Abstract
Background and Aims
Pinus kwangtungensis is a five-needled pine, inhabiting isolated mountain tops, cliffs or slopes in the montane areas of southern China and northern Vietnam. Global warming and long-term deforestation in southern China threaten its existence and genetic integrity, and this species is listed as vulnerable in the China Species Red List. However, the level and distribution of genetic diversity in this vulnerable species are completely unknown. In this paper, the genetic diversity and structure are examined using paternally inherited plastid markers to shed light on its evolutionary history and to provide a genetic perspective for its conservation.
Methods
By means of direct sequencing, a new polymorphic fragment containing a minisatellite site was identified within the plastid genome of P. kwangtungensis. Using the minisatellite site along with five SNPs (one indel and four substitutions) within the same fragment, the population genetic structure and pollen flow were analysed in 17 populations of P. kwangtungensis in southern China.
Key Results
Analysis of 227 individuals from 17 populations revealed ten haplotypes at the minisatellite site. The haplotype diversity at species level was relatively high (0·629). Genetic diversity of each population ranged from 0 to 0·779, and the western populations harboured more genetic variation than the eastern and Hainan populations, although the former appeared to have experienced a bottleneck in recent history. Population subdivision based on this site was high (FST = 0·540 under IAM; RST = 0·677 under SMM). Three major clusters (eastern, western and Hainan) were identified based on a neighbor-joining dendrogram generated from genetic distances among the populations. The genetic structures inferred from all the polymorphic sites and the SNPs were in concordance with that from the minisatellite site.
Conclusions
The results suggest that there are at least three refugia for P. kwangtungensis and that populations in these refugia should be treated as separate evolutionarily significant units or conservation units. The high diversities in the western populations suggest that these were much larger in the past (e.g. glacial stages) and that the shrinking population size might have been caused by recent events (e.g. deforestation, global warming, etc.). The western populations should be given priority for conservation due to their higher genetic diversity and limited population sizes. It is concluded that the newly found minisatellite may serve as a novel and applicable molecular marker for unravelling evolutionary processes in P. kwangtungensis.
Key words: Pinus kwangtungensis, minisatellite, population genetics, conservation
INTRODUCTION
In recent decades, powerful insights have been gained into the ecology, history and evolution of organisms from analyses of organelle DNA at inter- and intraspecific levels (Avise, 2004). Compared with mitochondrial DNA in animals, however, the nucleotide substitution rate in the widely used plastid DNA of plants is 10–100 times lower (Zurawski et al., 1984; Wolfe et al., 1987). Detecting plastid sequence variation can be difficult, especially in taxa with long generations (Provan et al., 2001). This is especially so in conifers, and the silent (synonymous and non-coding) mutation rate of plastid genomes in pines is 20 times slower than that of angiosperms (Ann et al., 2007). However, empirical studies have demonstrated that some regions of the plastid genome contain variable number tandem repeats (Nakamura et al., 1987). These regions generally show high levels of polymorphism, rendering them useful in population genetics (Weber and May, 1989; Edwards et al., 1992).
Two categories of variable number tandem repeats are recognized depending on the number of bases in the repeated sequences: microsatellites (repeat unit of 1–5 bp) and minisatellites (6–64 bp) (Estoup and Angers, 1998; Avise, 2004). In contrast to plastid microsatellites, minisatellites are less widespread in plastid genomes (Tautz and Schlotterer, 1994; Jarne and Lagoda, 1996), although they are common in the nuclear genomes of eukaryotes (Jeffreys et al., 1985). Plastid minisatellites have been used less in population genetics relative to plastid microsatellites (Avise, 2004). However, a few studies indicated that plastid minisatellites have a mutation rate similar to that for nuclear microsatellites and possibly higher than for plastid microsatellites. They may thus represent more powerful markers for studying intraspecific relationships in plants (Cozzolino et al., 2003b).
Hypervariable markers such as minisatellites may have a mutation rate greater than the coalescent process or may be prone to recurrent or parallel mutations that lead to size homoplasy, and these factors may limit the utility of such markers (Cozzolino et al., 2003b). Ideally, a combination of markers with different mutation rates should be used to elucidate both the historical and more recent processes for a species (Schaal et al., 1998). If sufficient variation owing to nucleotide substitutions and insertions/deletions (indels) can be detected, these polymorphisms may be useful controls for evaluating the utility of hypervariable markers. So far, however, few direct comparisons between minisatellites and nucleotide substitutions and indels have been made.
