Abstract
Background and Aims
Populus alba is a thermophilic forest tree present in the Mediterranean basin. Its habitat is highly fragmented and its distribution range has been subject to long-term human interference, resulting in debate surrounding whether certain populations are native or exotic in origin. In particular, populations from the islands of Corsica and Sardinia are of uncertain origin. While populations of P. alba mainly reproduce sexually, clonal reproduction is also common. The aims of this study were to locate and molecularly characterize the poorly studied island populations of P. alba and compare these with samples from various spatial scales, in order to provide information on the genetic structure and phylogeography of this species. This information will provide evidence on whether the species is native to Sardinia, which is important for the development of conservation strategies.
Methods
DNA extracts were obtained from the following P. alba trees: 159 from Sardinia, 47 from Ticino regional park (northern Italy), 15 acquired from an Italian Germoplasm Bank (IRC; Italian Reference Collection) and 28 from the Mediterranean basin (MB). Genetic polymorphisms were revealed at nuclear and chloroplast DNA (cpDNA) microsatellite loci, both at the island scale (Sardinia) and at broader scales, for comparative assessment of the genetic and genotypic diversity and phylogeography.
Key Results
Based on nuclear microsatellite loci, Sardinian white poplar consists of a small number of genets (26), each of which is represented by several ramets. Despite the uniqueness of the Sardinian haplotypes and the very low value of genetic diversity at the cpDNA level (vK = 0·15), the HT (0·60) and the AR (3·61) values, estimated at the nuclear level for Sardinia, were comparable with those of the other populations and collections.
Conclusions
The uniqueness of the cpDNA haplotypes, the prevalence of clonality and the restricted number of genets recorded suggest that Sardinian white poplar could be a floristic relict of the native flora of the island, which has spread through available habitats on the island mainly by means of vegetative propagation and human activities.
Key words: Populus alba, Sardinia, genets, ramets, phylogeography, native forest species
INTRODUCTION
Island systems have long served as natural laboratories for evolutionist biologists (Darwin, 1859; Emerson, 2002). Despite their frequent small size, islands usually contain a range of habitats suitable for the colonization of an array of plants and animals. Once an island is colonized, population differentiation among islands, or continents, can be facilitated by isolation (Juan et al., 2000).
The degree of isolation of island and mainland populations, as well as other historical and demographic factors will impact patterns of genetic diversity within and among populations. The combination of a wide geographic range and the presence of dispersed populations makes it possible for a plant species to harbour a certain level of genetic variability. This genetic richness can be present in the form of either allelic variability or allelic uniqueness of some populations (Petit et al., 1998). These genetic characters can be used to define the origin and to evaluate the phylogeography of a species (Petit et al., 2005). The occurrence of peculiar allelic combinations, absent from other areas of the distribution range, may reflect both past demographic and selective processes, and may contribute to a population's ability to adapt to environmental changes or particular conditions, affecting the species' ability to survive and spread in a particular habitat (Meloni et al., 2006).
Many perennial plants combine sexual reproduction and clonal propagation as population regeneration mechanisms (Abrahamson, 1980; Peterson and Jones, 1997). In some clonal species the relative success of sexual vs. clonal recruitment can vary in response to ecological and genetic factors that favour one regeneration mechanism over the other (Eckert, 2002). The demographic balance between sexual and clonal recruitment is likely to have important consequences for clonal diversity and genetic structure of plant populations (Ellstrand and Roose, 1987; Eckert and Barrett, 1993). Clonal reproduction (Alfonso-Corrado et al., 2004; Storme et al., 2004; Baali-Cherif and Besnard, 2005; Honnay and Bossuy, 2005) can be defined as the generation of new individuals genetically identical to the ancestral organism, through binary fission (bacteria) or mitosis (eukaryotic cells) (King and Stansfield, 1990). In particular, clonal reproduction is a strategy commonly employed by early generation hybrids (since they often show a low level of fertility) in order to increase the number of flowering ramets per genet (Burke et al., 2000; Silvertown, 2008). Ramets are defined as clonally produced parts of a plant with their own roots and a potentially independent existence, whereas a genet is comprised of all ramets arising from a single seed (Silvertown and Lovett Doust, 1993). Clonal growth enables a genet to distribute itself across an area ranging from a few centimetres to hundreds of metres (Cook, 1985; Horak et al., 1987).
Clonal growth is a common characteristic, with varying importance, of members of the genus Populus (Arens et al., 1998; Winfield et al., 1998; Storme et al., 2004; Suvanto and Latva-Karjanmaa, 2005). For instance, a single American aspen (Populus tremuloides) clone consisting of 47 000 ramets has been reported to cover an area of 43 ha (Kemperman and Barnes, 1976). Populus alba, a forest tree species that is phylogenetically closely related to American aspen (Dickmann, 2001), can reproduce asexually via cuttings, root-suckers or cladoptosis (Dickmann, 2001), although, in natural populations, its reproduction is described as mainly sexual (Sabatti et al., 2001; Fossati et al., 2004; Lexer et al., 2005). Populus alba, also referred to as European white poplar, is widely distributed in floodplain forests throughout northern Africa, southern Europe and central Asia (Tchou, 1948). It is resistant to insect pests, and fungal and bacterial pathogens, and tolerates diverse environmental stresses such as drought, wind, salinity and high temperatures (Dickmann, 2001).
White poplar is considered native to the continental Mediterranean basin, as demonstrated recently by the discovery of fossil remains in Southern France (Roiron et al., 2004). In contrast, its status on Mediterranean islands is controversial. For example, it was considered of uncertain status on the island of Corsica (Briquet, 1910), native on Sardinia and Sicily (Camarda and Valsecchi, 1985; Greuter et al., 1989; Pignatti, 1998) and was not assessed on most of the other islands.
Nevertheless, no critical analysis according to the criteria of Webb (1985) has been performed to confirm the putative native status of white poplar on Sardinia. Mediterranean islands are commonly subject to plant invasions (Hulme et al., 2008) due to the long-term interaction between humans and their environment. In fact, during the course of human occupation, the introduction of alien plants and animals is a very common event (Gil et al., 2004) and the range of many forest tree species has been impressively modified by human activities through the ages (Richardson, 1998).
