Abstract
Background and Aims
European white oaks (Quercus petraea, Q. pubescens, Q. robur) have long puzzled plant biologists owing to disputed species differentiation. Extensive hybridization or shared ancestry have been proposed as alternative hypotheses to explain why genetic differentiation between these oak species is low. Species delimitation is usually weak and often shows gradual transitions in leaf morphology. Hence, individual identification may be difficult, but remains a critical step for both scientific work and practical management.
Methods
Multilocus genotype data (five nuclear microsatellites) were used from ten Swiss oak stands for taxon identification without a priori grouping of individuals or populations, using model-based Bayesian assignment tests.
Key Results
Three groups best structured the data, indicating that the taxonomical signal was stronger than the spatial signal. Most individuals showed high posterior probabilities for either of three genetic groups that were best circumscribed as taxonomical units. The assignment of a subset of trees, whose taxonomic status had been previously characterized in detail, supported this classification scheme.
Conclusions
Molecular-genetic assignment tests are useful in the identification of species status in critical taxon complexes such as the European white oaks. Such an approach is of practical importance for forest management, e.g. for stand certification or in seed trade to trace the origin of forest products.
Key words: Assignment test, Bayesian inference, multilocus genotype, nuclear microsatellites, Quercus sp., species complex
INTRODUCTION
Species from the widespread temperate forest tree genus Quercus have long been used as model systems for studying hybridization in plants (Stebbins et al., 1947; Kremer et al., 1993; Howard et al., 1997; Craft et al., 2002; Ishida et al., 2003; Petit et al., 2003; Tovar-Sánchez and Oyama, 2004; González-Rodríguez et al., 2005). Particularly in the European white oaks, including Quercus petraea, Q. pubescens and Q. robur, numerous studies have applied morphological (Kremer et al., 2002a), ecophysiological (Ponton et al., 2002), palaeoecological (Brewer et al., 2002), and a broad variety of molecular methods (Zanetto et al., 1994; Bodénès et al., 1997b; Kremer et al., 2002b; Mariette et al., 2002a; Petit et al., 2002; Scotti-Saintagne et al., 2004; Muir and Schlötterer, 2005) to elucidate the evolution of this taxon complex. These studies embraced spatial scales ranging from the continental to the local stand level.
Despite this multitude of studies, it is still debated how relevant natural hybridization and introgression are, and the degree to which these processes affect species differentiation and, hence, taxon identification (Lexer et al., 2006; Muir and Schlötterer, 2006). For the case of Q. petraea and Q. robur, there are both arguments in favour of extensive hybrid formation leading to nuclear capture (Petit et al., 2003) or of a generally low species differentiation as a consequence of shared ancestral polymorphism (Muir and Schlötterer, 2005). Gugerli et al. (2007) argue that the number of effectively reproductive hybrids within local populations is low owing to small-scale assortative mating, low frequencies of natural interspecific cross-fertilizations, short-distance seed dispersal, and local selection favouring those taxonomically ‘pure’ seedlings that grow in their maternal habitat.
Molecular studies in Q. petraea and Q. robur have shown that species differentiation at neutral loci is generally low but heterogeneous. For example, only 2 % of 2800 random amplified polymorphic DNA (RAPD) markers showed significant differentiation between the two species (Bodénès et al., 1997a). Likewise, frequency differences in only four out of 155 reliable amplified fragment length polymorphisms (AFLPs) exhibited >10 % interspecific genetic differentiation in six mixed populations from across Europe (Mariette et al., 2002b). This suggests that only few genomic regions are involved in species separation, and two recent mapping studies confirmed this assumption (Saintagne et al., 2004; Scotti-Saintagne et al., 2004). Even though no diagnostic molecular marker could be identified to date, it is possible to find distinct taxonomic signals in molecular data sets when choosing either large numbers of nuclear markers or else specifically selected, highly differentiating markers (Muir et al., 2000; Muir and Schlötterer, 2005; Gugerli et al., 2007). For the species pair Q. petraea and Q. pubescens, studies of isozymes and nuclear microsatellites (simple sequence repeats, nSSRs) generally agree that introgression plays an important role, which is corroborated by leaf morphological analyses and pollination experiments (Müller, 1999; Bruschi et al., 2000). As such, species identification in the European white oaks is not straightforward and often requires carefully evaluating multiple characters and molecular markers.
To date, studies on European white oaks have relied on a priori taxon assignment based on leaf morphological characters. Here, the reverse approach, namely to test if trees or populations may be reliably assigned to either taxon without prior classification, was used (Craft et al., 2002; Aldrich et al., 2003). Specifically, the potential of identifying the taxonomic composition of European white oak stands was explored using assignment tests based on individual multilocus genotypes. First, we validated whether homogeneity of genetic clusters represents putative species or simply indicates geographical origin. Next, individuals were assigned to one of three clusters based on the posterior probabilities from assignment tests. Finally, an evaluation was carried out to see if the multilocus genotype assignment of the individuals from a mixed stand of Q. petraea and Q. robur corresponds to their taxonomic identification based on local allele frequencies and leaf morphology as previously characterized by Gugerli et al. (2007).
From the results it is concluded that Bayesian assignment methods are valuable for species identification in species complexes, that few nuclear markers may be sufficient for taxon assignment if appropriately selected for differentiation power, and that such a method is also useful for practical purposes such as the certification of forestry products.
