Abstract
Background and Aims
Iron deficiency is one of the most common nutritional disorders in plants, especially in fruit trees grown in calcareous soil. Malus baccata is widely used as an apple rootstock in north China and is highly resistant to low temperatures. There are few studies on iron absorption by this species at the molecular level. It is very important to understand the mechanism of iron uptake and transport in such woody plants. As a helpful tool, the aim of the present study was the cloning and functional analysis of NRAMP (natural resistance-associated macrophage protein) genes from the apple tree in relation to trafficking of micronutrients (Fe, Mn and Cd).
Methods
Reverse transcription-PCR (RT-PCR) combined with RACE (rapid amplification of cDNA ends) was adopted to isolate the full-length NRAMP1 cDNA. Southern blotting was used to test gene copy information, and northern blot was used to detect the gene's expression level. Complementation experiments using the yeast mutant strains DEY1453 and SLY8 were employed to confirm the iron- and manganese-transporting ability of NRAMP1 from apple, and inductively coupled plasma (ICP) spectrometry was used to measure Cd accumulation in yeast. NRAMP1–green fluorescent protein (GFP) fusion protein was used to determine the cellular localization in yeast.
Key Results
A 2090 bp cDNA was isolated and named MbNRAMP1. It encodes a predicted polypeptide of 551 amino acids. MbNRAMP1 exists in the M. baccata genome as a single copy and was expressed mainly in roots. MbNRAMP1 rescued the phenotype of yeast mutant strains DEY1453 and SLY8, and also increased Cd2+ sensitivity and accumulation. MbNRAMP1 expression in yeast was largely influenced by iron status, and the expression pattern of MbNRAMP1–GFP varied with the environmental iron nutrition status.
Conclusions
MbNRAMP1 encodes a functional metal transporter capable of mediating the distribution of ions as well as transport of the micronutrients, Fe and Mn, and the toxic metal, Cd.
Key words: Apple, cadmium, iron-regulated transporter, iron, manganese, NRAMP1, Malus baccata
INTRODUCTION
During evolution, plants have developed unique ways to acquire essential nutritional elements and exclude or extract toxic metals from the soil (Lahner et al., 2003). Insight into the pathway by which metals accumulate in plants will have a dramatic impact on both plant and human health. Identifying metal transporters is an important step in this pathway. Previous studies showed that metal transporters regulated by iron can transport a variety of divalent metals, suggesting that iron regulation is important for a plant's response to environmental metals status (Bereczky et al., 2003; Sharma et al., 2004). Iron deficiency-induced plant chlorosis in young leaves is a major global agricultural problem (Römheld and Marschner, 1986; Ling et al., 1999). Molecular characterization of transporter genes involved in iron mobilization in plants exposed to iron deficiency has become an urgent task. To date, two classes of transporters, Irt/Zip and Dmt/Nramp, involved in iron uptake have been identified in plants (Eide et al., 1996; Belouchi et al., 1997; Curie et al., 2000; Guerinot et al., 2000; Thomine et al., 2000; Connolly et al., 2002; Vert et al., 2002; Bereczky et al., 2003; Kaiser et al., 2003).
NRAMP (natural resistance-associated macrophage protein) consists of a group of highly conserved integral membrane proteins and exists widely in bacteria, fungi, plants and animals (Gruenheid et al., 1995; Cellier et al., 1996, 2001; Williams et al., 2000). Previous reports showed that NRAMPs exhibit functional divergence and somewhat broad substrate specificity in different species. LeNRAMP1 from Lycopersicon esculentum has a root-specific expression and is regulated by iron status (Bereczky et al., 2003). AtNRAMP1, AtNRAMP3 and AtNRAMP4 encode functional plant metal transporters and are expressed at higher levels upon low iron supply in plants (Curie et al., 2000; Thomine et al., 2000). AtNRAMP1 was reported as a metal transporter and complements the growth defect of the yeast Fe(II) transport mutant DEY1453 (Curie et al., 2000; Thomine et al., 2000). AtNRAMP3 acts as a metal transporter exporting metals from the vacuolar compartment to the cytosol in association with the vacuolar membrane (Thomine et al., 2003). AtNRAMP3 and AtNRAMP4 mobilize vacuolar Fe stores and are crucial for seed germination (Lanquar et al., 2005). TjNRAMP4, identified from an Ni-hyperaccumulating plant, Thlaspi japonicum, transports specifically Ni but not Zn, Cd or Mn (Mizuno et al., 2005).
Malus baccata is widely used as an apple rootstock in north China (Han and Shen, 1991) and is highly resistant to low temperatures (up to –45°C). At present, nothing is known about the cellular localization or regulation of NRAMP homologues in woody plants. Are these proteins in woody plants functionally similar to the already identified NRAMP homologues in plants, or do they act in unique metal transport pathways? To address this issue, the MbNRAMP1 gene from an apple tree (M. baccata) was cloned and functionally characterized. The role of NRAMP1 was also clarified using subcellular localization with MbNRAMP1 fused to green fluorescent protein (GFP) in yeast.