On the basis of the above considerations and since no previous genetic, phylogeographical or ecological research has been conducted on white poplar of Sardinia, a molecular survey was carried out describing patterns of genetic variation and phylogeography of the whole island, using additional samples from the species range for comparative purposes.
MATERIALS AND METHODS
Study area and sample collection
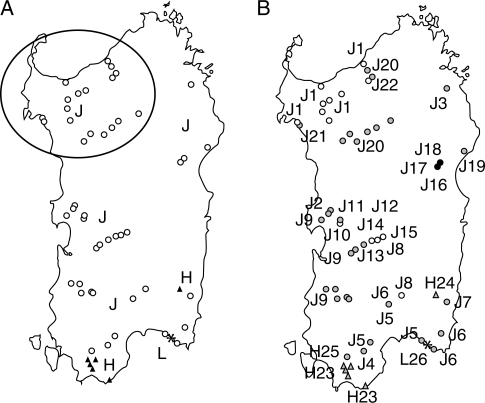
Sardinia, located between 39° and 41°N latitude, is the second largest island (24 000 km2) in the Mediterranean basin, with all the typical climate, land use and landscapes features common to this region. Sardinian samples of P. alba were obtained in two field seasons, in 2004 and 2005. During the first field survey (2004), an exploratory sampling was performed following the approach of Lowe et al. (2004a). A total of 93 white poplar trees were sampled from 30 sites covering an area of about 8000 km2 at the north end of the island. The location of sampled trees was recorded by GPS, and leaf material was collected wherever the species was found to be naturalized (i.e. not obviously cultivated; Fig. 1A). In the absence of data on the genetic structure of the island populations, it was assumed a priori that each site represented a local population or sub-population, according to the distance between collecting sites (>15–20 km) and/or the presence of geographical barriers. At each site, a population consisted of a few hundreds to several hundred thousand individuals. At each sampling site, between two and five specimens were collected. Field observations, including morphological assessment of gender, as well as preliminary results of genetic [simple sequence repeat (SSR)] analyses suggested clonal structure, and indicated that a second collection should be conducted to cover the whole island, and that 1–2 samples per site could be collected without loss of information. Consequently, during the second sampling season (2005), 66 additional individuals were collected from 50 sites (Fig. 1), to obtain broader insights into P. alba population genetics on Sardinia. It was assumed a priori that each additional site corresponded to a single genet.
Fig. 1.
(A) Distribution map of the three cpSSR haplotypes (J, H and L) detected in the island of Sardinia. The circles indicate the location of the 30 sites where 93 white poplar individuals were sampled in 2004. The location of the 50 sites (66 individuals) sampled in 2005 is also reported. (B) Distribution map of the 26 genets detected in Sardinia in the 80 surveyed sites. Genets codes are reported in Table 1.
Additional materials were collected for broad-scale comparisons. The broadest sample is denoted MB (Mediterranean basin) and includes 28 specimens (including samples from Corsica, Rhodes, Camarguein the South of France, Sicily, Lesvos and Macedonia). Fifteen samples are denoted IRC (Italian Reference Collection), and include samples from riparian river systems from northern, central and southern Italy (e.g. Bormida river Northern Italy, Vomano Central Italy and Sele Southern Italy; see Supplementary Information Table S1, available online). A natural population located along the Ticino river Regional Park in North Italy includes 47 individuals that were previously characterized for molecular polymorphisms (Fossati et al., 2004).
Molecular analysis
For all samples, DNA was extracted from 20 mg of dried leaves, using the DNeasy Plant Mini kit (Qiagen, Germany), following the manufacturer's instructions.
The entire set of 159 Sardinian white poplar individuals and the other populations and collections (MB, IRC and Ticino) were scored for a total of five nuclear and three chloroplast (cp) SSRs. The nuclear SSRs, being co-dominant markers, are useful for population genetics, whereas the cpSSRs are the most informative markers for phylogeography (Lowe et al., 2004b).
Five unlinked SSR nuclear loci (WPMS05, WPMS14, WPMS15, WPMS18 and WPMS20; Cervera et al., 2001; Gaudet et al., 2008), originally identified in P. nigra (Schoot et al., 2000; Smulders et al., 2001), were surveyed. These five highly polymorphic SSRs were sufficient to discriminate nearly 100 % of poplar individuals in previous studies (Dayanandan et al., 1998; Fossati et al., 2003, 2005). Polymerase chain reaction (PCR) conditions, scoring and recording were described by Fossati et al. (2004).
For the cpSSR analysis, a polymorphic sub-set (ccmp2, ccmp6 and ccmp10) of the original set of cpSSRs described by Weising and Gardner (1999) was used. The PCRs were performed in 15 µL volumes containing 1–10 ng of DNA, 1× PCR Green reaction buffer (Promega), 200 µm of each dNTP, 0·75 U of Taq polymerase (GoTaq, Promega), 1·5 mm MgCl2 and 0·4 µm of each primer. The forward primers were labelled with FAM, HEX or TMR. The PCR cycle was: denaturation at 94 ºC for 5 min followed by 30 cycles of 94 ºC for 30 s, 50 ºC for 30 s, 72 ºC for 30 s and a final extension at 72 ºC for 10 min. The PCR products were separated by capillary electrophoresis, with a 400 bp size standard, using the MegaBACE automated sequencer (Amersham, GE Healthcare). Alleles were sized using Fragment Profiler version 1·2 (Amersham, Uppsala, Sweden).
Nuclear SSR data
The clonal diversity index [calculated as G/N where G is the number of genotypes observed and N is the total number of individuals sampled in a population(s)], estimating genotypic diversity, and evenness, measuring the relative abundance (percentage frequency) of the different genotypes making up the genotype richness of the sampled area, were computed for Sardinian white poplar. Both indices were calculated only in the case of the first sampling (2004), because no assumption (based on the preliminary data produced) on the genetic structure of the Sardinian white poplar was made.
The psex index was calculated in order to estimate the probability that identical genotypes are the result of sexual reproduction rather than of clonal reproduction within the naturally reproduced population of the island of Sardinia (n = 159 individuals), using the option available in the Software package GenClone 2·0 (http://si-wagner.ualg.pt/ccmar/maree/software.php?soft=genclon). A detailed and comprehensive description of the mathematical formula of psex is available at the following web site: http://si-wagner.ualg.pt/ccmar/maree/pdf/Readme.GenClone2.0.pdf.