MATERIALS AND METHODS
Study species
The three study taxa Quercus petraea (Matt.) Liebl., Q. pubescens Willd., and Q. robur L. of section Quercus or white oaks (Nixon, 1993) are important tree species of temperate, low-elevation broadleaved woodlands. Quercus petraea and Q. robur are widespread in western and central Europe, occurring from Mediterranean to subboreal areas. The natural range of Q. pubescens is more restricted, with a centre in southern and south-eastern Europe (Tutin et al., 1993). In accordance with their ecological amplitudes, two or all three species may occur in the same locality.
The taxon pairs Q. petraea–Q. robur and Q. petraea–Q. pubescens are considered interfertile (Rushton, 1993; Bacilieri et al., 1996; Müller, 1999; Petit et al., 2003), but the frequency of naturally occurring hybrids is still debated (Bruschi et al., 2000; Petit et al., 2003; Muir and Schlötterer, 2005). The taxa further share the same type of breeding system characterized by monoecy, anemophily and a combination of baleo- and zoochory.
Field sampling
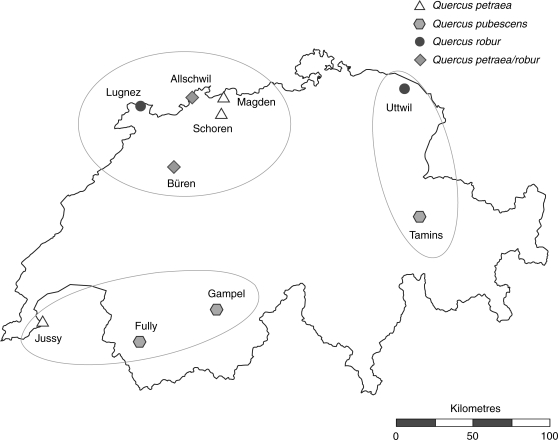
Sampling comprised ten oak stands from Switzerland (Fig. 1, Table 1). The selected stands were previously categorized based on leaf morphological analyses as Q. petraea (three stands), Q. robur (two), mixed Q. petraea and Q. robur (two) and Q. pubescens (two; Finkeldey and Mátyás, 2003). The morphological assignment of stands was based on 100 leaves randomly collected from the ground across each sampling range. Leaves were measured and data analysed following Kremer et al. (2002a) as described in Finkeldey and Mátyás (2003). Within each stand, fresh leaf tissue samples were collected from 32 trees in 2003, respecting a minimum distance of 30 m between sampled individuals. In the mixed stand of Büren (Q. petraea and Q. robur), where all adult oak trees had already been morphologically assessed and genotyped (Gugerli et al., 2007), a random sample of 32 individuals was drawn from the existing data set. The taxon composition of this subset was representative of the entire stand, i.e. taxon frequencies did not significantly differ (data not shown).
Fig. 1.
Location of Quercus spp. stands sampled in Switzerland. Symbols refer to Q. petraea, Q. robur, Q. pubescens or Q. petraea/robur mixed populations based on leaf morphology (according to Finkeldey and Mátyás, 2003). Encircled stands were grouped as south-western, north-western and eastern in AMOVA (Table 2).
Table 1.
Taxonomic status and genetic diversity estimates of ten stands of Quercus spp. from Switzerland, based on five nuclear microsatellite markers
| Taxonomic status |
Diversity estimates |
||||
|---|---|---|---|---|---|
| Stand | n | Leaf morphology | Microsatellites | Ar | He |
| Jussy | 31 | petraea | petraea | 11·8 | 0·868 |
| Magden | 26 | petraea | petraea | 11·6 | 0·849 |
| Schoren | 31 | petraea | petraea/robur | 13·1 | 0·889 |
| Allschwil | 32 | petraea/robur | petraea/pubescens | 12·5 | 0·893 |
| Büren | 32 | petraea/robur | petraea/robur | 13·6 | 0·838 |
| Lugnez | 32 | robur | robur | 11·8 | 0·796 |
| Uttwil | 32 | robur | robur | 12·0 | 0·792 |
| Fully | 32 | pubescens | petraea/robur | 13·2 | 0·878 |
| Gampel | 27 | pubescens | pubescens | 12·9 | 0·890 |
| Tamins | 30 | pubescens | petraea/pubescens | 11·5 | 0·876 |
The taxonomic status of stands was based on leaf morphology (Finkeldey and Mátyás, 2003) or on multilocus genotypes (see Results).
n, sample size for nuclear microsatellites, Ar, mean allelic richness, He,expected heterozygosity.
DNA isolation and microsatellite analysis
Following the laboratory procedures described in Gugerli et al. (2007), five nuclear dinucleotide microsatellite loci were analysed: QpZAG9, QpZAG104 (Steinkellner et al., 1997), QrZAG30, QrZAG96 (Kampfer et al., 1998) and MSQ13 (Dow et al., 1995). According to Barreneche et al. (2004), these loci are on separate linkage groups. The loci proved to be informative in a test on the differentiation potential between Q. petraea and Q. robur over their entire range (P. Goicoechea et al., Neiker, Vitoria; unpubl. data) and on the local scale (Gugerli et al., 2007). Two multiplex PCRs (QpZAG9, QpZAG104 and MSQ13; QrZAG30 and QrZAG96) were run separately for fragment length analysis on an ABI3100-Avant automated capillary sequencer (Applied Biosystems, Rotkreuz, Switzerland), including an internal size standard ROX400HD (Applied Biosystems). GeneScan 3·1 and Genotyper 2·5 (Applied Biosystems) were used for genotyping.