MATERIALS AND METHODS
Plant growth
For root induction, sterilized, semi-lignified Malus baccata (L.) Borkh shoot-tip cuttings were cultured in MS medium with all components except Fe and vitamins at half strength. Robust and healthy looking rooted cuttings were grown in the solution as previously described (Han et al., 1994) for about 1 month. For micronutrient deficiency treatment, two-thirds of the plants were transplanted into media with a low (4 µm Na2Fe-EDTA) iron concentration but with adequate concentrations of other nutrient elements.
MbNRAMP1 isolation and sequences analysis
Total RNA was extracted separately from M. baccata roots and leaves. First-strand cDNA was synthesized from mRNA using SuperScript™III reverse transcriptase (Invitrogen). Degenerate primers (forward 5′-GTD ATG CCR CAY AAT CTN TTC YTB CAC TC-3′; reverse 5′-GCY TCW GGW CAR AGC TCN AC-3′) corresponding to the conserved motifs of NRAMP1 in plants were used to amplify specific DNA fragments from cDNA. The PCR product amplified with Ex Taq DNA polymerase (TaKaRa, Japan) was cloned into the T-cloning vector pGEM®-T Easy Vector. Both 3′ and 5′ end fragments were obtained according to the 3′- and 5′-Full RACE Core Sets (TaKaRa). The complete MbNRAMP1 cDNA containing the open reading frame was amplified from the first-strand cDNA using a forward primer (5′-ATA CAC ATC CTA CAG ATT CAA ATT-3′) and a 3′ site adaptor primer. Sequence alignment analysis was performed with CLUSTALW (http:// www.ebi.ac.uk/clustalw) and BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). The dendrogram was produced with MEGA3.
Southern and northern blot analysis
For Southern blot analysis, 15 µg samples of M. baccata genomic DNA were digested overnight with BamHI or HindIII, separated by electrophoresis on a 0·8 % agarose gel, transferred to a nylon membrane, and bound to the membrane by UV cross-linking. For northern blot analysis, total RNA was separately extracted from roots and leaves. A 10 µg aliquot of total RNA was separated by electrophoresis on an 18 % denaturing 1× MOPS/1 % (w/v) agarose gel containing formaldehyde and transferred onto a nylon membrane (Sambrook et al., 2001). The RNA was fixed to the membrane by UV cross-linking. Full-length cDNAs were labelled by [α-32P]dCTP as probes. Hybridization was conducted for 16 h at 65°C. After washing in 5× SSC/0·1 % SDS at 65°C for 2× 15 min and 0·1× SSC/0·1 % SDS at 70°C for 3× 15 min, the filters were exposed to phosphor screen (Perkin Elmer™ Lifesciences).
Functional expression of MbNRAMP1 in yeast
The yeast strains used in this study were wild types 190 and DEY1457 (MATα can1 his3 leu2 trp1 ura3 ade6), and yeast mutant strains DEY1453 (fet3-2::His3/ fet3-2::His3, fet4-1::LEU2/fet4-1::LEU2, ade2/ + ura3/ura3 trp1/trp1 leu2/leu2 his3/his3 can1/can1) and smf1 (smf1Δ, SLY8; MATα his3 ade2 leu2 trp1 ura3 smf1::HIS3). To grow the yeast cells, a yeast extract–peptone–dextrose (YPD) medium or synthetic dextrose (SD) medium was used (Gietz and Woods, 2002). Yeast cells were grown on YPD before transformation and on SD medium containing amino acid supplements without tryptophan and 2 % glucose (SD-Trp) after transformation. A synthetic chelator of iron and manganese, BPDS (bathophenanthroline disulfonic acid disodium salt), and EGTA were added separately to the SD-Trp medium to deprive yeast mutant strains of Fe or Mn during rescue experiments.
The MbNRAMP1-coding region was inserted into the NdeI and BamHI sites of pGBKT7 (Clontech) behind the ADH1 promoter, yielding pGBKT7-MbNRAMP1. The plasmid was introduced into the yeast according to the method of Gietz and Woods (2002). The transformed cells were selected on solid SD-Trp medium overnight and cultured in liquid SD-Trp medium overnight. The overnight cultures of transformed cells were each diluted to OD 1·00, 0·10 and 0·01 at 600 nm. Diluted transformed cells (5 µL) were then spotted onto SD-Trp media plates. For the DEY1453 growth test experimental culture, the SD-Trp medium was supplemented with 10 µm iron (II) citrate (20 : 1 sodium citrate:FeSO4) with (Fe deficient) or without 30 µm BPDS. For the SLY8 rescue experiment, the medium was adjusted to pH 6·0 with 50 mm MES and 20 mm EGTA (Mn deficient). Photographs of the yeast colonies were taken after 3 d growth at 30°C.