Nei's H values were computed using the GenAlEx package (Peakall and Smouse, 2006), the mean number of alleles per locus (AR) was calculated using FSTAT 2·9·3 (Goudet, 1995). Nei's H values and the AR index were computed only in the case of the Sardinian and Ticino populations.
Chloroplast SSR data
Haplotypes were defined as distinct combinations of size variants at the three cpSSRs, and a minimum spanning tree (MST) was constructed using the software MINSPNET (http://cmpg.unibe.ch/software.htm) to visualize the minimum number of size differences (bp) between haplotypes. To reconstruct the tree, the genetic distance (dxy) between haplotypes i and y was calculated as follow:
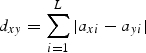 |
where axi and axy are the sizes of the microsatellite of the ith locus for the haplotypes x and y, L being the number of analysed loci (Excoffier and Smouse, 1994).
The level of polymorphism within populations was estimated as haplotypic richness AS (El Mousadik and Petit, 1996) using the software FSTAT.
We also estimated haplotype diversity (hS, vK and vS) based on haplotype frequencies, both for each population and collection and within the species according to Pons and Petit (1996) either ignoring distances between haplotypes (hS), or taking them into account (vK and vS). Weights, defined as distances between the haplotypes, were considered as the proportion of polymorphic alleles between haplotypes and used to calculate vK, vS and NST (Pons and Petit, 1996). Haplotypic diversity (hS, hT, vS and vT) and differentiation (GST and NST) statistics were calculated using HaploNst (Pons and Petit, 1996), for the entire population of white poplar and for the entire population but with the Sardinian population removed. Differentiation among populations was computed from unordered and ordered alleles (GST and NST, respectively) and the contribution of allele phylogeography to total differentiation was assessed with a permutation method (Pons and Petit, 1996). This involves permutation of distances between pairs of haplotypes, and therefore a significant NST > NST permuted (i.e. NST > GST) reflects that closely related haplotypes are found together in polymorphic populations more often than expected by chance, which can be interpreted as the existence of a significant genetic structure within the species (Pons and Petit, 1996). All computations were performed using the software PERMUT available at the following web site: http://www.pierroton.inra.fr/genetics/labo/Software/Permut/, weighting distances between haplotypes by their frequencies (default option).
RESULTS
Clonal structure of Sardinian white poplar
In most of the surveyed localities hundreds to thousands of P. alba ramets of different ages were found, derived from a single genet, of the same gender, forming, in some cases, linear formations of some kilometres along river systems, or covering large areas of several hectares. These observations are in sharp contrast to previous experimental results and observations on the Italian continental populations (Fossati et al., 2004), which are always composed of individuals of both genders. Moreover, this pattern of sex distribution in the Sardinian P. alba supported the possibility of clonal spread through cladoptosis and root-suckers of a single genet forming stands of thousands of ramets.
Within Sardinia, all five nuclear SSRs assayed were polymorphic. Computational analysis showed the presence of 26 genets (Table 1) out of 159 Sardinian specimens (three genets out of 93 individuals in the first sampling in 2004), highlighting the clonal genetic structure of the Sardinian population (Fig. 1).
Table 1.
Genets identified within the Sardinian white poplar and evenness of the entire collection and of the first sampling (2004)
| No. | Genet code | No. of ramets, entire collection | Evenness, entire collection ( %) | No. of ramets, 2004 sampling | Evenness, 2004 sampling ( %) | Gender |
|---|---|---|---|---|---|---|
| 1 | J1 | 75 | 47·17 | 66 | 70·96 | NR |
| 2 | J2 | 1 | 0·63 | NR | ||
| 3 | J3 | 1 | 0·63 | NR | ||
| 4 | J4 | 1 | 0·63 | NR | ||
| 5 | J5 | 5 | 3·14 | NR | ||
| 6 | J6 | 5 | 3·14 | NR | ||
| 7 | J7 | 1 | 0·63 | Male | ||
| 8 | J8 | 3 | 1·89 | Male | ||
| 9 | J9 | 12 | 7·55 | Male | ||
| 10 | J10 | 2 | 1·26 | Female | ||
| 11 | J11 | 1 | 0·63 | Female | ||
| 12 | J12 | 1 | 0·63 | Female | ||
| 13 | J13 | 1 | 0·63 | Male | ||
| 14 | J14 | 2 | 1·26 | Male | ||
| 15 | J15 | 1 | 0·63 | Male | ||
| 16 | J16 | 2 | 1·26 | NR | ||
| 17 | J17 | 1 | 0·63 | NR | ||
| 18 | J18 | 1 | 0·63 | NR | ||
| 19 | J19 | 1 | 0·63 | NR | ||
| 20 | J20 | 26 | 16·35 | 23 | 24·73 | NR |
| 21 | J21 | 1 | 0·63 | NR | ||
| 22 | J22 | 4 | 2·52 | 4 | 4·30 | Female |
| 23 | H23 | 7 | 4·4 | NR | ||
| 24 | H24 | 1 | 0·63 | NR | ||
| 25 | H25 | 1 | 0·63 | NR | ||
| 26 | L26 | 2 | 1·26 | NR |
NR, not recorded.
Clonal diversity of the Sardinian white poplar in the case of the first sampling (2004) was extremely low (0·032) due to the limited number of genets (three) over the high number of collected trees (93). Evenness was calculated as the percentage of the genets over the total number of sampled trees for the entire collection (2004 + 2005) and for the first (2004) sampling (Table 1).
The 26 genets from Sardinia show only three cpSSR haplotypes (J, H and L; see below, and Fig. 2B). The genets J1, J9, J20, H22, for example, form stands of up to thousands of ramets spread throughout an area of >4000 km2, 1700 km2, 300 km2 and 100 km2, respectively. The other white poplar genets detected in Sardinia are based on a more limited number of ramets which exhibit local clustering, covering areas of up to several square kilometres, with distribution ranges that do not overlap each other (Fig. 1).