Statistical analyses
In the few cases where more than two fragments were amplified per locus (cf. Dzialuk et al., 2007), that particular locus was coded as missing in the respective individual. For analyses, only those individuals were considered for which unambiguous data were obtained for at least three nSSR loci, leaving a total of 305 individuals (Table 1).
Mean allelic richness (Ar, using rarefaction) and mean expected heterozygosity (He) were calculated for each stand using Fstat 2·9·3·2 (Goudet, 1995) to characterize local genetic diversities.
Two types of assignment tests were run with the allelic data (a) to allocate individuals to genetic clusters and (b) to associate the resulting clusters to the three oak taxa. In the first assignment test, for which Structure 2·2 was used (Pritchard et al., 2000; Falush et al., 2003), an attempt was made to find the optimal grouping of individuals without prior classification. The program was run for K = 1–10 expecting K = 3 (i.e. three species) to be the optimal cluster number. Chosen for the present study were 50 000 burnin periods and 50 000 Monte Carlo Markov Chain repetitions after burnin, selecting the admixture model option (uniform prior alpha = 1·0 for all populations) with correlated frequencies (prior mean FST = 0·1 equal for all populations) and no a priori assignment to local populations (POPINFO off). Five iterations were run at each level of K, using mean assignment probabilities per individual over these runs. From the results of the most likely Structure run (K = 3; see Results), individuals were grouped with a mean assignment probability >0·8 for either of the clusters into arbitrary taxonomic units. Hence, three artificial taxonomic clusters of trees (hereafter termed ‘synthetic populations’) were obtained, irrespective of their origin. These synthetic populations were considered as representative of any of the three taxa studied (Q. petraea, Q. pubescens and Q. robur), accepting a certain degree of ambiguity in these genetic clusters owing to the relatively low assignment probability threshold (0·8).
The second type of assignment test was performed to confirm that the above clustering indeed represented separate oak species, specifically testing Q. petraea and Q. robur. This assignment test, implemented in Arlequin 3·11 (Excoffier et al., 2005), calculates the maximum likelihood of multilocus genotypes to belong to either of two entities, given their respective allele frequencies (Paetkau et al., 1997; Waser and Strobeck, 1998). Accordingly, formerly identified individuals from the stand Büren, whose species membership had previously been determined (Gugerli et al., 2007), were assigned to either of the synthetic Q. petraea or Q. robur populations (see above), excluding the Büren individuals from the respective synthetic populations. Because the majority of Büren individuals had previously been identified as Q. robur, all individuals from this stand were a priori combined with the Q. robur synthetic population for calculating underlying allele frequencies of groups in the test. However, the reverse grouping, i.e. all Büren genotypes associated with Q. petraea, did not affect the subsequent assignment results (data not shown). One should note that the Büren individuals showed nearly perfect congruence between the taxon assignments based on leaf morphology and multilocus genotype (Gugerli et al., 2007), precluding circular reasoning.
Analyses of molecular variance (AMOVAs) were also run with Arlequin, with 10 000 permutations for significance testing in order to find the maximum among-group variation (ΦCT), suggesting the biologically most meaningful population grouping in analogy to SAMOVA (Dupanloup et al., 2002). Stands were grouped either as belonging to Q. petraea, Q. pubescens, Q. robur, or being mixed, or only ‘pure’ stands were classified in three groups. Both types of AMOVAs were run with taxonomic assignment of stands based on leaf morphology (Finkeldey and Mátyás, 2003), based on the above genetic assignment test, respectively (Table 1), or based on their geographic locations (three groups: south-western, north-western and eastern; Fig. 1).
RESULTS
Though not previously applied to Q. pubescens, all loci amplified well in all three taxa. Even QrZAG30, despite its complex sequence described for Q. petraea and Q. robur (Gugerli et al., 2008), revealed electrophoretic patterns that could be readily scored also in Q. pubescens. With the five nuclear nSSRs genotyped, a total of 171 alleles was detected, with individual loci revealing 15–50 alleles. Mean allelic richness (Ar) per stand ranged from 11·5 to 13·6, and expected heterozygosities (He) were high with values >0·792 (Table 1).