Transporting abilities of Cd2+
The wild yeast strain, DEY1457, transformed with an empty vector, pGBKT7, or pGBKT7 containing MbNRAMP1, was grown for 36 h starting from an OD of 0·01 in 20·0 mL of SD-Trp medium supplemented with 10 µm CdCl2. Cells were allowed to grow for 36 h in 20 mL of the culture and the OD was measured at 0, 24 and 36 h. After 36 h, cells were collected and washed by centrifugation with 50 mm Tris–HCl, pH 6·5 and 10 mm EDTA. Cell pellets were digested in 200 µL of 5:2 nitric acid:perchloric acid at 80°C for 1 h following the methods established by Li and Kaplan (1998). After digestion, samples were diluted to 1·0 mL with deionized water and then analysed with a Perkin-Elmer inductively coupled plasma (ICP) atomic absorption spectrometer.
Localization of GFP fusion protein in yeast
Enhanced green fluorescent protein (EGFP), excised from pEGFP (Clontech) using BamHI and PstI, was cloned into the corresponding sites of pGBKT7, yielding pGBKT7-GFP. The MbNRAMP1 open reading frame region without a stop codon was integrated into pGBKT7-GFP to express a fusion protein, MbNRAMP1–GFP, yielding pGBKT7-NRAMP1 fused to GFP. The pGBKT7-NRAMP1-GFP was firstly transformed into yeast mutants DEY1453 and SLY8 to test if MbNRAMP1–GFP can rescue the phenotype of mutant strains. Then plasmids pGBKT7-NRAMP1-GFP and pGBKT7-GFP were transformed into the yeast strain, DEY1457 (MATα can1 his3 leu2 trp1 ura3 ade6) individually. Positive colonies selected on SD-Trp solid medium were each inoculated into the SD-Trp liquid medium (normal growth condition) or SD-Trp liquid medium containing either 30 µm BPDS or 20 mm EGTA at 30°C with shaking (160 r.p.m.). After 2 d of growth, cells were observed with Leica Confocal Systems TCS SP2 (Germany), for localization of GFP fusion protein fluorescence (excitation, 488 nm wavelength; emission measured between 505 and 520 nm).
RESULTS
Isolation, cloning and sequence analysis of the MbNRAMP1 cDNA from M. baccata
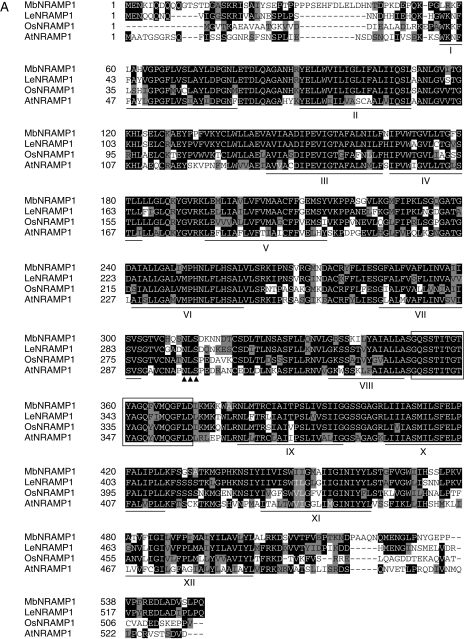
A 2090 bp cDNA was isolated using reverse transcription-PCR (RT-PCR), followed by 5′ and 3′ RACE (rapid amplification of cDNA ends) analysis and determination of the sequence of one NRAMP homologue through BLAST at the NCBI website. The sequence of the NRAMP cDNA was designated as MbNRAMP1 and was filed in the GenBank databases under the accession number AY724413. The sequence was cloned into pGEM-T easy vector for subsequent analysis. The deduced amino acid sequence shows that MbNRAMP1 encodes a polypeptide of 551 amino acids with a predicted molecular mass of 59·7 kDa. The MbNRAMP1 was 76 % identical with LeNRAMP1 (accession no. AY196091) of L. esculentum, 68 % identical with OsNRAMP1 (accession no. L41217·1) of Oryza sativa and 58 % identical with AtNRAMP1 (accession no. AAF36535) of Arabidopsis thaliana. Figure 1A shows the alignment of amino acid sequences of MbNRAMP1 and other NRAMP1 homologues in other plant species.
Fig. 1.
