, since the ecological conditions faced by the northern (lower altitudes, flat meadows with permanently damp soils, or margins of semi-permanent lagoons) and southern populations (higher altitudes and slopes, and soils flooded by running water from springs) may be considerably different. The fact that two out of the three different ‘gene pools’ revealed by STRUCTURE are more predominant in populations located at the northern limit of the distribution range seem to support this hypothesis.
Geographical structuring of genetic variation
Two contrasting patterns of genetic variation seem to be operating at different spatial scales over the distribution area of N. longispathus. Although genetic differentiation among populations tends to increase with increasing geographical distance across the whole geographical range of the species, the fact that this relationship explains only approx. 18 % of the genetic differentiation among populations points to a more complex scenario. A detailed analysis of this relationship reveals that at small spatial scales (<4 km) there exists a gene-flow-genetic drift equilibrium, and genetic differentiation is closely related to geographical distance. However, at greater distances drift prevails over gene flow, suggesting that isolation by distance does not apply at these greater spatial scales. It is remarkable that, despite the contrasting approaches, piecewise regression and spatial autocorrelation analyses provided closely coincident views of geographical genetic structuring in N. longispathus.
Different features of species' reproductive biology can explain these findings. The main pollinators of N. longispathus are small, solitary bees, which generally have restricted movement capacities (see Fenster, 1991). In addition, the species lacks specialized seed dispersal mechanisms and most seeds are dispersed by gravity at a close distance from the maternal plant. Nevertheless, occasional long-distance endozoochorous dispersal by large mammals and secondary seed movement into adjacent areas by water cannot be ruled out, as plants frequently grow on river margins. Therefore, it might be expected that pollen and propagule movements of N. longispathus should be relatively frequent among nearest populations, but exceptional among populations that are beyond a certain distance threshold. As a consequence of its specialized habitat, N. longispathus populations are separated by broad expanses of inhospitable landscape. This scenario would further act to constrain and disrupt pollination and seed dispersal processes, rendering genetic drift more influential as distance between populations increases.
In other plant species in which isolation by distance is detected across broad geographical scales, the relationship between geographical and genetic distances have also revealed different patterns of gene flow operating at different spatial scales (e.g. Wolf et al., 2000; Gonzales and Hamrick, 2005). In Hymenocallis coronaria, Markwith and Scanlon (2007) explicitly compared genetic structure at different geographical scales; significant isolation by distance occured only at distances <16 km, whereas non-equilibrium conditions were suggested to prevail at distances >56 km. Narcissus longispathus and H. coronaria would thus fit into the ‘Case IV’ scenario of Hutchison and Templeton (1999), in which migration/drift equilibrium occurs over small distances, whereas divergence between more-distantly separated populations is relatively more influenced by drift. According to these authors, this scenario would be expected in a recently invaded region, not yet at equilibrium, in which dispersal is just localized between neighbouring populations. Given sufficient time and conditions of stability, isolation-by-distance equilibrium between gene flow and genetic drift would be achieved across the whole region. In N. longispathus, other possible scenarios that would produce this pattern can not be ruled out. The observed pattern could be a consequence of the natural discontinuity of permanently humid habitats in Mediterranean mountains, and thus stable over time. Alternatively, recent human-mediated disturbances of humid habitats commonly occurring in the study region (draining, over-pumping of ground water, flow alteration, reforestation, grazing) may have enhanced fragmentation and the loss of potential habitats, leading to the disequilibrium between migration and drift observed at greater distances. It is not possible to discriminate among these possibilities with the data currently available.