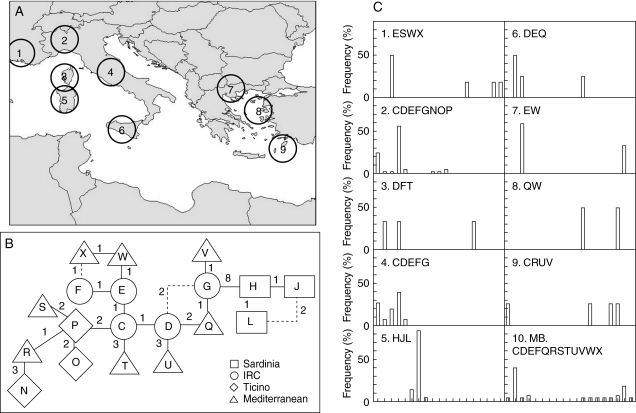
Fig. 2.
(A) Location of collection sites in the Mediterranean study area: (1) Camargue, France; (2) Ticino Regional Park, Italy; (3) Corsica, France; (4) IRC, Italian Reference Collection; (5) Sardinia, Italy; (6) Sicily, Italy; (7) Macedonia, Greece; (8) Lesvos, Greece; (9) Rhodes, Greece. (B) Minimum spanning tree of the 19 cpDNA haplotypes found in the studied area. The major links between haplotypes are represented as bold lines. Other possible links identified are shown as dotted lines. Superimposed on the tree are the number of mutational steps between haplotypes. Populations from Sardinia, IRC, Ticino and Mediterranean basin are as indicated. (C) Graphs showing the relative frequencies of the 19 observed haplotypes in the nine collection sites and in the Mediterranean basin (MB) reported in the text and in Table 3.
The psex index estimated for each single collected white poplar tree of the Sardinian population was equal to zero, indicating that the probability of sampling an identical individual derived from sexual reproduction is highly unlikely, confirming the importance of asexual reproduction in Sardinia.
Broad-scale comparisons and minimum spanning tree
Considering the different origin and features of the four white poplar sample sets used in this study (Sardinia, Ticino, IRC and MB), it is only possible to make general comparisons between the Sardinian, Ticino and the two white poplar collections (IRC and MB). In particular the MB and IRC collections clearly are not populations, and studying them as such might be somewhat misleading. Considering Sardinian white poplar as a single population is not realistic either, given the size of the island and the clonal structure that was exhibited. Despite these difficulties in comparing the data sets from the different collections, it was nevertheless possible to draw meaningful conclusions regarding the molecular and genetic characterization of white poplar populations in Sardinia. The average number of alleles (Na, Table 2) per locus in Sardinian white poplars is 5·0 (range 2–7) and this is similar to that found in the other collections and populations. The mean allelic richness is 3·61 and this is only slightly different from that recorded for the Ticino samples (4·17; Table 2) (additional information on genetic diversity of the samples is available online as Supplementary Information, Table S2). WPMS loci have a total number of 25 alleles in the Sardinian white poplar, and two private alleles were recorded (allele 301 at locus WPMS05 and allele 190 at locus WPMS15). Furthermore, locus WPMS20 is polymorphic both in Sardinia and in the MB collection, while it is monomorphic in Ticino, the IRC sample set and in two other white poplar Italian populations (S. Castiglione, unpubl. res.). The two recorded alleles (219 and 222) of locus WPMS20 have frequencies of 0·15 and 0·85 for Sardinia compared with 0·02 and 0·98 in the MB collection. More precisely, the rarest allele (219) is known from a single individual outside Sardinia, collected on the Italian island of Sicily.
Table 2.
Summary of the genetic diversity, based on the five analysed nuclear SSR loci (WPMS) of each population or collection
| Sardinia | Ticino | IRC | MB | |
|---|---|---|---|---|
| Na | 5·00 | 6·00 | 3·60 | 6·00 |
| He | 0·60 | 0·57 | – | – |
| Ho | 0·70 | 0·55 | – | – |
| AR | 3·61 | 4·17 | – | – |
| Pa | 2 | 6 | 0 | 4 |
| Na,total | 25 | 30 | 18 | 30 |
| HT | 0·60 | 0·57 | – | – |
Na, number of observed alleles per locus; He, expected heterozygosity; Ho, observed heterozygosity; Pa, private alleles numberper population or collection; Na,total, total number of alleles; HT, total heterozygosity of the analysed populations; AR, and allelic richness corrected for sample size using nine individuals for the Sardinia and Ticino populations.
The observed heterozygosity (Ho, Table 2) is higher than expected (He) for each locus in the Sardinian white poplar and also higher than in the Ticino population.
CpSSR analysis revealed a total of 19 haplotypes (K) across all the investigated populations and collections. Haplotypic richness (AS) in P. alba is quite low in the case of the Sardinian population (AS = 1·60), higher for the Ticino population (AS = 2·77) and very high for the MB collection (AS = 4·21; Table 3). The Sardinian population has a very low number of haplotypes (K = 3), which are not present in any of the other populations and collections assessed. The Ticino population and the MB collection are much more polymorphic (K = 8 and K = 12, respectively) and the IRC collection has an intermediate value (K = 5) of haplotypes. The low level of polymorphisms is also reflected by the low number of total alleles (Na,total = 5) in Sardinia when compared with those of the Ticino population and in the MB collection (Na,total = 9 and 13, respectively; Table 3: information on allele sizes of each cpSSR is available online as Supplementary Information, Table S3).
Table 3.
Summary of the haplotype diversity of each population or collection
| Sardinia | Ticino | IRC | MB | |
|---|---|---|---|---|
| K | 3 | 8 | 5 | 12 |
| Na | 1·66 | 3 | 2 | 4·66 |
| Na,total | 5 | 9 | 6 | 13 |
| AS | 1·6 | 2·77 | 2 | 4·21 |
| vk | 0·15 | 0·3 | 0·36 | 0·44 |
K, Number of haplotypes found in populations and collections; Na, mean allele number; Na,total, total number of alleles; AS, haplotypic richness corrected for sample size using 14 individuals; and vk, population diversity.