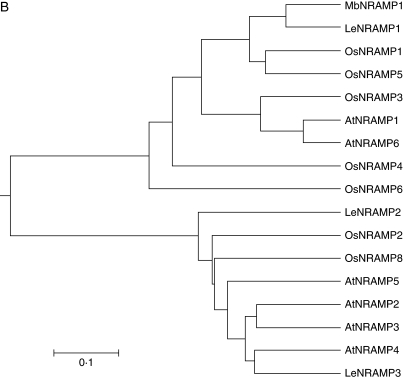
Sequence conservation in plant NRAMP family transporter proteins. (A) Multiple alignment of deduced amino acid sequences of NRAMP1 transporter proteins of M. baccata (MbNRAMP1, accession no. AY724413), L. esculentum (LeNRAMP1, accession no. AY196091), O. sativa (OsNRAMP1, accession no.L41217·1) and A. thaliana (AtNRAMP1, accession no. AAF 36535) with the ClustalW program; residues were shaded with Boxshade. Black and grey backgrounds indicate identical residues and conservative substitutions. respectively. Lines under the sequences indicate the positions of the transmembrane domains. The consensus transport motif between transmembrane domains VIII and IX is boxed. The arrowheads which showed putative N-linked glycosylation sites presented in picture of figure 1A. (B) Dendrogram showing the amino acid sequence of the NRAMP proteins from M. baccata, L. esculentum, O. sativa and A. thaliana produced with MEGA3.
From Fig. 1A, it was found that all conserved features of NRAMP described previously (Supek et al., 1996; Curie et al., 2000) were present in the predicted MbNRAMP1 sequence, such as 12 transmembrane domains (TMs), putative N-linked glycosylation sites and a consensus transporter motif (CTM) located in the fourth intracellular loop between TMs VIII and IX at amino acids 351–371. In bacteria, this motif (CTM) is believed to interact with ATP-coupling subunits; however, its function is unknown and it is obviously different from the nucleotide-binding fold of ABC transporters (Gunshin et al., 1997). Phylogenic analysis of MbNRAMP1 with three Lycopersicon, eight Oryza and six Arabidopsis NRAMPs revealed that MbNRAMP1 and LeNRAMP1 clustered in a small group and are most related (Fig. 1B).
DNA and RNA hybridization analysis of the MbNRAMP1 gene
The frequency of occurrence of the MbNRAMP1 gene was confirmed with Southern blots of DNA digested with either BamHI or HindIII probed with MbNRAMP1. A single strong band was present (Fig. 2), suggesting the existence of a single copy of MbNRAMP1 in the M. baccata genome.
Fig. 2.

Southern blot of MbNRAMP1 in M. baccata plants. Blots were probed with MbNRAMP1. Each lane contains 15 µg of M. baccata genomic DNA digested with either BamHI (lane 1) or HindIII (lane 2).
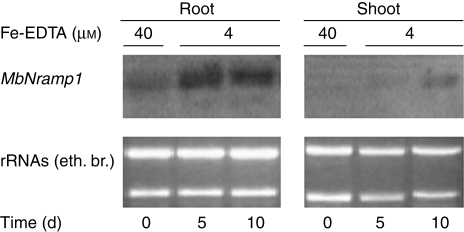
To investigate the expression pattern of MbNRAMP1's response to the iron status, total RNA was extracted from roots and shoots of plants which were grown separately under nutrient-sufficient (0 day, as control) and Fe2+-deficient conditions (4 µm Fe2+) for 5 and 15 d. The RNA was then analysed by northern blotting with MbNRAMP1 as a probe (Fig. 3). Expression of MbNRAMP1 was markedly increased in roots under iron deficiency, and its mRNA accumulation patterns differed with the induction time. Under adequate nutrition, the MbNRAMP1 transcripts were very faint in roots, and nearly invisible in shoots (Fig. 3). The highest level of MbNRAMP1 transcripts was observed in roots which had been grown under Fe deficiency for 5 d. Although the transcripts level in roots was slightly decreased at day 15 compared with day 5, it was still higher than the control. In shoots, the MbNRAMP1 mRNA was very weakly detected in seedlings grown under low iron conditions (Fig. 3).
Fig. 3.
Expression of MbNRAMP1 mRNA as regulated by iron status. Northern blots in the upper figure indicated the temporal expression of MbNRAMP1 in M. baccata plants in control (0 day) and iron-deficient conditions for the different times indicated. The corresponding ethidium bromide-stained rRNAs are shown as a loading control.
M. baccata NRAMP1 gene complements yeast mutants defective in Mn and Fe transport
The high sequence homology between MbNRAMP1 and other NRAMPs at the putative protein level, as well as the observation that MbNRAMP1 was indeed regulated by iron status suggests that MbNRAMP1 might encode a functional metal NRAMP transporter. Previous reports on NRAMP family transporters revealed that they transport metals such as Fe and Mn (Curie et al., 2000; Bereczky et al., 2003). Here, the Fe and Mn transport abilities of MbNRAMP1 were also investigated using yeast mutants defective in Mn and Fe transport.