Our STRUCTURE results also suggest the existence of genetic structuring at a spatial scale that is commesurate with the whole distribution range of the species. The results illustrate the capacity of the Bayesian approach implemented in STRUCTURE to draw inferences about the uppermost hierarchical level of genetic structure (Evanno et al., 2005) even with low mutation markers such as allozymes. Our results must be interpreted cautiously, however, because of the small power of these markers to correctly estimate the true number of populations (Waples and Gaggiotti, 2006). The results of STRUCTURE can be sensitive to the sampling scheme when there is clinal variation in allele frequencies (Pritchard et al., 2000). It is unlikely, however, that our results reflect an artifact of this type, as we have sampled about two-thirds of all known populations of the species and sampling encompasses the whole distribution range. For this reason, we are confident that the STRUCTURE results actually reflect the broad-scale organization of genetic diversity throughout the distribution range of N. longispathus. At this broad geographical scale, it was possible to estimate that in N. longispathus the number of panmictic units or ‘gene pools’ (K) from which the studied populations seem to derive is three. The findings also suggest that in the central and south-eastern portions of its range, admixture is more frequent and consequently populations should be less differentiated; whereas at the periphery of the distribution range more differentiated populations appear, which derive predominantly from just one of the different ‘gene pools’ recognized by STRUCTURE (RAJ, PES, OSO and HOR). Based on our current knowledge, it is difficult to suggest which historical factors may have produced this pattern of geographical structuring of genetic variation but the results do suggest that, at least within the last few generations, different patterns of gene flow have been operating at different spatial scales throughout the whole distribution range of the species.
In conclusion, despite being a narrow endemic with a small distribution, very restrictive habitat requirements and fragmented, relatively isolated populations, N. longispathus exhibits high levels of genetic diversity at both the population and species levels. Genetic diversity is geographically structured, but with isolation-by-distance occurring only over very short ranges, and drift playing the predominant role at distances beyond 4 km. Superimposed on this, there is a broad-scale genetic pattern characterized by the existence of three distinct gene pools at peripheral locations of the distribution and extensive admixture in the central parts of the distribution range. The results illustrate the importance of sampling populations over a broad range of spatial scales so that the risks of generalizing spurious patterns over an entire species or environment can be minimized.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at http://aob.oxfordjournals.org/ and gives allele frequencies for 14 polymorphic loci for the 27 populations of N. longispathus surveyed in this study.
ACKNOWLEDGEMENTS
We are grateful to Alfredo Benavente, Juan Pedro Martín and Amelia Garrido for their invaluable help in locating the Narcissus longispathus populations; Rocío Requerey and Marina García for conducting most of the laboratory work; Joaquín Muñoz, Bill Cole and Rafael Albaladejo for advice on electrophoresis and assistance in the interpretation of some results; Javier Fuertes for unpublished information on Narcissus phylogeny; two anonymous referees for comments on the manuscript; and Consejerías de Medio Ambiente, Junta de Andalucía and Junta de Castilla-La Mancha for authorizing this work and providing invaluable facilities in Sierras de Cazorla, Las Villas, Segura and Alcaraz regions. Support for this work was provided by grants RNM-156 and P06-RNM-01627 from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, and CGL2006–01355 from the Spanish Ministerio de Educación y Ciencia. MM was supported by a postdoctoral contract from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía.