Pinus kwangtungensis is a five-needled pine that inhabits isolated mountain tops, cliffs or slopes in montane areas (700–1900 m of altitudinal range) of southern China (including Guangdong, Hainan, Hunan and Guizhou Provinces and Guangxi Zhuang Autonomous Region) and northern Vietnam (Fu et al., 2000). It is listed as vulnerable in the China Species Red List (Wang and Xie, 2004). It is primarily distributed in the Nanling mountains (south Hunan, north Guangdong and north-east Guangxi) and eastern Yungui Plateau mountains (north-west Guangxi and south-east Guizhou). Population sizes are often moderate to large and some populations, such as Ruyang and Jiufeng, may have tens of thousands of individuals according to field observations. However, global warming has threatened the existence of populations in the south and low-elevation areas because of the preference of the species for temperate or cool environments. Moreover, long-term deforestation in south China may contribute further to the population decline of P. kwangtungensis. For example, only three mature individuals survive on a cliff in Leye county, Guangxi, where natural vegetation has been almost extirpated by farmers. All these factors pose intense threats to the continued existence of this species and the integrity of its natural gene pool. However, the genetic structure and gene flow among populations, which are relevant to the long-term persistence of this vulnerable species, are completely unknown.
Development of bisaccate pollen in conifers is thought to be a significant adaptation to wind dispersal (Ledig, 1998). Liepelt et al. (2002) provided a striking example in the high levels of pollen flow in Abies alba between geographically isolated refugia. For most conifer species, the plastid genome is inherited paternally and transmitted by pollen (Petit et al., 2005), making plastid markers powerful tools for investigating pollen flow. Through comprehensive searches in the plastid genome of P. kwangtungensis, a new plastid minisatellite together with four substitutions and one indel were characterized within the same fragment. These findings provide an excellent opportunity to elucidate intraspecific relationships in P. kwangtungensis and evaluate the utility of the novel minisatellite in population genetics.
With the novel minisatellite and four substitutions and one indel within the same fragment, the following specific questions were addressed in this paper. (a) What are the level and distribution of the plastid variation in P. kwangtungensis throughout its natural range in southern China? Is there a high level of pollen flow between populations as expected? (b) Is this hypervariable marker suitable for population genetic analysis? (c) What are the implications of the results for the conservation of this vulnerable pine?
MATERIALS AND METHODS
Fresh needles of Pinus kwangtungensis Chun ex Tsiang were collected from 17 populations across southern China (Table 1). Except for Vietnam, the sampled populations covered most of the recorded sites of P. kwangtungensis based on herbarium specimens at the Chinese National Herbarium (PE), South China Botanical Garden Herbarium (IBSC), Kunming Institute of Botany Herbarium (KUN) and Guangxi Institute of Botany Herbarium (GXIB). All samples were dried in silica gel and stored at −20 °C until they were processed. Populations were defined as groups of trees spaced about 50–100 m apart on a mountain.
Table 1.
Sample locations, estimated population sizes, sample sizes and plastid haplotype frequencies in 17 populations of Pinus kwangtungensis
| Plastid haplotype |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Population | Lat. (N) | Long. (E) | Alt. (m) | N | Population size | Nh | Genetic diversity | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 |