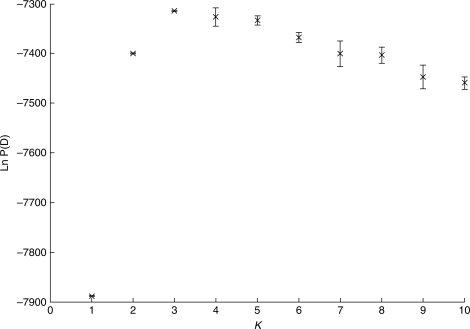
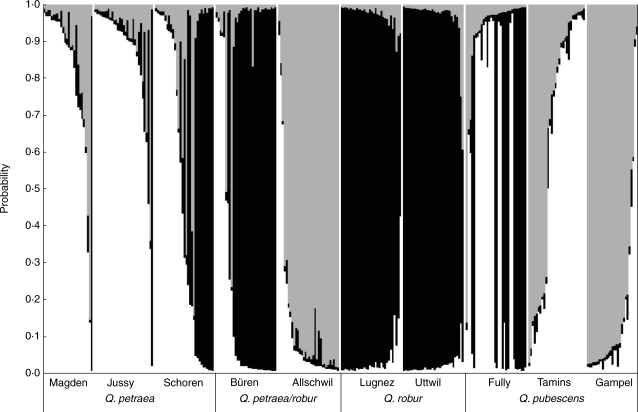
The assignment of individual multilocus genotypes into K = 1–10 genetic clusters indicated that K = 3 represented the most likely grouping according to the model assumptions and based on the criteria suggested by Pritchard et al. (2000) (Fig. 2). These criteria are (a) maximum posterior probability, Ln P(D) and (b) low variation among runs of the same K. The present data nicely fit these requirements for K = 3, and convergence of results was visually confirmed. The majority of genotypes (83·9 %) had a posterior probability >0·8 to belong to either of these three clusters (Fig. 3). Of the 47 genotypes (15·4 %) with missing data at one or two loci, 23·4 % showed ambiguous taxon assignment. The 49 trees (16·1 %) with individual posterior probability <0·8 were rather equally distributed among the ten stands. When a higher threshold (0·9) was applied, the number of trees with ambiguous assignment increased to 95 (31·1 %).
Fig. 2.
Mean (from five runs) and standard deviation of posterior probabilities, Ln P(D), as a function of K = 1–10 clusters obtained from Structure analysis (Pritchard et al., 2000; Falush et al., 2003), using nuclear microsatellite data of ten Quercus spp. stands from Switzerland.
Fig. 3.
Results of the assignment test for individuals from ten Quercus spp. stands from Switzerland using Structure (Pritchard et al., 2000; Falush et al., 2003) with K = 3 clusters. Each individual is represented by a vertical bar, with taxon assignment probabilities for Quercus petraea (white), Q. robur (black) or Q. pubescens (grey). Stands are arranged along the x-axis with their taxonomic classification based on leaf morphology by Finkeldey and Mátyás (2003). Within stands, individuals are sorted based on their posterior probabilities.
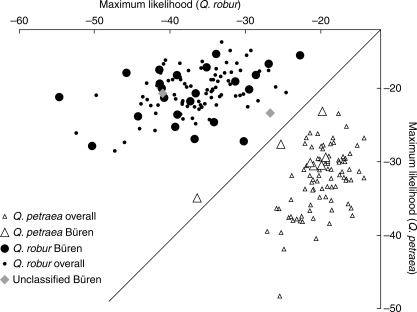
In the case of the mixed stand Büren, all individuals had previously been identified as Q. petraea, Q. robur or putative hybrids (‘unclassified’), based on leaf morphology as well as multilocus genotypes (Gugerli et al., 2007). This information was used to see whether the Structure clusters had been correctly associated with the respective oak taxa. Therefore, two synthetic populations, comprising all individuals assigned to either Q. petraea or Q. robur from nine stands, were compiled (see Materials and methods) and these were used for a second assignment test of the 32 Büren individuals randomly selected for the present study. All trees from stand Büren, except for one individual previously identified as Q. petraea, were assigned to their respective synthetic populations (Fig. 4). The two previously ‘unclassified’ trees from stand Büren were both associated with the Q. robur group (Fig. 4), which corresponded to the taxon assignment inferred by Gugerli et al. (2007).
Fig. 4.
Assignment of individual trees from the mixed stand Büren (Quercus petraea, Q. robur) to either of two synthetic populations composed from nine Quercus spp. stands from Switzerland (for details see text). Figure amended from (Gugerli et al., 2005).
In all four types of groupings used in the AMOVAs, most of the total genetic variation was assigned to the within-population level (90·9–94·3 %, P < 0·001; Table 2). Among-group variation (ΦCT) was higher when considering only pure stands rather than including mixed ones, and groups assigned on the basis of nSSRs differentiated more clearly than when classified according to leaf morphology (Table 2). The highest percentage of among-group genetic variation resulted when only pure stands as identified by multilocus genotype assignment were included (Table 2). When grouping stands according to their geographic locations (Fig. 1), the resulting among-group variation did not significantly differ from zero (ΦCT = –0·008, P > 0·05). Values of overall population differentiation (ΦST) followed the same rank order as the one for ΦCT (Table 2).
Table 2.