For evaluation of the contribution of the island of Sardinia to haplotype diversity, differentiation and clonal structure in the study area, several parameters were calculated (Tables 3 and 4) either including or omitting the Sardinian white poplars (Table 4). The vK values, estimated for each population (Table 3), show, once again, that the haplotype diversity of the Sardinian population is the lowest (vK = 0·15), and is only half of that calculated for the other studied populations and collections (Ticino vK = 0·30, IRC vK = 0·36 and MB vK = 0·44). Total hT and vT and within-population haplotype diversity (hS and vS) for white poplar, either including or omitting the Sardinian population, is generally quite high (Table 4). Differentiation among populations and collections is not pronounced when the alleles are both ordered (NST = 0·091) and unordered (GST = 0·165), when Sardinia is excluded from the cpSSR analysis (Table 4). In contrast, when the analysis also includes the Sardinian population, there is much clearer differentiation (NST > GST; NST Permut = 0·325 with P < 0·01; Table 4). This result can be interpreted as evidence of genetic structure within Sardinian white poplar.
Table 4.
Chloroplast marker diversity (haplotypic diversities h and v) and differentiation (GST, NST) statistics from white poplar populations in the Mediterranean basin including all populations and collections, or excluding the Sardinian population
| Populations | hS | hT | GST | vS | vT | NST | NST (perm)* | NST > NST (perm)* |
|---|---|---|---|---|---|---|---|---|
| Overall population without Sardinia | 0·700 | 0·838 | 0·165 | 0·742 | 0·817 | 0·091 | – | – |
| Overall population | 0·624 | 0·928 | 0·327 | 0·440 | 0·986 | 0·554 | 0·325 | P < 0·01 |
Subscripts: S, within populations; T, total population.
*Permutation test NST > NST (perm) (1000 permutations) according to Pons et al. (1996).
The MST visually describes the phylogeographic relationships among the 19 haplotypes in the study area (Fig. 2B). The three haplotypes observed in Sardinia are highly similar, with only one and two mutational steps separating haplotype L from H and J, respectively, but the Sardinian haplotype J is separated from the rest of the Mediterranean basin (IRC, MB and Ticino) by at least eight mutational steps. In contrast, all the other haplotypes of the Mediterranean basin are separated from one another by only 1–3 mutational steps.
DISCUSSION
Clonal structure in Sardinian white poplar
Nuclear and cpSSR analysis has demonstrated that Sardinian white poplar comprises a few genets (26, see Table 1), and their ramets, in some cases, form huge, monoclonal stands of the same gender (Table 1), which are geographically clustered and cover very large areas (e.g. see genet J1 in Fig. 1). The field and molecular surveys (2004–2005) provide evidence for abundant vegetative propagation. This is confirmed by the remarkable scarcity of fertile seed production (detected in only one of 80 sampling sites covering the whole island) and the complete absence of seedlings in all sampled sites. Moreover, semi-natural areas, in particular those which have been abandoned by man, or are no longer devoted to agricultural and grazing activities, are often colonized by white poplar through vegetative spread. It seems that in Sardinia this alternative strategy to sexual reproduction for white poplar is successful. This clonal structure was totally unexpected on the basis of the previous synecological, taxonomic and floristic information available. Certainly, on the basis of current knowledge, one of the white poplar genets discovered in Sardinia is among the largest genets described to date in nature for the genus Populus, as it consisted of thousands and thousands of ramets. Until now, the most spectacular example of a poplar monoclonal stand is that cited by Kemperman and Barnes (1976). In the case of P. nigra and other forest trees, there are several examples in the literature of large local and spatially limited clones (Arens et al., 1998; Winfield et al., 1998; Alfonso-Corrado et al., 2004; Barsoum et al., 2004), suggesting that vegetative reproduction varies among species, but also depends on the dynamics of the river or site under study (Barsoum et al., 2004).
Native status of Sardinia white poplar
The pattern of cpSSRs and nuclear SSRs is informative when Sardinian white poplar is compared with all the other populations and collections. In fact, cpSSRs present in Sardinian samples show the presence of three different and private haplotypes H, J and L. These three haplotypes are well separated from the other haplotypes known from the Mediterranean basin, as clearly illustrated by the MST (Fig. 2B). This suggests that Sardinia may have acted as a glacial refugium. Sardinian white poplar was certainly present in the floral refugia in the Mediterranean basin at the end of the last ice age. It co-existed with other forest taxa including other species of Salicaceae, which share common habitat preferences (e.g. P. nigra). Several tree species, including poplar, occurred locally in small suitable sites within the Mediterranean region, but also at the southern edge of the cold and dry steppe–tundra area in eastern, central and south-western Europe (Ray and Adams, 2001; Petit et al., 2003; Cottrell et al., 2005). In particular, on the basis of cpDNA data, it has been hypothesized that Sardinia could have been a primary refugium for one white oak species (Petit et al., 2002) during the last glacial era. This hypothesis is supported by the presence in white oak of an endemic haplotype in Sardinia and Corsica that has not migrated to northern parts of Europe (Petit et al., 2002). Populus alba may have followed the same recolonization routes, beginning from the Southern refugia (the south of Spain, Italy and the Balkans; Hewitt, 1999) used by many other poplar species such as P. tremula and P. nigra (Petit et al., 2003; Cottrell et al., 2005). With the exception of the Sardinian haplotypes, white poplar spread from these glacial refugia to colonize the Mediterranean basin and other sites in Europe, as also demonstrated by its presence in the South of France based on paleobotanical data (Roiron et al., 2004). Therefore, a reasonable explanation for the presence of white poplar on the island, as the present molecular data would suggest, is that P. alba is a relict species of the Sardinian flora. The catastrophic climatic changes, due to the last glaciation, caused a dramatic reduction of the native white poplar populations in Sardinia, resulting in the survival of only a few genets. The absence of sexual reproduction might be linked to asynchrony of floral phenology of the remaining clones, an unbalanced sex ratio or the limited number of surviving genets. Surviving individuals are those that were able to switch to vegetative propagation, later aided by human activity that further contributed to the spread of the surviving genets around the whole island. Information derived from interviews with local Sardinian farmers has allowed the putative routes of local spread by vegetative propagation to be proposed. For centuries, farmers have exchanged cuttings to establish trees used for fencing, shade and ornamental purposes. As early as 1859 Moris described the species as locally present across the whole island of Sardinia and, in 1910, Briquet argued that white poplar was not native to Corsica, while he described the species as much more abundant and naturally regenerated in Sardinia. Today, it is a widespread component of the flora of Sardinia, thriving in a range of habitats: from those that are semi-natural to those that are highly anthropic and fragmented.