| Eastern | Longsheng (LS) | 25°36·241′ | 109°54·843′ | 1326 | 15 | 1000–3000 | 2 | 0·3429 ± 0·1278 | 3 | 12 | ||||||||
| Jinxiu (JX) | 24°10·424′ | 110°14·583′ | 1128 | 15 | 1000–5000 | 2 | 0·5333 ± 0·0515 | 8 | 7 | |||||||||
| Gongcheng (GC) | 24°51·459′ | 111°06·840′ | 1158 | 15 | 1000–5000 | 2 | 0·4190 ± 0·1132 | 11 | 4 | |||||||||
| Rongshui (RS) | 25°00·000′ | 109°12·000′ | 1100 | 15 | 5000–10000 | 3 | 0·4476 ± 0·1345 | 1 | 3 | 11 | ||||||||
| Lingui (LG) | 25°40·306′ | 110°05·836′ | 1169 | 15 | 500–1000 | 2 | 0·1333 ± 0·1123 | 14 | 1 | |||||||||
| Maoershan (MES) | 25°42·000′ | 110°16·480′ | 1508 | 15 | 500–1000 | 2 | 0·1333 ± 0·1123 | 1 | 14 | |||||||||
| Xinning (XN) | 26°27·209′ | 111°01·448′ | 1297 | 15 | 1000–5000 | 3 | 0·5905 ± 0·1060 | 4 | 2 | 9 | ||||||||
| Jianghua (JH) | 24°40·721′ | 111°33·347′ | 1063 | 15 | 1000–2000 | 1 | 0 | 15 | ||||||||||
| Ruyang (RY) | 24°53·951′ | 113°01·229′ | 1559 | 15 | >20000 | 3 | 0·3619 ± 0·1448 | 2 | 1 | 12 | ||||||||
| Jiufeng (JF) | 25°21·866′ | 113°26·123′ | 1376 | 15 | >20000 | 1 | 0 | 15 | ||||||||||
| Yingde (YD) | 24°10·120′ | 113°26·123′ | 1050 | 15 | 1000–3000 | 1 | 0 | 15 | ||||||||||
| Total | 0·4388 ± 0·0322 | |||||||||||||||||
| Hainan | Wuzhishan (WZS) | 18°53·826′ | 109°42·000′ | 1833 | 10 | 3000–5000 | 1 | 0 | 10 | |||||||||
| Yinggeling (YGL) | 18°57·021′ | 109°24·478′ | 1440 | 15 | 3000–5000 | 2 | 0·5143 ± 0·0690 | 6 | 9 | |||||||||
| Total | 0·3800 ± 0·0913 | 6 | 19 | |||||||||||||||
| Western | Libo (LB) | 25°36·217′ | 107°42·641′ | 978 | 20 | 1000–2000 | 6 | 0·7789 ± 0·0646 | 1 | 1 | 5 | 8 | 3 | 2 | ||||
| Leye (LY) | 24°51·459′ | 106°28·051′ | 1433 | 3 | 3 | 2 | 0·6667 ± 0·3143 | 2 | 1 | |||||||||
| Duan (DA) | 24°02·675′ | 108°14·363' | 576 | 9 | <200 | 3 | 0·7222 ± 0·0967 | 4 | 2 | 3 | ||||||||
| Longzhou (LZ) | 22°22·120′ | 106°51·000′ | 585 | 5 | 5 | 3 | 0·7000 ± 0·2184 | 1 | 3 | 1 | ||||||||
| Total | 0·8138 ± 0·0403 | |||||||||||||||||
| Total | 227 | 0·6293 ± 0·0296 | 4 | 1 | 1 | 2 | 6 | 14 | 46 | 3 | 128 | 22 | ||||||
Abbreviations: Lat., latitude; Long., longitude; Alt., altitude; N, number of sample size; Nh, number of haplotypes; H, haplotype
Genomic DNA was extracted from about 50 mg of the silica-gel-dried needles following a modified 2xCTAB protocol (Doyle and Doyle, 1987). Sequence variation in the plastid genome was screened using universal primers (see Table S1 in Supplementary Information, available online) and specific primers for P. kwangtungensis (Table S2 in Supplementary Information, available online). Through this survey, one polymorphic fragment containing a minisatellite and several substitutions and one indel were found. The forward and reverse primers for amplifying the screened fragment were 5′-AAAGATTCGGATACTCTCAAA-3′ and 5′-TCTTCCCATGAGTTCTTCGTC-3′, respectively. DNA amplification was performed in a T1 thermocycler (Biometra, Germany) as follows: 4 min at 94 °C followed by 11 cycles with a 0·5 °C descending series of annealing temperatures each cycle (60–55 °C), and 26 cycles with an annealing temperature of 54 °C. Denaturation was performed at 94 °C (1 min) and extension at 72 °C (2 min) in all cycles, with a final 10 min extension at 72 °C to end the reaction. Reactions were carried out in a volume of 10 µL containing 1·0 mmol L−1 MgCl2, 0·25 µmol L−1 dNTP, 10 × buffer, 1·25 µmol L−1 primer, 0·5 U Taq DNA polymerase and 10 ng DNA template.
Amplification products were used for direct sequencing. Sequencing reactions were conducted with a newly designed primer (5′-CAAGAGCGGAAAAAGATAGAG-3′ ) using the DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech), following the manufacturer's protocol. Sequencing was done on a Megabase 1000 automatic DNA sequencer (Amersham Pharmacia Biotech), after the sequencing reaction product was purified through precipitation with 95 % ethanol and 3 M sodium acetate (pH 5·2).