Analyses of molecular variance (AMOVAs) based on five nuclear microsatellites of ten Swiss oak stands grouped as pure and mixed stands (Quercus petraea, Q. robur, Q. pubescens and mixed Q. petraea/robur) or pure stands only (both groupings were done on the basis of either leaf morphology or microsatellite data)
| Data basis for taxon assignment |
|||||||
|---|---|---|---|---|---|---|---|
| Leaf morphology |
Microsatellites |
||||||
| Grouping | Source of variation | d.f. | SS | Percentage variation | d.f. | SS | Percentage variation |
| Pure and mixed stands | Among groups (ΦCT) | 3 | 43·2 | 2·1 n.s. | 3 | 54·2 | 3·9** |
| Among populations within groups | 6 | 42·4 | 3·6*** | 6 | 31·4 | 2·2*** | |
| Within populations | 600 | 1277·5 | 94·3*** | 600 | 1277·5 | 93·9*** | |
| ΦST | 0·057*** | 0·061*** | |||||
| Pure stands | Among groups (ΦCT) | 2 | 36·9 | 3·5* | 2 | 49·6 | 7·3*** |
| Among populations within groups | 5 | 28·4 | 2·6*** | 3 | 13·6 | 1·8 *** | |
| Within populations | 474 | 1005·7 | 93·8*** | 354 | 744·2 | 90·9*** | |
| ΦST | 0·062*** | 0·091*** | |||||
Taxonomic assignment of stands to the respective groups follows the one given in Table 1.
d.f., degrees of freedom, SS, sums of squares.
*P < 0·05; **P < 0·01; ***P < 0·001; n.s., not significant.
The Structure assignment revealed a taxonomic identification of the ten studied oak stands which sometimes differed from that of the same ten stands as based on leaf morphology (Finkeldey and Mátyás, 2003). For two of the Q. petraea populations (Magden and Jussy), the two Q. robur populations (Lugnez and Uttwil), one of the Q. pubescens populations (Gampel), and the mixed stand Büren, the two assignments corresponded well (Fig. 3). In contrast, the mixed Q. petraea/Q. robur stand Allschwil showed a multilocus genotype pool fitting to Q. pubescens with few Q. petraea individuals. Likewise, Schoren as a Q. petraea population additionally indicated Q. robur trees and some influence of Q. pubescens, while two of the Q. pubescens populations revealed either Q. petraea contributions (Tamins) or even dominant Q. petraea/Q. robur composition (Fully) (Table 1 and Fig. 3).
DISCUSSION
In the present study on white oak stands of Switzerland (Quercus petraea, Q. pubescens and Q. robur), taxonomic assignment based on multilocus genotypes of nSSRs appeared reliable for a majority of individuals tested, though analysing only five loci. Unlike previous attempts in the literature that deal with genetic differentiation in closely related white oak taxa, the present approach did not rely on a priori classification of individuals or stands. Only posterior association of genetic clusters to taxa, based on previous analyses of detailed molecular and leaf morphological data for Quercus petraea and Q. robur (Gugerli et al., 2007), was used for respective classification, inferring that the third cluster represented Q. pubescens. It was further demonstrated that five loci may be sufficient to reach good separation of taxa even within a geographically limited area. Despite some limitations in the present study (no individual morphological data, loci selected for discrimination power between two of the three taxa studied), great potential is seen in the application of multilocus genotype assignments for further clarification of species status in white oaks or in other taxon complexes (Anderson and Thompson, 2002), but also for their use in certification of forestry products.
Several lines of arguments are provided that support the above approach for taxon assignment in the closely related European white oaks: (a) the most likely clustering in Structure with K = 3 corresponded to the number of taxa involved in the analysis; (b) a large majority of individual genotypes showed high posterior probability (>0·8) for either of these three clusters; (c) high success rate of assignment of individuals whose taxon was independently identified based on multivariate leaf morphology and multilocus genotypes; and (d) highest among-group variation when only using pure populations in molecular assignment. These aspects are further elucidated in the subsequent paragraphs.
First, it is shown that selecting three clusters in the Structure analysis best groups individuals based on their multilocus genotypes (Fig. 2). This finding is in line with the taxonomical rather than spatial association of the oak individuals studied, which is considered as the main pre-assumption to illustrate that the grouping approach is biologically reasonable.
Secondly, the clear assignment of individuals to either of the three clusters, irrespective of their stand of origin, also indicates the relevance of the present results. Since a general level of posterior probability cannot reasonably be set (Manel et al., 2005), 0·8 was considered as an adequate threshold of assignment (thresholds range between 0·6 and 0·9, depending on the question asked; e.g. Rosenberg et al., 2001; Craft et al., 2002; Werth et al., 2007). Only 16·1 % of the 305 trees analysed fell below this threshold (Fig. 3). Almost one-third of these low-probability assignments concerned individuals of Q. pubescens populations, for which the selected loci might not be optimal with regard to species discrimination. In addition, even genotypes with data at only three or four nSSR loci fitted well into one of the three clusters. These figures may indicate a low rate of natural hybridization, at least between Q. petraea and Q. robur, though five loci is insufficient to provide reliable estimates of hybridization rates (Vähä and Primmer, 2006). On the other hand, the five loci largely provided clear taxon assignment for most trees, even when the posterior probability threshold was increased to 0·9. Given that the chosen loci are not necessarily best suited for discriminating Q. pubescens from its congeners because the markers were originally developed for Q. petraea, Q. robur and Q. macrocarpa (Dow et al., 1995; Steinkellner et al., 1997; Kampfer et al., 1998), this is considered a promising resolution of taxonomic identity.