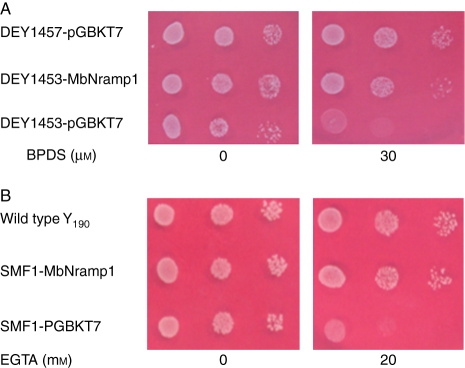
The Fe-transporting ability of the transporter MbNRAMP1 was inferred from the growth of Saccharomyces cerevisiae mutant strain DEY1453 on the medium supplemented with 0 and 30 µm BPDS. DEY1453, a fet3fet4 double mutant, is defective in both low- and high-affinity iron uptake systems and highly sensitive to iron limitation (Eide et al., 1996). Heterologous expression of MbNRAMP1 restored the growth of the yeast mutant strain DEY1453 on iron-depleted media. As shown in Fig. 4A, the MbNRAMP1-transformed DEY1453 mutant strain grew better than the DEY1453 mutant strain transformed with the empty vector in all treatments. Compared with the wild-type yeast, the growth of the DEY1453 yeast strain with MbNRAMP1 expression was the same on the medium without BPDS. When 30 µm BPDS was added to the medium, the growth of the DEY1453 mutant strain transformed with the empty vector was substantially impaired, but cells transformed with MbNRAMP1 grew very well, like the wild-type yeast (DEY1457), although they were slightly retarded at 30 µm BPDS compared with their growth on the medium without BPDS.
Fig. 4.
Functional analysis of MbNRAMP1 in yeast strains. (A) Effect of MbNRAMP1 supplementation on the yeast mutant DEY1453 grown in an iron-deficient medium. Yeast cells were spotted on synthetic defined medium at pH 4·2 containing 10 µm iron (III) citrate in the absence or presence of 30 µm BPDS. (B) Effect of MbNRAMP1 supplementation on the yeast mutant SMF1 grown in a manganese-deficient medium. Yeast cells were spotted on synthetic defined medium at pH 6·0 in the absence or presence of 20 mm EGTA. The OD values are 0·1, 0·01 and 0·001 from left to right.
SLY8, a yeast smf1 mutant deficient in a broad range metal transporter, NRAMP homologue (Liu et al., 1999), was used to test the Mn-transporting ability of MbNRAMP1. SLY8 containing MbNRAMP1 or an empty control vector and a positive control wild-type yeast, Y190, were spotted at different dilutions on SD-Trp medium or in the presence of the divalent metal chelator EGTA at 20 mm (Fig. 4B). All the yeast strains grew as well as the wild-type yeast on the medium without EGTA. At 20 mm EGTA, the MbNRAMP1-transformed smf1 mutant strain, similarly to the wild-type yeast, grew better than the empty vector-transformed mutants, but the smf1 mutant strain transformed with an empty vector was clearly impaired. Therefore, MbNRMP1 encodes a functional metal transporter able to restore growth of the yeast Fe uptake mutant, DEY1453, and the Mn-defective mutant, SLY8, in corresponding metal-deficient media.
M. baccata NRAMP1 Cd2+-transporting ability
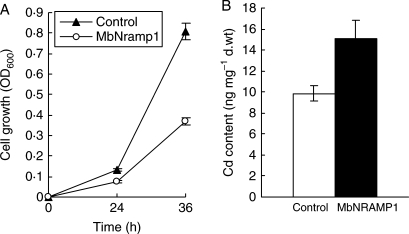
Cadmium is a toxic heavy metal transported across plant membranes via metal transporters (Thomine et al., 2000; Mizuno et al., 2005). A previous study reported that AtNRAMPs can transport Cd2+. To clarify whether the MbNRAMP1 transporter is involved in Cd2+ transport, the influence of MbNRAMP1 expression on Cd2+ toxicity was investigated in yeast. During liquid medium culture, 10 µm CdCl2 strongly impaired the growth of wild-type yeast expressing MbNRAMP1 compared with those transformed with an empty vector. As shown in Fig. 5A, the yeast expressing MbNRAMP1 showed strong growth repression, and the OD of this yeast strain reached less than half (0·3702 ± 0·03) the value of the control (0·8093 ± 0·05) after 36 h cultivation with 10 µm CdCl2. This result indicated that the MbNRAMP1 enhanced Cd2+ sensitivity in yeast. To elucidate further the Cd2+ transport activity of MbNRAMP1, the measurement of Cd2+ content after 36 h showed that the yeast strain transformed with the MbNRAMP1 gene displayed increased Cd2+ accumulation compared with the empty control vector (Fig. 5B). The Cd2+ content in yeast with MbNRAMP1 gene expression increased to 1·5 times (15·09 ± 1·69) that of the control (9·87 ± 0·70) (Fig. 5B).
Fig. 5.
MbNRAMP1 expression increases Cd2+ sensitivity and accumulation in yeast. (A) Pre-cultured cells were grown starting from an OD of 0·01 supplied with a new medium containing 10 µm CdCl2 and cultured at 30°C with shaking (160 r.p.m.). Cell growth was determined as the OD600 after 24 and 36 h of culture. Plotted values are averages of five independent experiments. (B) Cells were harvested after 36 h of culture, and the Cd content in yeast was analysed with a Perkin-Elmer ICP atomic absorption spectrometer. Values are shown as means ± s.e., n = 5.