APPENDIX
Characteristics and descriptive genetic parameters for the 27 Narcissus longispathus populations sampled for this study, listed by increasing latitude. See Fig. 1 for locations
| Isolationb |
Genetic parameter estimatesc |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | Acronym | Plants sampled | Population sizea | Dist1 (km) | Dist2 (km) | P99 (%) | A | Ho | He | FIS |
| Barranco la Canal | CAN | 25 | S | 8·3 | 41·0 | 26·3 | 1·32 | 0·117 | 0·124 | 0·057 |
| Fuente Cueva del Horno | HOR | 30 | M | 2·8 | 33·2 | 31·6 | 1·32 | 0·102 | 0·093 | –0·108 |
| Tornillos Gualay – A | TGA | 30 | M | 2·8 | 30·9 | 42·1 | 1·42 | 0·161 | 0·143 | –0·135 |
| Nava San Pedro | NSP | 31 | S | 1·1 | 27. 6 | 31·6 | 1·32 | 0·055 | 0·104 | 0·468 |
| Barranco Guadalentín | GTN | 31 | L | 2·0 | 27·3 | 31·6 | 1·32 | 0·104 | 0·105 | 0·015 |
| Valdetrillo – bajo piedra | VTP | 34 | M | 0·1 | 27·1 | 47·4 | 1·47 | 0·113 | 0·131 | 0·140 |
| Valdetrillo – fuente | VTF | 33 | M | 0·1 | 27·1 | 52·6 | 1·53 | 0·108 | 0·114 | 0·052 |
| Valdecuevas | VCU | 30 | L | 2·2 | 26·4 | 31·6 | 1·32 | 0·099 | 0·095 | –0·046 |
| Majal de las Carrascas | MCA | 30 | L | 2·5 | 26·6 | 36·8 | 1·42 | 0·140 | 0·125 | –0·128 |
| Fuente del Perro | PER | 30 | M | 1·9 | 27·5 | 21·1 | 1·21 | 0·049 | 0·046 | –0·078 |
| La Cabrilla | CAB | 33 | L | 4·0 | 27·4 | 31·6 | 1·32 | 0·091 | 0·094 | 0·031 |
| Arroyo del Valle | VAL | 32 | L | 1·9 | 28·0 | 26·3 | 1·26 | 0·087 | 0·092 | 0·055 |
| Fuente los Clavelones | CLA | 36 | S | 2·2 | 29·3 | 31·6 | 1·32 | 0·080 | 0·084 | 0·040 |
| Fuente de la Reina | REI | 34 | L | 2·6 | 25·9 | 42·1 | 1·42 | 0·115 | 0·120 | 0·043 |
| Cuevas Bermejas | CBE | 34 | M | 3·0 | 25·7 | 36·8 | 1·37 | 0·118 | 0·118 | –0·001 |
| Barranco los Centenares | CEN | 34 | S | 13·0 | 28·5 | 21·1 | 1·21 | 0·050 | 0·054 | 0·090 |
| Cortijo de la Traviesa | CTR | 33 | M | 1·6 | 28·6 | 52·6 | 1·58 | 0·123 | 0·147 | 0·168 |
| Collado Perenoso | CPE | 34 | L | 1·6 | 28·2 | 52·6 | 1·58 | 0·113 | 0·141 | 0·200 |
| Carrales | CAR | 33 | S | 7·4 | 30·8 | 26·3 | 1·26 | 0·075 | 0·091 | 0·183 |
| Fuente de la Jordana | JOR | 31 | L | 5·4 | 37·5 | 36·8 | 1·42 | 0·105 | 0·114 | 0·081 |
| Arroyo del Tejuelo | TEJ | 38 | L | 1·2 | 40·4 | 47·4 | 1·47 | 0·127 | 0·128 | 0·006 |
| Navalcaballo | NCA | 34 | M | 1·2 | 40·2 | 47·4 | 1·47 | 0·150 | 0·151 | 0·003 |
| Arroyo de Navalasna | NAS | 30 | S | 3·3 | 42·4 | 47·4 | 1·47 | 0·202 | 0·184 | –0·098 |
| Fuente del Oso – Siles | OSO | 38 | M | 7·9 | 52·0 | 42·1 | 1·47 | 0·116 | 0·115 | –0·015 |
| Villaverde de Guadalimar | GMA | 30 | S | 7·9 | 56·1 | 47·4 | 1·53 | 0·135 | 0·157 | 0·143 |
| Fuente de la Raja | RAJ | 30 | M | 2·5 | 77·2 | 52·6 | 1·58 | 0·123 | 0·165 | 0·258 |
| Nacimiento Río Pesebre | PES | 20 | S | 2·5 | 77·0 | 36·8 | 1·37 | 0·124 | 0·118 | –0·053 |
| Mean over all populations | 38·2 | 1·40 | 0·111 | 0·117 | 0·054 | |||||
| Species level d | 68·4 | 2·10 | 0·110 | 0·139 | – | |||||
aPopulation size classes: S, <200 flowering ramets; M, 200–1000 flowering ramets; L >1000 flowering ramets.