Thirdly, when the present method was tested with the data from individually characterized trees from stand Büren (Gugerli et al., 2007), 31 of the 32 trees were classified as belonging to the same taxon as in the previous study. The multilocus genotype of the single Q. petraea individual misclassified showed an only slightly higher likelihood for Q. robur than for Q. petraea (Fig. 4). Note that individuals of the Büren population were excluded when calculating the allele frequencies of the synthetic populations to prevent circular reasoning. Of the two previously ‘unclassified’ individuals, that were associated with Q. robur by multilocus genotype and spatial analyses in Gugerli et al. (2007), both were again assigned to Q. robur (Fig. 3). These observations were taken as support for taxonomic clustering of the ten study stands based on the first assignment test, further corroborating the respective association of genetic clusters (synthetic populations) and taxa (Table 1 and Fig. 3).
Fourthly, the clearest separation was obtained in AMOVA, i.e. the highest among-group partitioning of genetic variation (ΦCT), when grouping stands according to the present assignment based on nSSR multilocus genotypes. Alternatively grouping stands according to their geographical proximity resulted in an among-group partitioning of genetic variation that was not significantly different from zero. This evidence shows that the taxon-specific signal of differentiation is stronger than the spatially induced differentiation in the present data set, suggesting that gene flow is lower among taxa than among populations.
Final evidence comes from regional oak occurrence in Switzerland. In several cases, the former taxonomic classification of the study stands, based on leaf morphology (Finkeldey and Mátyás, 2003), did not correspond to the clustering obtained with the assignment of multilocus genotypes in the present study. These discrepancies may be best explained by the distribution of oak species in Switzerland. Schoren is a mesic site in an area for which mixing of Q. petraea and Q. robur is reported (Brodtbeck et al., 1997). The proximity of the Jura mountains, where Q. pubescens s.l. occurs, suggests that the Q. petraea population Allschwil may well be associated with Q. pubescens (Brodtbeck et al., 1997). The stand Fully is situated in the central Alpine Rhone valley, where Q. pubescens has one of its main areas of distribution in Switzerland. However, both other oak species also occur in this region and intermixing is possible (Gams, 1927; Braun-Blanquet, 1961; Werner, 1988). Finally, Tamins is located in the central Alpine valley of the Rhine, which is at the northern brink of the occurrence of Q. pubescens in Switzerland. It is likely that Q. petraea, the most abundant of the three oak species in this area (Müller, 1999), had an introgressive effect on this population. There is even the argument that Q. pubescens cannot be found in its pure form north of the Alps (Müller, 1999). Taken together, the present genetic assignment of stands to the three oak species is in accordance with their occurrence in Switzerland and with the literature, though detailed analyses at the individual level would be required to substantiate the coincidence of morphology and molecular data. We further advocate to thoroughly test the applicability of sets of nSSRs to clarify the status of Q. pubescens in Switzerland and elsewhere (but see Bruschi et al., 2000). Since it was only possible to associate Q. petraea and Q. robur to particular genetic clusters based on previous results, the concluding proof that the third cluster represents Q. pubescens is also pending.
There were still some individuals with low posterior probabilities for one of the three clusters in the assignment test (Fig. 3). This is taken as an indication that the composition of some genotypes could be the result of hybridization and introgression. However, only diagnostic markers for the three species would unambiguously identify such hybrids and allow us to better characterize species compositions in white oak forests. Unfortunately, such diagnostic molecular markers are not available to date. Furthermore, because of the low intensity of within-genome sampling in the present study it is not possible to infer properly the degree of admixture among the three taxa studied.
From the present study, it is concluded that assignment methods using multilocus genotypes have great potential to identify species composition, genetic admixture and putative hybridization. Given adequate regional sampling and the selection of differentiated loci, single stands or seed lots could be readily characterized. Forest managers are in favour of maintaining pure oak stands, in particular when seed will be harvested for propagation. They argue that hybrid formation in mixed stands may hamper regeneration success – assuming that fitness is reduced in hybrids compared with offspring from intraspecific crosses. If aiming at selecting pure stands, assignment tests as proposed here provide decision support towards the selection of suitable stands for management purposes. However, one must consider that long-distance gene flow via pollen may still lead to hybrid seeds even in pure stands. Furthermore, the present approach may be useful to identify and certify seed and other forestry products such as wood (Deguilloux et al., 2004).
ACKNOWLEDGEMENTS
The authors thank Sonia Angelone and Annie Diarra for help with the sampling. Antoine Kremer, Rémy Petit, Giovanni ‘Beppe’ G. Vendramin, Christian Lexer as the handling editor and anonymous reviewers critically read and improved earlier versions of the manuscript. We also thank the local forest services for allowing us to collect leaves in their stands. Funding is gratefully acknowledged from the Swiss Agency of the Environment (BAFU); European Commission (OAKFLOW project, QLK5-2000–00960); and State Secretariat for Education and Research, Switzerland (SER, 99·0838) to F.G.