Localization of MbNRAMP1 protein in yeast
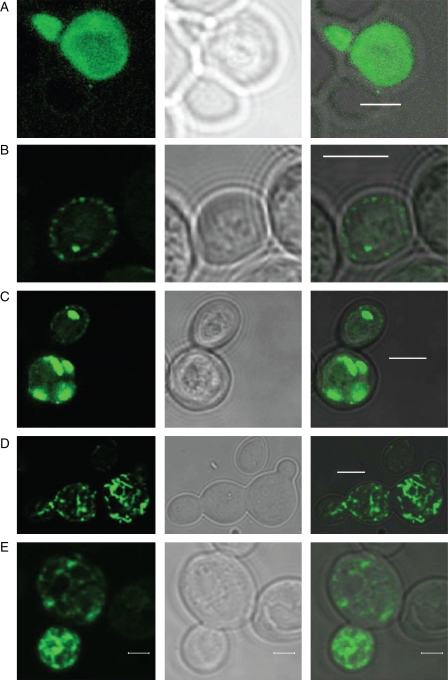
The subcellular localization of MbNRAMP1 was examined using MbNRAMP1–GFP in yeast. MbNRAMP1–GFP complements yeast mutant strains DEY1453 and SLY8. Generally, only about 3 % of cells expressed MbNRAMP1–GFP in normal SD medium. Autofluorescence was not detected in untransformed yeast cells. However, the number of cells increased up to 50–52 % when 30 µm BPDS or 30 µm BPDS and 20 mm EGTA were added to the growth medium, and the fluorescence of fusion proteins was even more readily detected in the stained cells. The number of stained cells was marginally increased up to 8 % in the presence of only 20 mm EGTA (Table 1). Different staining patterns were observed when GFP was fused with MbNRAMP1 protein (Fig. 6B–E), whereas fluorescence was fairly homogeneous when GFP was expressed in its free form without the fused MbNRAMP1 protein (Fig. 6A). Under normal growth conditions, among cells exhibiting GFP fluorescence, in approx. 66 % of the cells, MbNRAMP1–GFP was mainly expressed in peripheral vesicles (Fig. 6B), and also a small amount in bright intracellular vesicles seen as irregular structures (about 34 % among cells exhibiting MbNRAMP1–GFP; see Fig. 6C). Upon BPDS or BPDS and EGTA supplementation, MbNRAMP1–GFP localization was unevenly spread across the whole cell (Fig. 6D, E), and about 10 % of the cells appeared to be unequally accumulated on the surface of a vacuole-like structure (Fig. 6E). In summary, the MbNRAMP1 protein expression and localization patterns are regulated by iron, and to a lower extent manganese, supply in yeast. MbNRAMP1 was predominantly localized in peripheral vesicles and, to a lesser extent, in intracellular vesicles under normal growth conditions. However, in an iron-deficient environment, the protein was unevenly localized across cells and less frequently accumulated on the vacuole-like structure. When iron deprivation ocurred, a large amount of MbNRAMP1 protein was synthesized to ensure cells survive the iron-depleted phase, and was distributed throughout the whole cell.
Table 1.
Effect of metal nutrition conditions on expression of MbNRAMP1–GFP in yeast
| Proportion of each staining pattern among cells exhibiting GFP fluorescence in each condition |
|||||
|---|---|---|---|---|---|
| Culture conditions | Cell staining (%) | Peripheral vesicles | Intracellular vesicles | Whole cells | Vacuole-like structure |
| SD-B0-E0 | 3 ± 1 | 66 ± 4 | 34 ± 4 | nd | nd |
| SD-B30-E0 | 50 ± 4 | nd | nd | 100 | 10 ± 2 |
| SD-B30-E20 | 52 ± 4 | nd | nd | 100 | 11 ± 3 |
| SD-E20 | 8 ± 2 | nd | nd | 100 | 10 ± 3 |
Values are means ± s.e. n = 4.
nd, not detected; SD-B0-E0, SD medium without BPDS and EGTA; SD-B30-E0, SD medium with 30 µm BPDS; SD-B30-E20, SD medium with 30 µm BPDS and 20 mm EGTA; SD-E20, SD medium with 20 mm EGTA.
Fig. 6.
Localization of MbNRAMP1–GFP in yeast. Upper case letters show different GFP staining patterns: left, GFP; middle, differential interference contrast (DIC) of the same fields; right, GFP with DIC superimposed. (A) Free GFP staining as control in normal growth conditions; homogenous fluorescence. (B–E) Examples of MbNRAMP1–GFP localization patterns in yeast. MbNRAMP1 exhibits a shift in cellular localization with the iron status. (B) MbNRAMP1–GFP staining in peripheral vesicles, under normal growth conditions. (C) MbNRAMP1–GFP is localized in brightly fluorescent intracellular vesicles seen as irregular structures, under normal growth conditions. (D) MbNRAMP1–GFP staining in the whole cell in the presence of BPDS and EGTA. (E) MbNRAMP1–GFP staining in the whole cell, and unequally accumulated on the vacuole-like structure surface in the presence of BPDS and EGTA. Scale bars: (A–D) = 4 µm; (E) = 2 µm.