DNA sequences were aligned with ClustalX1·81 (Thompson et al., 1997) and the alignnent was refined manually. The haplotypes characterized by the minisatellite were plotted on a map using ArcGIS 8·3. Total genetic diversity (HT) and within-population diversity (HS) were calculated with ARLEQUIN 2000 (Schneider et al., 2000). The level of genetic differentiation among populations was assessed using the FST estimator assuming the infinite allele model (IAM) (Kimura and Crow, 1964), and the estimator RST assuming the stepwise-mutation model (SMM) (Otha and Kimura, 1973). Statistical significance of the observed values was tested using 1000 permutations. These procedures were performed with ARLEQUIN 2000. Based on the genetic distance matrix (pairwise FST or RST) among the 17 populations, neighbor-joining (NJ) dendograms among populations were generated using MEGA version 2·1 (Kumar et al., 2001).
To provide a comparison for evaluating the utility of the minisatellite, the population genetics of P. kwangtungensis was also analysed using the four substitutions and one contiguous indel [the substitutions and indel are referred to as single nucleotide polymorphisms (SNPs) following the definition of Zhu et al. (2003)] and all the polymorphic sites within the same fragment, separately. The indel was coded as 0/1 according to its absence or presence. The population genetic parameters based on the SNPs and all polymorphic sites were also calculated using ARLEQUIN 2000. An NJ dendogram among populations was also generated using MEGA version 2·1 based on the genetic distance matrix (pairwise FST) of all polymorphic sites.
For evaluating the influence of recent events (e.g. global warming and deforestation) on the demography of P. kwangtungensis, the program Bottleneck version 1·2 (Piry et al., 1999) was used to determine whether populations have experienced a recent reduction in their effective population sizes. The Wilcoxon's one-tailed test rather than the sign test was adopted, since the former is more powerful when analyzing data with less than ten polymorphic sites (Piry et al., 1999). Given that no mutational model of the minisatellite had been depicted, the estimations were conducted under the assumption of the SMM and the IAM.
RESULTS
The DNA sequences of the screened region were determined for 17 populations of P. kwangtungensis, corresponding to 98102–99543 bp of the P. koraiensis plastid genome. The whole region varied in length from 711 to 883 bp with six polymorphic sites. These polymorphisms included four substitutions, one 52-bp indel and one minisatellite consisting of several 15-bp tandem repeats. All haplotype sequences were deposited in GenBank databases under the accession numbers EF114102–EF114115.
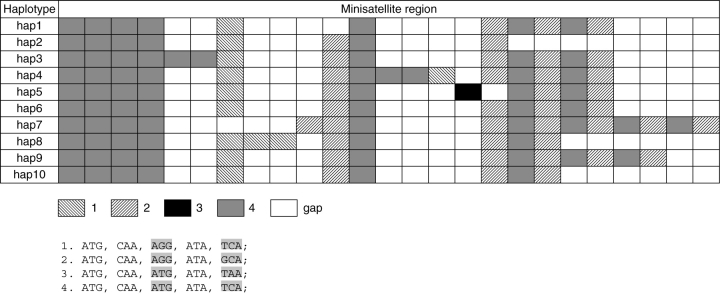
The minisatellite sequences had seven length variants ranging from 120 to 240 bp. Four motifs (i.e. tandem repeated units: 1, 2, 3 and 4 in Fig. 1) were detected in the tandem repeat region. Only one base substitution separated motif 1 and motif 2, motif 1 and motif 4 and motif 3 and motif 4. The motif combination pattern in the minisatellite was (4)4–6 + (1)0–3 +(2)0–2 + (4)1–3 + (1)0–1 + (2)0–1 + (3)0–1 + (42)0–4 (Fig. 1), where the subscripts are the repeated number of motifs. The BLAST survey of nucleotide sequences (GenBank database) indicated that the 15-bp tandem repeats were homologous to the plastid ORF95 located at 98834 bp of P. koraiensis. Although the function of the protein encoded by this open reading frame (ORF) is unclear, it was assumed that the tandem repeats represent a non-functional domain of a primary amino acid sequence, because no related phenotypic differences have been noted among the individuals. Ohyama et al. (1986) even found that the analogous ORF was totally absent from the plastid genome of Marchantia polymorpha.