Evaluation of Webb's criteria for determining native status
As mentioned, Sardinian P. alba is, in some cases, locally regarded as a cryptogenic species (sensu Carlton, 1996), due to its diverse status in different Mediterranean floras. Webb (1985) proposed eight criteria for determining whether a species is native (not all of them applicable to every species), and although very seldom is any one of them decisive, when several point in the same direction one is justified in accepting the composite evidence as reasonably conclusive. In the present research all the eight criteria proposed by Webb (fossil evidence, historical evidence, habitat, geographical distribution, frequency of known naturalization, genetic diversity, reproductive pattern and possible means of introductions) were assessed, although the focus was mainly on genetic diversity and reproductive patterns. This is partly because, in the absence of fossil or historical evidence, molecular tools allow comparison of Sardinian P. alba with populations from other Mediterranean localities, thus excluding some possible routes of introduction. Furthermore, although striking examples can be found of disjunct but undoubtedly native distributions, more or less continuous distributions are much more common for species with native status. In the Sardinian survey the habitats where P. alba thrives were also recorded; growing extensively in more or less natural habitats it is likely to be native, which is in agreement with the operational logic suggested by Webb (1985). Finally, the prevalence of clonal spread may be regarded as a symptom of non-nativeness. In fact, Webb stated that it seemed reasonable to assume that most native plants are capable of reproducing, at least in part, by seed, and that if a plant reproduces entirely vegetatively it can legitimately be suspected of being an alien. He did, however, acknowledge that there are, of course, some exceptions and that the converse is not true, as many aliens reproduce entirely by seed. However, it has been significantly demonstrated that, despite the prevalence of clonal spread, the genetic diversity of Sardinian P. alba supports its original native status in the island.
Broad-scale comparison
The clonal diversity index, estimated here for the first time in Sardinian white poplar, demonstrates, once again, the clonal structure and the very low level of genotypic diversity of the Sardinian population, while measures of evenness show the prevalence of a few genets over the whole territory. In contrast, the clonal diversity index achieves the maximum possible value (a) in the case of the Ticino river population, in which all sampled individuals are unique genets, even though the Ticino population is very restricted to a much smaller area (only 100 km2, compared with Sardinia at 24 000 km2). Moreover, analysis of cpSSR diversity and differentiation, at the broad scale, is strongly influenced by the inclusion or omission of the Sardinian samples in the analysis. The GST value is lower than NST when Sardinian data are included, thereby indicating the presence of a clear genetic structure (Pons and Petit, 1996), probably associated with the vegetative reproduction, while the situation is exactly the opposite when the Sardinian data are omitted (GST > NST, Table 4).
The present results concerning the Mediterranean basin region further support the unique character of Sardinian white poplar and its native status. The HT and AR values estimated at the nuclear level for the Sardinian white poplar are comparable with those of the other analysed population (Ticino). These indices suggest that Sardinian white poplar maintains a fairly high level of genetic variability regardless of its vegetative propagation. However, the possibility cannot be excluded that such a high level of genetic diversity could be due to recent pollen flow from outside Sardinia, although this seems unlikely.
Taken together, these data can also be regarded as further support for the native status of white poplar in Sardinia, vs. a hypothetical human introduction in the past. In fact similar genetic studies on vegetatively reproducing plant species introduced beyond their native range by human activities, and now regarded as established aliens, show a totally contrasting pattern of genetic diversity (Fay et al., 1999; Hollingsworth and Bailey, 2000; Gil et al., 2004; Williams et al., 2005).
The uniqueness and isolation of the Sardinian white poplar cpSSR haplotypes, as well as the presence of private alleles at nuclear loci, provide further support for the native status of the species. However, because of the distinct nature of the Sardinian samples, the present molecular results do not allow a clear introduction route to be proposed. In contrast, two other large islands, Sicily and Corsica, show one unique haplotype each, and an evident phylogeographic relationship with the continental populations (e.g. with Ticino, IRC and Camargue; Fig. 2). In fact, haplotype D is fairly widespread in the Mediterranean basin (Camargue, Macedonia, Sicily, Corsica, Ticino and IRC; Fig. 2), but absent from Sardinia.
Concluding remarks
White poplar represents an important native species of the riparian plant communities (Populetalia albae Br.-Bl. 1931) in Sardinia and in the Mediterranean basin.
At present the species is not under threat of serious decline in Sardinia, and there is even some evidence that it is vegetatively colonizing new areas as a result of land abandonment. Nevertheless, the unique nature of the relict Sardinian genets represents an important germplasm resource that should be managed for in situ conservation. Therefore, in accordance with advice issued for other native forestry species, the introduction of white poplar from external sources should not be permitted without careful consideration of the possible adverse consequences. This is particularly true if introductions are planned for riparian and other sensitive habitats. Molecular tools proved, once again, to be very powerful for assessing biodiversity at the genetic level and have provided useful insights into the origin and status of this cryptogenic species.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at www.aob.oxfordjournals.org/ and consists of the following. Table S1, registry codes and provenances of the white poplar clones examined in this study; Table S2, details of the genetic diversity for the five SSR loci analysed in the study for each population or collection; and Table S3, a list of haplotypes identified in each population or collection analysed.
ACKNOWLEDGEMENTS
This research was supported by the Italian Ministry of Environment, Land and Sea Protection (Ministero dell'Ambiente e della Tutela del Territorio e del Mare), within the research project ‘Research and development in biotechnology applied to the protection of the environment, in collaboration with the Popular Republic of China’. We are grateful to G. Tsurlis, G. Domina, P. Dalias, G. Zuccarelli, A. Brunu, L. Carta, M. Manca and A. Solinas for invaluable assistance with sampling; C. Jarvis, M. Spencer and P. V. Arrigoni for helpful discussions; S. Gomarasca for laboratory support; J. Cottrell (Forestry Research, Northern Research Station, Roslin, UK) and A. Spada (Department of Biology, University of Milan, Italy) for fruitful discussions on an earlier version of the paper and language revision; Genexpress Laboratory (Dipartimento di Biotechnologia Agricoltura, Universitá di Firenze, Sesto Fiorentino, Italy) for the incomparable work done in fingerprinting the white poplar populations; the two anonymous reviewers and the handling editor J. Whitton who helped us to improve the manuscript greatly.