LITERATURE CITED
- Aldrich PR, Parker GR, Michler CH, Romero-Severson J. Whole-tree silvic identifications and the microsatellite genetic structure of a red oak species complex in an Indiana old-growth forest. Canadian Journal of Forest Research. 2003;3:2228–2237. [Google Scholar]
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- Barreneche T, Casasoli M, Russell K, Akkak A, Meddour H, Plomion C, et al. Comparative mapping between Quercus and Castanea using simple-sequence repeats (SSRs) Theoretical & Applied Genetics. 2004;108:558–566. doi: 10.1007/s00122-003-1462-2. [DOI] [PubMed] [Google Scholar]
- Bodénès C, Joandet S, Laigret F, Kremer A. Detection of genomic regions differentiating two closely related oak species Quercus petraea (Matt.) Liebl. and Quercus robur L. Heredity. 1997;a 78:433–444. [Google Scholar]
- Bodénès C, Labbé T, Pradère S, Kremer A. General vs. local differentiation between two closely related white oak species. Molecular Ecology. 1997;b 6:713–724. [Google Scholar]
- Braun-Blanquet J. Die inneralpine Trockenvegetation. Stuttgart: Gustav Fischer; 1961. [Google Scholar]
- Brewer S, Cheddadi R, de Beaulieu JL, Reille M, Contributors D. The spread of deciduous Quercus throughout Europe since the last glacial period. Forest Ecology and Management. 2002;156:27–48. [Google Scholar]
- Brodtbeck T, Zemp M, Frei M, Kienzle U, Knecht D. Flora von Basel und Umgebung. Mitteilungen der Naturforschenden Gesellschaften beider Basel. 1997;2:9–543. 1980–1996 Teil 1. [Google Scholar]
- Bruschi P, Vendramin GG, Bussotti F, Grossini P. Morphological and molecular differentiation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. (Fagaceae) in northern and central Italy. Annals of Botany. 2000;85:325–333. [Google Scholar]
- Craft KJ, Ashley MV, Koenig WD. Limited hybridization between Quercus lobata and Quercus douglasii (Fagaceae) in a mixed stand in central coastal California. American Journal of Botany. 2002;89:1792–1798. doi: 10.3732/ajb.89.11.1792. [DOI] [PubMed] [Google Scholar]
- Deguilloux M-F, Pemonge M-H, Petit RJ. DNA-based control of oak wood geographic origin in the context of the cooperage industry. Annals of Forest Science. 2004;61:97–104. [Google Scholar]
- Dow BD, Ashley MV, Howe HF. Characterization of highly variable (GA/CT)n microsatellites in the bur oak, Quercus macrocarpa. Theoretical & Applied Genetics. 1995;91:137–141. doi: 10.1007/BF00220870. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Dzialuk A, Chybicki I, Welc M, Sliwinska E, Burczyk J. Presence of triploids among oak species. Annals of Botany. 2007;99:959–964. doi: 10.1093/aob/mcm043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkeldey R, Mátyás G. Genetic variation of oaks (Quercus spp.) in Switzerland. 3. Lack of impact of postglacial recolonization history on nuclear gene loci. Theoretical & Applied Genetics. 2003;106:346–352. doi: 10.1007/s00122-002-1002-5. [DOI] [PubMed] [Google Scholar]
- Gams H. Von den Follatères zur Dent des Morcles. Beiträge zur geobotanischen Landesaufnahme. 1927;15:1–760. [Google Scholar]
- González-Rodríguez A, Arias DM, Oyama K. Genetic variation and differentiation of populations within the Quercus affinis–Quercus laurina (Fagaceae) complex analyzed with RAPD markers. Canadian Journal of Botany. 2005;83:155–162. [Google Scholar]
- Goudet J. FSTAT (version 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Gugerli F, Brodbeck S, Holderegger R. Die unerträgliche Leichtigkeit, eine reine Eiche zu sein. Blattmorphologie und genetischer Fingerabdruck als unterschiedlich einsetzbare Bestimmungshilfen. Informationsblatt Forschungsbereich Landschaft. 2005;63:1–5. [Google Scholar]
- Gugerli F, Walser J-C, Dounavi K, Holderegger R, Finkeldey R. Coincidence of small-scale spatial discontinuities in leaf morphology and nuclear microsatellite variation of Quercus petraea and Q. robur in a mixed forest. Annals of Botany. 2007;99:713–722. doi: 10.1093/aob/mcm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugerli F, Brodbeck S, Holderegger R. Insertions–deletions in a microsatellite flanking region may be resolved by variation in stuttering patterns. Plant Molecular Biology Reporter. 2008;26:255–262. [Google Scholar]
- Howard DJ, Preszler RW, Williams J, Fenchel S, Boecklen WJ. How discrete are oak species? Insights from a hybrid zone between Quercus grisea and Quercus gambelii. Evolution. 1997;51:747–755. doi: 10.1111/j.1558-5646.1997.tb03658.x. [DOI] [PubMed] [Google Scholar]
- Ishida TA, Hattori K, Sato H, Kimura MT. Differentiation and hybridization between Quercus crispula and Q. dentata (Fagaceae): insights from morphological traits, amplified fragment length polymorphism markers, and leafminer composition. American Journal of Botany. 2003;90:769–776. doi: 10.3732/ajb.90.5.769. [DOI] [PubMed] [Google Scholar]
- Kampfer S, Lexer C, Glössl J, Steinkellner H. Characterization of (GA)(n) microsatellite loci from Quercus robur. Hereditas. 1998;129:183–186. [Google Scholar]
- Kremer A, Savill PS, Steiner KC, editors. Genetics of oaks. Paris: Elsevier; 1993. [Google Scholar]