Fig. 1.
Schematic representation of the ten haplotypes found in the minisatellite site, showing the number, type, interspersion pattern and alignment of the repeat units. The different repeat motifs of the tandem repeat region are represented by four different boxes. White boxes represent gaps introduced for alignment and correspond to indels of the repeated motifs.
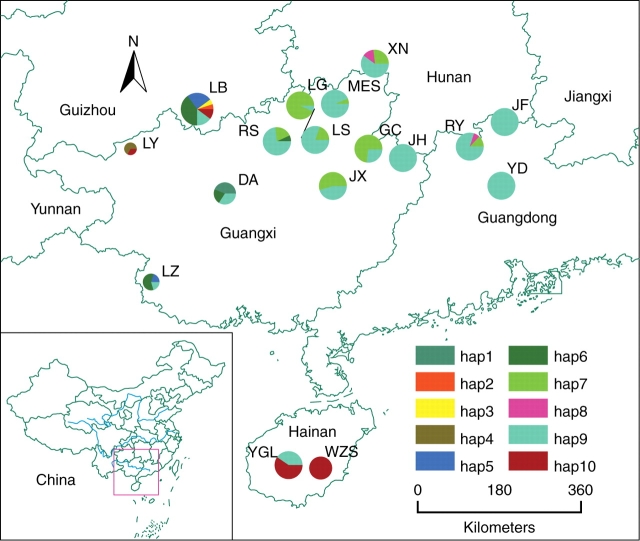
Ten haplotypes were characterized by the minisatellite. The distribution of these haplotypes is shown in Table 1 and Fig. 2. Hap9 was the most widespread haplotype, only absent in populations LY and WZS, possibly due to their small sample sizes (three and ten individuals, respectively). However, other haplotypes were much more geographically localized. Several haplotypes were restricted to one or two populations. For example, hap4 was restricted to LY, hap1 to DA, hap2 and hap3 to LB and hap8 to RY and XN. Some haplotypes were regionally widespread, e.g. hap6 was found in several western populations (LB, DA and LZ) and hap7 was found in eastern populations (LS, JX, GC, RS, LG, MES, XN and RY). The distribution of hap10 was quite different from the others, occupying four geographically distant populations (LB, LY, YGL and WZS), possibly due to long-distance pollen flow or convergent evolution of the minisatellite.
Fig. 2.
Distribution of ten plastid minisatellite haplotypes within and among populations of Pinus kwangtungensis. Pie-chart sizes are proportional to the corresponding sample sizes.
The distribution pattern of the 14 haplotypes identified by all polymorphic sites (Fig. S1 in Supplementary Information, available online) was similar to that of the ten haplotypes characterized by the minisatellite (Fig. 1), except without a range-wide haplotype. Due to only four haplotypes being identified by the SNPs, the distribution of these haplotypes was simple (Fig. S2 in Supplementary Information, available online). However, this map is similar to the other two haplotype maps, partitioning all populations into three groups depending on the dominant haplotype in each group, namely the western group (LB, LY, LZ and DA) dominated by hapII, the eastern group (LS, JX, GC, RS, LG, MES, XN, JH, RY, JF and YD) by hapIV and the Hainan group (WZS and YGL) by hapIII.
A relatively high level of minisatellite genetic diversity (HT = 0·629) was detected across all P. kwangtungensis populations. Haplotype diversity varied greatly at the population level, ranging from 0 to 0·779 (Table 1). Western populations (LB, LZ, LY and DA) contained two to six haplotypes, resulting in high genetic diversity (0·667–0·779, HT = 0·814). Eastern and Hainan populations had fewer haplotypes (one to three) and thus showed low genetic diversity (0–0·590, HT = 0·439 for the eastern populations; 0–0·514, HT = 0·380 for the Hainan populations). The t-test (Nei, 1987) confirmed that the difference was statistically significant between the western and the eastern populations (t = 3·874, P < 0·001) and the western and the Hainan populations (t = 4·347, P < 0·001).
The genetic diversity (HT = 0·665) in P. kwangtungensis revealed by all the polymorphic sites was only slightly higher than that for the minisatellite alone (HT = 0·629), indicating that most of the variation resides in the minisatellite site. This fact is more obvious when considering the genetic diversity (HT = 0·380) detected by the SNPs. The population-level diversity for all polymorphic sites had a similar pattern to that for the minisatellite: higher diversity in western populations than in the eastern and Hainan ones (data not shown). However, the genetic diversity detected by SNPs was much less variable among populations because of a paucity of SNP haplotypes (data not shown).