bDegree of geographical isolation of the population as estimated with Dist1, distance to nearest population (km) and Dist2, mean distance to the rest of the populations (km).
cP99, percentage of polymorphic loci (0·99 criterion); A, mean number of alleles per locus; Ho, mean observed heterozygosity; He, gene diversity or expected heterozygosity (unbiased estimate; Nei, 1978); and FIS = Wright's inbreeding coefficient. Statistically significant multilocus inbreeding coefficients (P < 0·05) are in bold type.
dComputed by combining the data for all individuals (n = 858) into a single sample.
LITERATURE CITED
- Aparicio A, Albaladejo RG, Ceballos CL. Genetic differentiation in silicicolous Echinospartum (Leguminosae) indicated by allozyme variability. Plant Systematics and Evolution. 2002;230:189–201. [Google Scholar]
- Arnaud-Haond S, Teixeira S, Massa SI, Billot C, Saenger P, Coupland G, Duarte CM, Serrão EA. Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Molecular Ecology. 2006;15:3515–3525. doi: 10.1111/j.1365-294X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- Barrett SCH, Kohn JR. Genetic and evolutionary consequences of small population size. In: Falk DA, Holsinger KE, editors. Genetics and conservation of rare plants. New York: Oxford University Press; 1991. pp. 3–30. [Google Scholar]
- Barrett SCH, Cole WW, Herrera CM. Mating patterns and genetic diversity in the wild daffodil Narcissus longispathus (Amaryllidaceae) Heredity. 2004;92:459–465. doi: 10.1038/sj.hdy.6800441. [DOI] [PubMed] [Google Scholar]
- Blanchard JW. Narcissus: a guide to wild daffodils. Pershore. Worcestershire, UK: Alpine Garden Society; 1990. [Google Scholar]
- Durka W. Genetic diversity in peripheral and subcentral populations of Corrigiola litoralis L. (Illecebraceae) Heredity. 1999;83:476–484. doi: 10.1038/sj.hdy.6886000. [DOI] [PubMed] [Google Scholar]
- Eckstein RL, O'Neill RA, Danihelka J, Otte A, Köhler W. Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Molecular Ecology. 2006;15:2367–2379. doi: 10.1111/j.1365-294X.2006.02944.x. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Pashley CH, Burke JM, McCauley DE. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Molecular Ecology. 2006;15:2345–2355. doi: 10.1111/j.1365-294X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB. Gene flow in Chamaecrista fasciculata (Leguminosae) I. Gene dispersal. Evolution. 1991;45:398–405. doi: 10.1111/j.1558-5646.1991.tb04413.x. [DOI] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany. 2000;87:783–792. [PubMed] [Google Scholar]
- Gonzales E, Hamrick JL. Distribution of genetic diversity among disjunct populations of the rare forest understory herb, Trillium reliquum. Heredity. 2005;95:306–314. doi: 10.1038/sj.hdy.6800719. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices. 2001. version 2·9·3 http://www2.unil.ch/popgen/softwares/fstat.htm .