- Kremer A, Dupouey JL, Deans JD, Cottrell J, Csaikl U, Finkeldey R, et al. Leaf morphological differentiation between Quercus robur and Quercus petraea is stable across western European mixed oak stands. Annals of Forest Science. 2002;a 59:777–787. [Google Scholar]
- Kremer A, Kleinschmit J, Cottrell J, Cundall EP, Deans JD, Ducousso A, et al. Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? Forest Ecology and Management. 2002;b 156:75–87. [Google Scholar]
- Lexer C, Kremer A, Petit RJ. Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Molecular Ecology. 2006;15:2007–2012. doi: 10.1111/j.1365-294X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Gaggiotti OE, Waples RS. Assignment methods: matching biological questions with appropriate techniques. Trends in Ecology & Evolution. 2005;20:136–142. doi: 10.1016/j.tree.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Mariette S, Cottrell JE, Csaikl UM, Goikoechea P, König A, Lowe AJ, et al. Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (Matt.) Liebl. and Q. robur L. stands. Silvae Genetica. 2002;a 51:72–79. [Google Scholar]
- Mariette S, Le Corre V, Austerlitz F, Kremer A. Sampling within the genome for measuring within-population diversity: trade-offs between markers. Molecular Ecology. 2002;b 11:1145–1156. doi: 10.1046/j.1365-294x.2002.01519.x. [DOI] [PubMed] [Google Scholar]
- Muir G, Schlötterer C. Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.) Molecular Ecology. 2005;14:549–561. doi: 10.1111/j.1365-294X.2004.02418.x. [DOI] [PubMed] [Google Scholar]
- Muir G, Schlötterer C. Moving beyond single-locus studies to characterize hybridization between oaks (Quercus spp.) Molecular Ecology. 2006;15:2301–2304. doi: 10.1111/j.1365-294X.2006.02912.x. [DOI] [PubMed] [Google Scholar]
- Muir G, Fleming CC, Schlötterer C. Species status of hybridizing oaks. Nature. 2000;405:1016. doi: 10.1038/35016640. [DOI] [PubMed] [Google Scholar]
- Müller B. Zürich: ETH Zürich; 1999. Variation und Hybridisierung von Quercus pubescens. PhD Thesis. [Google Scholar]
- Nixon KC. Infrageneric classification of Quercus (Fagaceae) and typification of sectional names. Annales des Sciences Forestières. 1993;50:25s–34s. [Google Scholar]
- Paetkau D, Waits LP, Clarkson PL, Craighead L, Strobeck C. An empirical evaluation of genetic distance statistics using microsatellite data from bear (Ursidae) populations. Genetics. 1997;147:1943–1957. doi: 10.1093/genetics/147.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, et al. Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecology and Management. 2002;156:5–26. [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytologist. 2003;161:151–164. [Google Scholar]
- Ponton S, Dupouey J-L, Bréda N, Dreyer E. Comparison of water-use efficiency of seedlings from two sympatric oak species: genotype × environment interactions. Tree Physiology. 2002;22:413–422. doi: 10.1093/treephys/22.6.413. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MAM, et al. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics. 2001;159:699–713. doi: 10.1093/genetics/159.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton BS. Natural hybridization within the genus Quercus L. Annales des Sciences Forestières. 1993;50:73s–90s. [Google Scholar]
- Saintagne C, Bodénès C, Barreneche T, Pot D, Plomion C, Kremer A. Distribution of genomic regions differentiating oak species assessed by QTL detection. Heredity. 2004;92:20–30. doi: 10.1038/sj.hdy.6800358. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodénès C, et al. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q. petraea (Matt.) Liebl.] Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL, Matzke EB, Epling C. Hybridization in a population of Quercus marilandica and Q. ilicifolia. Evolution. 1947;1:79–88. [Google Scholar]
- Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, et al. Identification and characterization of (GA/CT)n-microsatellite loci from Quercus petraea. Plant Molecular Biology. 1997;33:1093–1096. doi: 10.1023/a:1005736722794. [DOI] [PubMed] [Google Scholar]
- Tovar-Sánchez E, Oyama K. Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. American Journal of Botany. 2004;91:1352–1363. doi: 10.3732/ajb.91.9.1352. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Burges NA, Chater AO, Edmonds JR, Heywood VH, Moore DM, et al. Flora Europaea. 2 edn. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Vähä J-P, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Waser PM, Strobeck C. Genetic signatures of interpopulation dispersal. Trends in Ecology & Evolution. 1998;13:43–44. doi: 10.1016/s0169-5347(97)01255-x. [DOI] [PubMed] [Google Scholar]
- Werner P. Etude botanique des Follatères (Dorénaz et Fully, Valais). III. Les Fôrets. Bulletin de la Murithienne; société valaisianne des sciences naturelles. 1988;106:101–117. [Google Scholar]
- Werth S, Gugerli F, Holderegger R, Wagner HH, Csencsics D, Scheidegger C. Landscape-level gene flow in Lobaria pulmonaria, an epiphytic lichen. Molecular Ecology. 2007;16:2807–2815. doi: 10.1111/j.1365-294X.2007.03344.x. [DOI] [PubMed] [Google Scholar]
- Zanetto A, Roussel G, Kremer A. Geographic variation of inter-specific differentiation between Quercus robur L. and Quercus petraea (Matt.) Liebl. Forest Genetics. 1994;1:111–123. [Google Scholar]