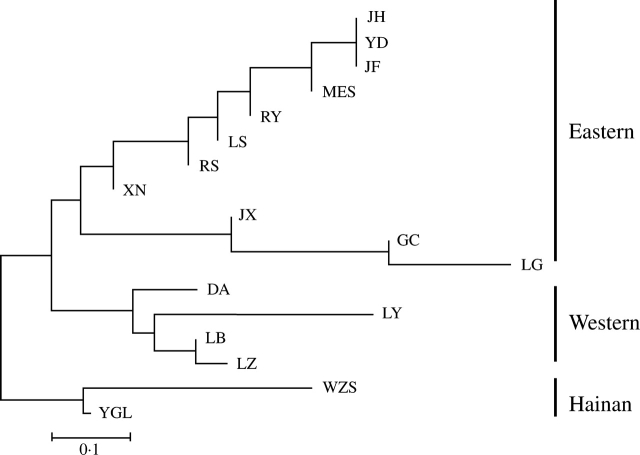
Two NJ dendrograms were generated from genetic distance (pairwise FST) among populations for the minisatellite (Fig. 3) and for all polymorphic sites (Fig. S3 in Supplementary Information, available online). The same three major clusters, eastern, western and Hainan, were identified, which was consistent with the distribution pattern of the four haplotypes identified by the SNPs. However, the topology of the NJ dendrogram based on the pairwise RST of the minisatellite was a little different from the haplotype distribution pattern, with Hainan populations grouping with the western or the eastern group. This is reasonable because the genetic distances differ slightly under different mutational models.
Fig. 3.
Neighbor-joining tree of populations of Pinus kwangtungensis based on pairwise FST. Three major groups, eastern, western and Hainan are indicated.
The levels of genetic differentiation among populations estimated from the minisatellite site under IAM and SMM were quite high (FST = 0·540; RST = 0·677), as was the genetic differentiation among populations (FST = 0·630) based on all polymorphic sites. The highest genetic structure was detected by the SNPs (FST = 0·926). Although there were differences among the values of genetic differentiation, a consistent pattern of clear genetic structure within P. kwangtungensis was revealed by the three data sets.
Wilcoxon's one-tail test showed that P. kwangtungensis populations in southern China deviated significantly from mutation-drift equilibrium (P = 0·002) under SMM, but not significantly under IAM (P > 0·05). This suggested that recent population bottlenecks in P. kwangtungensis might be possible. Further investigations determined that significant deviations from the mutation-drift equilibrium under both the IAM (P = 0·039) and SMM (P = 0·020) occurred in the western populations, but not in the eastern and Hainan populations (P > 0·05). These results were consistent with the field observation that western populations are shrinking demographically and the species-level deviation from mutation-drift equilibrium may result from regional population bottlenecks in the western populations.
DISCUSSION
Genetic diversity
The genetic diversity (HT = 0·629) in P. kwangtungensis detected by the minisatellite is slightly lower than the mean haplotype diversity (0·801) for 16 conifers based on plastid simple sequences repeats, but higher than that (0·556) for 19 conifers using plastid restriction fragment length polymorphism (RFLP; Petit et al., 2005). However, it was determined that genetic diversity for SNPs (indel and substitution) was only 0·380, which is much lower than the value of 0·556 detected in 19 conifer species using plastid RFLP (Petit et al., 2005). These results suggest that plastid diversity in P. kwangtungensis is lower than the average level of conifers. In addition, the lack of sequence variation in other fragments (Tables S1 and S2 in Supplementary Information, available online) also indicates that P. kwangtungensis does not possess abundant plastid genetic diversity. Therefore, the discovery of a highly variable minisatellite and several SNPs within the plastid genome is of great utility for population genetic studies in P. kwangtungensis.