REFERENCES
- Abrahamson WG. Demography and vegetative reproduction. In: Solbrig O, editor. Demography and evolution in plant populations. Oxford: Blackwell; 1980. pp. 89–106. [Google Scholar]
- Alfonso-Corrado C, Esteban-Jimenez R, Clark-Tapia R, Pinero D, Campos J, Mendoza A. Clonal and genetic structure of two Mexican oaks: Quercus eduardii and Quercus potosina (Fagaceae) Evolutionary Ecology. 2004;18:585–599. [Google Scholar]
- Arens P, Coops H, Jansen J, Vosman B. Molecular genetic analysis of black poplar (Populus nigra L.) along Dutch rivers. Molecular Ecology. 1998;7:11–18. [Google Scholar]
- Baali-Cherif D, Besnard G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Annals of Botany. 2005;96:823–830. doi: 10.1093/aob/mci232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum N, Muller E, Skot L. Variations in levels of clonality among Populus nigra L. stands of different ages. Evolutionary Ecology. 2004;18:601–624. [Google Scholar]
- Briquet J. Prodrome de la flore Corse. Lyon-F: Libraires-Édíteurs; 1910. [Google Scholar]
- Burke JM, Bulger MR, Wesselingh RA, Arnold ML. Frequency and spatial patterning of clonal reproduction in Louisiana iris hybrid populations. Evolution. 2000;54:137–144. doi: 10.1111/j.0014-3820.2000.tb00014.x. [DOI] [PubMed] [Google Scholar]
- Camarda I, Valsecchi F. Alberi e arbusti spontanei della Sardegna. Sassari, Italy: Edizioni Gallizzi; 1985. [Google Scholar]
- Carlton JT. Biological invasions and cryptogenic species. Ecology. 1996;77:1653–1655. [Google Scholar]
- Cervera M, Storme V, Ivens B, Gusmao J, Liu B, Hostyn V, et al. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. doi: 10.1093/genetics/158.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RE. Growth and development in clonal plant populations. In: Jackson JBC, Buss LW, Cook RE, editors. Population biology and evolution of clonal organisms. London: Yale University Press; 1985. pp. 259–296. [Google Scholar]
- Cottrell J, Krystufek V, Tabbener H, Milner A, Connolly T, Sing L, et al. Postglacial migration of Populus nigra L.: lessons learnt from chloroplast DNA. Forest Ecology and Management. 2005;206:71–90. [Google Scholar]
- Darwin CR. On the origin of species. London: John Murray; 1859. [Google Scholar]
- Dayanandan S, Rajora O, Bawa K. Isolation and characterization of microsatellites in trembling aspen (Populus tremuloides) Theoretical and Applied Genetics. 1998;96:950–956. [Google Scholar]
- Dickmann DI. An overview of the genus Populus. In: Dickmann DI, Isebrands JG, Eckenwalder JE, Richardson J, editors. Poplar culture in North America. Ottawa, Canada: NRC Research Press; 2001. pp. 1–42. [Google Scholar]
- Eckert CG. The loss of sex in clonal plants. Evolutionary Ecology. 2002;15:501–520. [Google Scholar]
- Eckert CG, Barrett SCH. Clonal reproduction and patterns of genotypic diversity in Decodon-Verticillatus (Lythraceae) American Journal of Botany. 1993;80:1175–1182. [Google Scholar]
- El Mousadik A, Petit RJ. Chloroplast DNA phylogeography of the argan tree of Morocco. Molecular Ecology. 1996;5:547–555. doi: 10.1111/j.1365-294x.1996.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. American Journal of Botany. 1987;74:123–131. [Google Scholar]
- Emerson BC. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Molecular Ecology. 2002;11:951–966. doi: 10.1046/j.1365-294x.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE. Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics. 1994;136:343–359. doi: 10.1093/genetics/136.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay M, Lledo M, Kornblum M, Crespo M. From the waters of Babylon? Populus euphratica in Spain is clonal and probably introduced. Biodiversity and Conservation. 1999;8:769–778. [Google Scholar]
- Fossati T, Grassi F, Sala F, Castiglione S. Molecular analysis of natural populations of Populus nigra L. intermingled with cultivated hybrids. Molecular Ecology. 2003;12:2033–2043. doi: 10.1046/j.1365-294x.2003.01885.x. [DOI] [PubMed] [Google Scholar]
- Fossati T, Patrignani G, Zapelli I, Sabatti M, Sala F, Castiglione S. Development of molecular markers to assess the level of introgression of Populus tremula into P. alba natural populations. Plant Breeding. 2004;123:382–385. [Google Scholar]
- Fossati T, Zapelli I, Bisoffi S, Micheletti A, Vietto L, Sala F, Castiglione S. Genetic relationships and clonal identity in a collection of commercially relevant poplar cultivars assessed by AFLP and SSR. Tree Genetics and Genomes. 2005;1:11–20. [Google Scholar]
- Gaudet M, Jorge V, Paolucci I, Beritognolo I, Scarascia Mugnozza G, Sabatti M. Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genetics and Genomes. 2008;4:25–36. [Google Scholar]
- Gil L, Fuentes-Utrilla P, Soto A, Cervera MT, Collada C. English elm is a 2000-year-old Roman clone. Nature. 2004;431:1053. doi: 10.1038/4311053a. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Greuter W, Burdet HM, Long G. Med-Cheklist. Genéve, Switzerland: Ed. des Conservatoire et Jardin botaniques de Genève; 1989. [Google Scholar]
- Hewitt GM. Post glacial recolonization of European Biota. Biological Journal of the Linnean Society. 1999;68:87–112. [Google Scholar]
- Hollingsworth ML, Bailey JP. Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed) Botanical Journal of the Linnean Society. 2000;133:463–472. [Google Scholar]
- Honnay O, Bossuy B. Prolonged clonal growth: escape route or route to extinction? Oikos. 2005;108:427–432. [Google Scholar]
- Horak M, Holt J, Ellstrand N. Genetic variation in yellow nutsedge (Cyperus esculentus) Weed Science. 1987;36:42–49. [Google Scholar]