DISCUSSION
Here, a member of the NRAMP family (MbNRAMP1) was cloned from a woody plant, M. baccata, and a functional analysis was developed based on its regulation by essential metals, toxic metal sensitivity and accumulation, and cellular localization. Sequence analysis (Fig. 1) showed that MbNRAMP1 is a member of the NRAMP family. As the NRAMP family genes were highly conserved during evolution, previous reports have indicated that NRAMP genes are widely distributed in mammals, yeasts and higher plants, and are known to be involved in a variety of processes, including metal transport and resistance to microbial infection (Vidal et al., 1993; Vidal and Gros, 1994; Cellier et al., 1996; Gunshin et al., 1997; Cohen et al., 2000). To date, several members of this family have been isolated from Arabidopsis, rice and tomato (Belouchi et al., 1997; Curie et al., 2000; Thomine et al., 2000; Bereczky et al., 2003). MbNRAMP1 is the first member of the NRAMP metal transporter family to be isolated from a woody plant.
From the dendrogram analysis, MbNRAMP1 showed the closest relationship with LeNRAMP1 from tomato, which was also only found in roots, and its transcript was enhanced under a low iron supply (Bereczky et al., 2003). Based on the results of northern blot experiments, it is concluded that MbNRAMP1 transcription is regulated by iron status in roots and shoots. However, MbNRAMP1 mRNA mainly accumulated in roots under Fe-deficiency stress and with a much more rapid and greater response than its accumulation in shoots (Fig. 3). This suggested that MbNRAMP1 is mainly involved in iron uptake and distribution in root cells.
Furthermore, the MbNRAMP1 cDNA restoration of the growth defect in the yeasts DEY1453 and SLY8 in metal-depleted media suggested that MbNRAMP1 was capable of iron and manganese acquisition (Fig. 4). Similar functions for NRAMP genes have also been found in Arabidopsis and tomato (Bereczky et al., 2003; Thomine et al., 2003). Although NRAMP transporters recognized a broad range of metal substrates (e.g. Cu, Cd, Mn and Fe), biological specificity may come from their regulation. For example, SMF1 and SMF2 were expressed only under manganese or iron starvation, whereas SMF3 was predominantly expressed under iron starvation (Portnoy et al., 2000). In plants, LeNRAMP1 and LeNRAMP3 are regulated by iron to a great extent (Bereczky et al., 2003). Linked with the results on MbNRAMP1–GFP in yeast, the number of cells exhibiting fused proteins greatly increased with iron deprivation. It is suspected that MbNRAMP1 may bear specific fragments of sequences as signals in response to environmental nutrition status, especially iron in yeast, but the precise part of the sequence reactive to the environmental iron status needs further clarification. Generation of plant cells transformed with MbNRAMP1–GFP will provide more exact details on iron-mediated post-transcriptional or post-translational regulation of this gene in plant cells.
To avoid the effects of toxic metals, plants separate essential nutrients from poisonous metals. Whereas most plant metal transporters identified to date have a broad substrate range, toxic metals share transport systems with harmless metals (Mizuno et al., 2005). NRAMP family transporters have a highly conserved region between TMs 8 and 9, called the CTM. This motif is also conserved in MbNRAMP and matches that of AtNRAMP1, except that one amino acid residue is different. As outlined in the Results, MbNRAMP1 isolated from apple seedling of M. baccata had Cd2+-transporting ability. However, the MbNRAMP1 transporter increased Cd accumulation in yeast cells, leading to poorer growth. Previous studies also showed that AtNRAMP1, 3 and 4 enhanced Cd2+ accumulation upon their expression in yeast (Thomine et al., 2000). Isolation of other members of the NRAMPs family in M. baccata will be needed to gain further information of their various contributions to metal transport by NRAMP family transporters.
In yeast, the NRAMP homologue SMF1 may play some roles in metal uptake and localize in parts of the plasma membrane (Liu and Culotta, 1999). SMF2 was observed in intracellular vesicles from where it may mobilize metals from intracellular stores (Portnoy, 2000). Under favourable metal conditions, SMF1 and SMF2 become targets of vacuolar degradation (Liu and Culotta, 1999; Portnoy et al., 2000). SMF3 was only detected in the vacuolar membrane where it may mobilize iron from vacuolar stores (Portnoy et al., 2000). LeNRAMP1 and LeNRAMP3 were mainly localized to vesicles upon manganese and iron deficiency. AtNRAMP3 and AtNRAMP4 targeting of the vacuole to mobilize vacuolar Fe stores is essential for Arabidopsis seed germination (Lanquar et al., 2005). MbNRAMP1 showed distinct cellular localization, since it was mainly distributed in the cytosol under iron-depleted conditions; SMF1 mutant complementation may occur via release of internal metals. Here the localization of MbNRAMP1 in yeast provides indirect information on how this protein functions in cells. Work is underway to identify the exact cellular localization in apple to improve our knowledge of the gene's function.