The minisatellite diversity varies greatly at the population level, showing no correlation with population size. The western populations harbour much more genetic diversity (HT = 0·814) than the eastern (HT = 0·439) and Hainan (HT = 0·380) populations, although the western populations are much smaller than the eastern and Hainan populations (Table 1). This result contradicts the theoretical expectation that larger populations possess higher genetic diversity in the absence of selection, recombination, migration or demographic differences (Kimura, 1983). The possibility of differentiated selection for this pattern can be excluded with certainty because of the newly found minisatellite with no functional constraint as discussed above. Apparently, recombination is also unlikely, because the plastid genome is inherited as one locus without recombination (Birky, 2001). More likely, the demographic differences among populations might contribute to uneven genetic diversity, because the Wilcoxon's one-tailed test showed that the western but not the eastern and Hainan populations have experienced a recent marked decline in population size. It is possible that the western populations may have been much larger in the past (e.g. in glacial stages) than at the present time. However, the causes of the population bottlenecks (e.g. deforestation, global warming, etc.) may have occurred recently, possibly after the last glacial maximum or human settlement. The duration of these events might be too transient to leave a strong genetic signature (i.e. genetic impoverishment due to genetic drift) on P. kwangtungensis (e.g. Bates, 2002).
Asymmetrical pollen flow caused by the southern or south-eastern monsoons from the Pacific Ocean might also explain the patterns observed. Pollination of P. kwangtungensis occurs in April–May, when the southern and south-eastern monsoons dominate the climate of southern China. These monsoons could potentially carry bisaccate pollen across the mountains in southern China from the south or south-east to the north or north-west. Therefore, western/northern populations might act as sink populations for the genes carried by the pollen. This pattern corresponds well to the theoretical migrant-pool model (Wade and McCauley, 1990), which describes a migratory pattern with colonists (pollen in this case) recruiting from a random sample of all the other populations. However, the fact that a frequent haplotype (hap7) in eastern populations is completely absent in the western populations compromises the hypothesis of asymmetrical pollen flow. Thus, justification of this hypothesis requires more evidence.
Genetic structure
Many studies have revealed low genetic differentiation in conifers based on paternally inherited plastid markers (e.g. Yandell, 1992; Jorgensen and Hamrick, 1997). The mean genetic differentiation (GST) for 37 conifer species for the paternally inherited markers was 0·165 (Petit et al., 2005). In contrast, genetic differentiation in P. kwangtungensis at the minisatellite site was extremely high (FST = 0·540 under IAM; RST = 0·677 under SMM), contradicting the expectation of a high inter-population gene flow mediated by bisaccate pollen. The analyses based on all the polymorphic sites and on the SNPs alone also showed a pronounced population structure in P. kwangtungensis (FST = 0·630 and 0·926, respectively). Only Pinus densata (GST = 0·533; Song et al., 2003) and Pinus muricata (GST = 0·875; Hong et al., 1993) were comparable to P. kwangtungensis among different pine species. Population genetic structure is determined by the patterns of historical relationships and genetic exchange among populations (Schaal et al., 1998). Thus the pronounced population structure of P. kwangtungensis could be the consequence of its special evolutionary history and limited on-going gene flow among the isolated and fragmented populations.
Hewitt (2000) indicated that in the southern temperate regions and in the tropics, the varied topography tended to subdivide species into populations surviving in different refugia, which had evolved independently during the Quaternary climate oscillations. The existence of Quaternary glaciation in the medium and low mountains of eastern China is a long-disputed issue, but temperatures were estimated to have been 5–13 °C lower in southern China during the last glacial maximum than at the present time (Shi et al., 1989). During the Quaternary, the montane habitat of P. kwangtungensis should have expanded and contracted, tracking the changing climate. Low temperatures during the cold periods may have caused migration of P. kwangtungensis both downwards (in the mountains) and southwards into several glacial refugia. As shown in Fig. 3, populations of P. kwangtungensis formed three deeply divergent clusters (see also the haplotype distribution in Fig. S2 in Supplementary Information, available online), suggesting that there might have been at least three glacial refugia for P. kwangtungensis during the late Quaternary. These refugia may have been located in the lowland areas south to the Nanling Mountains (north-east Guangxi and north-west Guangdong) and east Yungui Plateau Mountains (north-west Guangxi and south-east Guizhou) as well as the lowlands on Hainan Island. The idea of multiple refugia for plant species in southern China has also been suggested by other studies (Lu et al., 2001; Ge et al., 2002; Cheng et al., 2005; Shen et al., 2005; Wang and Ge, 2006). For example, Wang and Ge (2006) indentified four separate refugia for Cathaya argyrophylla, an endangered conifer of China analogous to P. kwangtungensis in ecological requirements and distribution pattern. Long-term isolation among multiple refugia may be the most likely explanation for the high genetic differentiation in P. kwangtungensis as well as in C. argyrophylla (Wang and Ge, 2006).