- Hulme PE, Brundu G, Camarda I, Dalias P, Lambdon P, Lloret F, et al. Assessing the risks to Mediterranean islands ecosystems from non-native plant introductions. In: Tokarska-Guzik B, Brock JH, Brundu G, Child LE, Pyšek P, Daehler CC, editors. Plant invasions: human perception, ecological impacts and management. Leiden, The Netherlands: Backhuys Publishers; 2008. pp. 39–56. [Google Scholar]
- Juan C, Emerson BC, Oromi P, Hewitt GM. Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends in Ecology and Evolution. 2000;15:104–109. doi: 10.1016/s0169-5347(99)01776-0. [DOI] [PubMed] [Google Scholar]
- Kemperman JA, Barnes BV. Clone size in American aspens. Canadian Journal of Botany. 1976;54:2603–2607. [Google Scholar]
- King R, Stansfield W. Encyclopedic dictionary of genetics. New York: Wiley; 1990. [Google Scholar]
- Lexer C, Fay M, Joseph J, Nica M, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): the role of ecology and life history in gene introgression. Molecular Ecology. 2005;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. [DOI] [PubMed] [Google Scholar]
- Lowe A, Harris S, Ashton P. Markers and sampling in ecological genetics. In: Lowe A, Harris S, Ashton P, editors. Ecological genetics: design, analysis, and application. a. Oxford: Blackwell Publishing; 2004. pp. 6–51. [Google Scholar]
- Lowe A, Harris S, Ashton P. Genetic diversity and differentiation. In: Lowe A, Harris S, Ashton P, editors. Ecological genetics: design, analysis, and application. b. Oxford: Blackwell Publishing; 2004. pp. 52–105. [Google Scholar]
- Meloni M, Perini D, Filigheddu R, Binelli G. Genetic variation in five Mediterranean populations of Juniperus phoenicea as revealed by inter-simple sequence repeat (ISSR) markers. Annals of Botany. 2006;97:299–304. doi: 10.1093/aob/mcj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris GG. Flora sardoa. Turin, Italy: Typ. Regia; 1859. [Google Scholar]
- Peakall R, Smouse PE. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CJ, Jones RH. Clonality in woody plant: a review and comparison with clonal herbs. In: de Kroon H, van Groenendael J, editors. The ecology and evolution of clonal plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 263–289. [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Petit RJ, Brewer S, Bordacs S, Burg K, Cheddadi R, Coart E, et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. Forest Ecology and Management. 2002;156:49–74. [Google Scholar]
- Petit RJ, Aguinagalde I, Beaulieu J, Bittkau C, Brewer S, Cheddadi R, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Pignatti S. Sguardo d'assieme sui boschi d'Italia. In: Pignatti S, editor. I boschi d'Italia. Sinecologia e biodiversità. Turin, Italy: UTET; 1998. pp. 105–123. [Google Scholar]
- Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N, Adams JM. A GIS-based vegetation map of the world at the last glacial maximum (25,000–15,000 BP) Internet Archaeology. 2001;11 [Google Scholar]
- Richardson DM. Forestry trees as invasive aliens. Conservation Biology. 1998;12:18–26. [Google Scholar]
- Roiron P, Ali AA, Guendon J-L, Carcaillet C, Terrla J-F. Preuve de l'indigénat de Populus alba L. dans le Bassin méditerranéen occidental. Comptes Rendus Biologies. 2004;327:125–132. doi: 10.1016/j.crvi.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Sabatti M, D'Ovidio R, Tanzarella O, Scarascia G. Assessment of geographic variation by RAPD markers among Italian open-pollinated progenies of Populus alba L. Genetic Resources and Crop Evolution. 2001;48:423–428. [Google Scholar]
- Schoot J, Pospiskova M, Vosman B, Smulders M. Development and characterization of microsatellite markers in black poplar (Populus nigra L.) Theoretical and Applied Genetics. 2000;101:317–322. [Google Scholar]
- Silvertown J. The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. International Journal of Plant Science. 2008;169:157–168. [Google Scholar]
- Silvertown JW, Lovett Doust J. Introduction to plant population biology. Oxford: Blackwell; 1993. [Google Scholar]
- Smulders M, Schoot J, Arens P, Vosman B. Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.) Molecular Ecology Notes. 2001;1:188–190. [Google Scholar]
- Storme V, Broeck A, Ivens B, Halfmaerten D, Slycken J, Castiglione S, et al. Ex-situ conservation of Black poplar in Europe: genetic diversity in nine gene bank collections and their value for nature development. Theoretical and Applied Genetics. 2004;108:969–981. doi: 10.1007/s00122-003-1523-6. [DOI] [PubMed] [Google Scholar]
- Suvanto LI, Latva-Karjanmaa T. Clone identification and clonal structure of the European aspen (Populus tremula L.) Molecular Ecology. 2005;14:2851–2860. doi: 10.1111/j.1365-294X.2005.02634.x. [DOI] [PubMed] [Google Scholar]
- Tchou Y-T. Etudes écologiques et phytosociologiques sur le forêts riveraines di Bas-Languedoc (Populetum albae) Vegetatio. 1948:4–384. [Google Scholar]
- Webb DA. What are the criteria for presuming native status? Watsonia. 1985;15:231–236. [Google Scholar]
- Weising K, Gardner R. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome. 1999;42:9–19. [PubMed] [Google Scholar]
- Williams DA, Overholt WA, Cuda JP, Hughes CR. Chloroplast and microsatellite DNA diversities reveal the introduction history of Brazilian peppertree (Schinus terebinthifolius) in Florida. Molecular Ecology. 2005;14:3643–3656. doi: 10.1111/j.1365-294X.2005.02666.x. [DOI] [PubMed] [Google Scholar]
- Winfield M, Arnold G, Cooper F, Ray M, White J, Karp A, et al. A study of genetic diversity in Populus nigra subsp. betulifolia in the Upper Severn area of the UK using AFLP markers. Molecular Ecology. 1998;7:3–10. [Google Scholar]