- Griffin SR, Barrett SCH. Genetic variation in Trillium erectum (Melanthiaceae), a widespread forest herb in eastern North America. Canadian Journal of Botany. 2004;82:316–321. [Google Scholar]
- Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer Associates Inc.; 1989. pp. 43–63. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 1996;351:1291–1298. [Google Scholar]
- Herrera CM. Floral biology, microclimate, and pollination by ectothermic bees in an early-blooming herb. Ecology. 1995;76:218–228. [Google Scholar]
- Herrera CM, Bazaga P. Adding a third dimension to the edge of a species' range: altitude and genetic structuring in mountainous landscapes. Heredity. 2008;100:275–285. doi: 10.1038/sj.hdy.6801072. [DOI] [PubMed] [Google Scholar]
- Herrera CM, Hernández-Bermejo E, Luque P, Benavente A. Narcissus longispathus Pugsley. In: Blanca G, Cabezudo B, Hernández-Bermejo E, Herrera CM, Molero Mesa J, Muñoz J, Valdés B, editors. Libro rojo de la flora silvestre amenazada de Andalucía. I. Especies en peligro de extinción. Sevilla: Consejería de Medio Ambiente, Junta de Andalucía; 1999. pp. 191–194. [Google Scholar]
- Honnay O, Coart E, Butaye J, Adriaens D, Van Glabeke S, Roldán-Ruiz I. Low impact of present and historical landscape configuration on the genetics of fragmented Anthyllis vulneraria populations. Biological Conservation. 2006;127:411–419. [Google Scholar]
- Honnay O, Adriaens D, Coart E, Jacquemyn H, Roldán-Ruiz I. Genetic diversity within and between remnant populations of the endangered calcareous grassland plant Globularia bisnagarica L. Conservation genetics. 2007;8:293–303. [Google Scholar]
- Hurtrez-Bousses S. Genetic differentiation among natural populations of the rare endemic Brassica insularis Moris: implications for conservation guidelines. Biological Conservation. 1996;76:25–30. [Google Scholar]
- Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Kang M, Jiang M, Huang H. Genetic diversity in fragmented populations of Berchemiella wilsonii var. pubipetiolata (Rhamnaceae) Annals of Botany. 2005;95:1145–1151. doi: 10.1093/aob/mci125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron JD. A comparison of levels of genetic polymorphism and self-compatibility in geographically restricted and widespread plant congeners. Evolutionary Ecology. 1987;1:47–58. [Google Scholar]
- Kimura M, Weiss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Leimu R., Mutikainen P. Population history, mating system, and fitness variation in a perennial herb with a fragmented distribution. Conservation Biology. 2005;19:349–356. [Google Scholar]
- Leimu R, Mutikainen P, Koricheva J, Fischer M. How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology. 2006;94:942–952. [Google Scholar]
- Lesica P, Allendorf FW. When are peripheral populations valuable for conservation? Conservation Biology. 1995;9:753–760. [Google Scholar]
- Lewis PO, Zaykin D. Genetic data analysis: computer program for the analysis of allelic data. 2001. version 1·1. Free program distributed by the authors over the internet at http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php .
- López-Pujol J, Bosch M, Simon J, Blanché C. Allozyme diversity of the two endemic Petrocoptis: P. montsicciana and its close related P. pardoi (Caryophyllaceae) Canadian Journal of Botany. 2001;79:1379–1389. [Google Scholar]
- López-Pujol J, Bosch M, Simon J, Blanché C. Population genetics and conservation priorities for the critically endangered island endemic Delphinium pentagynum subsp. formenteranum (Ranunculaceae) Biodiversity and Conservation. 2003;12:1937–1951. [Google Scholar]
- Markwith SH, Scanlon MJ. Multiscale analysis of Hymenocallis coronaria (Amaryllidaceae) genetic diversity, genetic structure, and gene movement under the influence of unidirectional stream flow. American Journal of Botany. 2007;94:151–160. doi: 10.3732/ajb.94.2.151. [DOI] [PubMed] [Google Scholar]
- Martín JP, Hernández Bermejo JE. Genetic variation in the endemic and endangered Rosmarinus tomentosus Huber-Morath & Maire (Labiatae) using RAPD markers. Heredity. 2000;85:434–443. doi: 10.1046/j.1365-2540.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Mathew B. Classification of the genus Narcissus. In: Hanks GR, editor. Narcissus and daffodil: the genus Narcissus. New York: Taylor and Francis; 2002. pp. 30–52. [Google Scholar]