ACKNOWLEDGEMENTS
We are grateful to David Eide (Columbia, MO) for providing the yeast strains, DEY1453 and DEY1457, and Catherine Curie (University of Montpellier, France) for the SLY8 strain. We are indebted to Dr Hongqing Ling (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), Dr Guixian Xia and Dr Borun Zhang (Institute of Microbiology, Chinese Academy of Sciences) for their help with and discussion of our subcellular localization experiments. We thank Professor A. Egrinya Eneji (College of Agronomy & Biotechnology, China Agricultural University) for the critical review of this paper. This work was financially supported by the National High-Tech 863 Program of the People's Republic of China 2006AA10Z1B6, the National Natural Science Foundation of China (No. 30671441) and Laboratory of Apple resources innovation and Genetic improvement (08-01-01-02).
LITERATURE CITED
- Belouchi A, Kwan T, Gros P. Cloning and characterization of the OsNRAMP family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Molecular Biology. 1997;33:1085–1092. doi: 10.1023/a:1005723304911. [DOI] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. Journal of Biological Chemistry. 2003;278:24697–24704. doi: 10.1074/jbc.M301365200. [DOI] [PubMed] [Google Scholar]
- Cellier M, Belouchi A, Gros P. Resistance to intracellular infections: comparative genomic analysis of NRAMP. Trends in Genetics. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- Cellier MFM, Bergevin I, Boyer E, Richer E. Polyphyletic origins of bacterial NRAMP transporter. Trends in Genetics. 2001;17:365–370. doi: 10.1016/s0168-9525(01)02364-2. [DOI] [PubMed] [Google Scholar]
- Cohen A, Nelson H, Nelson N. The family of SMF metal ion transporters in yeast cells. Journal of Biological Chemistry. 2000;275:33388–33394. doi: 10.1074/jbc.M004611200. [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. Expression of the IRT metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Jean MLE, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods in Enzymology. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Cellier M, Vidal S, Gros P. Identification and characterization of a second mouse Nramp gene. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- Guerinot ML. The ZIP family of metal transporters. Biochimica et Biophysica Acta. 2000;1465:190–198. doi: 10.1016/s0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- Han ZH, Shen T. Iron deficiency chlorosis: a literature review. Acta Horticulturae Sinica. 1991;18:323–328. [Google Scholar]
- Han ZH, Shen T, Korcak RF, Baligar VC. Screening for iron-efficient species in the genus Malus. Journal of Plant Nutrition. 1994;17:579–592. [Google Scholar]
- Kaiser BN, Sophie M, Castelli J, Thomson R, Lambert A, Bogliolo S. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant Journal. 2003;35:295–304. doi: 10.1046/j.1365-313x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nature Biotechnology. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LT, Kaplan J. Defects in the yeast high affinity iron transport system result in decreased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. Journal of Biological Chemistry. 1998;273:22181–22187. doi: 10.1074/jbc.273.35.22181. [DOI] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proceedings of the National Academy of Science, USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Culotta VC. Post-translation control of NRAMP metal transport in yeast. Journal of Biological Chemistry. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, Obata H. Cloning of three ZIP/NRAMP transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+- transport abilities. Plant Physiology and Biochemistry. 2005;43:793–801. doi: 10.1016/j.plaphy.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Portnoy ME, Liu XF, Culotta VC. Saccharomyces cerevisiae expresses three functional distinct homologues of the NRAMP family of metal transporters. Molecular and Cellular Biology. 2000;20:7893–7902. doi: 10.1128/mcb.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific system for iron phytosiderophores in roots of grasses. Plant Physiology. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, David WR. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sharma SS, Kaul S, Metwally A, Goyal KC, Finkemeier I, Dietz KJ. Cadmium toxicity to barley (Hordeum vulgare) as affected by varying Fe nutritional status. Plant Science. 2004;166:1287–1295. [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proceedings of the National Academy of Sciences, USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Wang RC, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to NRAMP genes. Proceedings of the National Academy of Sciences, USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelièvre F, Debrabieux E, Schroeder JI, Barlier-Brygoo H. AtNRAMP3, a multi-specific vacuolar metal transporter involved in plant responses to iron deficiency. Plant Journal. 2003;34:685–695. doi: 10.1046/j.1365-313x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gamard F. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Gros P. Resistance to infection with intracellular parasites – identification of a candidate gene. News in Physiological Sciences. 1994;9:178–183. [Google Scholar]
- Vidal S, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:460–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochimica et Biophysica Acta. 2000;1465:104–126. doi: 10.1016/s0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]