- Medrano M, Herrera CM, Barrett SCH. Herkogamy and mating patterns in the self-compatible daffodil Narcissus longispathus. Annals of Botany. 2005;95:1105–1111. doi: 10.1093/aob/mci129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendo M, Giménez E, Cano E, Gómez-Mercado F, Valle F. The endemic flora in the south of the Iberian Peninsula: taxonomic composition, biological spectrum, pollination, reproductive mode and dispersal. Flora. 2003;198:260–276. [Google Scholar]
- Neel MC, Ellstrand NC. Patterns of allozyme diversity in the threatened plant Erigeron parishii (Asteraceae) American Journal of Botany. 2001;88:810–818. [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. NewYork: Columbia University Press; 1987. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice HC, Lonn M, Rosquist G, Ihse M, Kindstrom M. Gene diversity in a fragmented population of Briza media: grassland continuity in a landscape context. Journal of Ecology. 2006;94:87–97. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz D. Seven forms of rarity. In: Synge H, editor. The biological aspects of rare plant conservation. London: Wiley and Sons; 1981. pp. 207–217. [Google Scholar]
- Ríos S, Rivera D, Alcaraz F, Obón de Castro C. Three new species of Narcissus L. subgenus Ajax Spach (Amaryllidaceae), restricted to the meadows and forests of south-eastern Spain. Botanical Journal of the Linnean Society. 1999;131:153–165. [Google Scholar]
- van Rossum F, Campos de Sousa S, Triest L. Genetic consequences of habitat fragmentation in an agricultural landscape on the common Primula veris and comparison with its rare congener, P. vulgaris. Conservation Genetics. 2004;5:231–245. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SE, Porth LS. A tutorial on the piecewise regression approach applied to bedload transport data. Fort Collins, CO: Department of Agriculture, Forest Service, Rocky Mountain Research Station; 2007. General Technical Report RMRS-GTR-189. [Google Scholar]
- Safriel UN, Volis S, Kark S. Core and peripheral populations, and global climate change. Israel Journal of Plant Sciences. 1994;42:331–345. [Google Scholar]
- Sainz Ollero H, Moreno Saiz JC. Flora vascular endémica española. In: Pineda FD, de Miguel JM, Casado MA, editors. La diversidad biológica de España. Madrid: Pearson Educación; 2002. pp. 175–195. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999;82:561–573. doi: 10.1038/sj.hdy.6885180. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Haufler CH, Darrow DC, Gastony GJ. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal. 1983;73:9–27. [Google Scholar]
- Stebbins GL. Rarity of plant species: a synthetic viewpoint. Rhodora. 1980;82:77–86. [Google Scholar]
- Thompson JD. Plant evolution in the Mediterranean. Oxford: Oxford University Press; 2005. [Google Scholar]
- Torres E, Iriondo JM, Pérez C. Genetic structure of an endangered plant, Antirrhinum microphyllum (Scrophulariaceae): allozyme and RAPD analysis. American Journal of Botany. 2003;90:85–92. doi: 10.3732/ajb.90.1.85. [DOI] [PubMed] [Google Scholar]
- Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- Webb DA. Amaryllidaceae. In: Tutin TG, Heywood VH, Burges NA, editors. Flora Europaea. vol. 5. Cambridge: Cambridge University Press; 1980. pp. 75–84. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Weeden NF. Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS, editors. Isozymes in plant biology. Portland, OR: Dioscorides Press; 1989. pp. 5–45. [Google Scholar]
- Wesche K, Hensen I, Undrakh R. Genetic structure of Galitzkya macrocarpa and G. potaninii, two closely related endemics of Central Asian mountain ranges. Annals of Botany. 2006;98:1025–1034. doi: 10.1093/aob/mcl182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AT, Howe RW, Hamrick JL. Genetic diversity and population structure of the serpentine endemic Calystegia collina (Convolvulaceae) in Northern California. American Journal of Botany. 2000;87:1138–1146. [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
- Young AG, Merriam HG, Warwick SI. The effects of forest fragmentation on genetic variation in Acer saccharum Marsh (sugar maple) populations. Heredity. 1993;71:277–289. [Google Scholar]