Furthermore, high-elevation/montane organisms were probably isolated in the mountain tops (‘sky islands’) during warm interglacials such as the present day (Dechaine and Martin, 2004, 2005). Dechaine and Martin (2004, 2005) deduced that high-elevation organisms in the Rocky Mountains could have experienced cycles of range expansion and fragmentation during climatic oscillations and exhibit a significant genetic structure. Hewitt (1996) also suggested that while leading edge populations were expanding north across Europe from southern glacial refugia, the mountains of southern Europe provided refugia during warm interglacials. The present distribution pattern of P. kwangtungensis may fit this model, because the Nanling Mountains and eastern Yungui Plateau mountains stretch almost continuously from west to east thus preventing this species from migrating northward. In contrast, most populations within glacial refugia moved towards the mountain tops during the present warm period. The high genetic differentiation may partially be the consequence of low-level pollen flow between the sky islands and subsequently enhanced genetic drift in these fragmented populations.
The plastid minisatellite as a genetic marker for P. kwangtungensis
During the last decade, several studies demonstrated that plastid minisatellites are useful markers in population genetics (e.g. Cozzolino et al., 2003a, b). In this study, one minisatellite site and several SNPs were found within the same fragment. This offers an unusual opportunity to evaluate the utility of the minisatellite, because indels and substitutions in the plastid genome rarely encounter homoplasy and are more likely to reflect the genuine evolutionary history than hypervariable markers such as microsatellites and minisatellites (Provan et al., 2001). The genetic structures of P. kwangtungensis based on the minisatellite, all polymorphic sites and the SNPs alone are essentially in accordance with each other, indicating that the minisatellite can unravel the authentic evolutionary history of P. kwangtungensis, although its mutational mechanism is complex (Haber and Louis, 1998; Buard et al., 2000).
Another two attributes of the minisatellite render this marker valuable in population genetic studies of P. kwangtungensis. First, the rate of evolution in the minisatellite is relatively high. The genetic diversity of P. kwangtungensis detected by the minisatellite is much higher than those found in other conifers by RFLPs, suggesting that the evolutionary rate of the minisatellite should be higher than those for point mutations and indels, which are usually detected by RFLPs. Comparison between the minisatellite and the SNPs within the same fragment provide further evidence that the minisatellite evolves much faster than SNPs. This characteristic is of particular value for P. kwangtungensis, because the point mutation or indel rate in the plastid genome is extremely low in pines (e.g. Ann et al., 2007). Secondly, the repeated unit of the minisatellite is relatively large (15 bp), allowing different haplotypes to be scored easily on agarose gels. This characteristic lends the minisatellite a technical advantage over microsatellites, because the later must be scored with polyacrylamide or sequencing gels.
Nevertheless, some caution should be taken when using this marker. First, minisatellites with this unusual type of plastid variation often do not provide useful information for population demographic inferences or for fine-scale phylogenetics and phylogeographic analysis (e.g. Faber and Stepien, 1998; Cozzolino et al., 2003b) because of the poor knowledge about their evolution mechanisms. Secondly, homoplasy might be one question of concern in the minisatellite when using the electrophoretic method to distinguish haplotypes, because size homoplasies were found between hap3 and hap9 and among hap5, hap6 and hap8. However, the size homoplasies may not be serious enough to compromise the utility of the minisatellite, because the present analyses based on the electrophomorphs (data not shown) transformed from the minisatellite sequences were compatible with those inferred from the sequences themselves.
Conservation implications and setting priorities
The genetic profile uncovered in this study has provided powerful insights into the genetic structure and evolutionary history of P. kwangtungensis. The results could have obvious implications for the conservation of this vulnerable species. First, three deeply divergent groups of populations were identified in this study, indicating the three groups have independent evolutionary histories (maybe due to the presence of three refugia) and deserve to be conserved as separate evolutionarily significant units or conservation units. Secondly, the western populations (LY, LZ, DA and LB) are worthy of being conserved as a priority because of their higher genetic variability. In addition, the western populations have experienced much stronger population shrinking than the eastern and Hainan ones in recent history, making their conservation an even greater priority, because small populations face more serious stochastic processes and are more prone to extinction than large, stable populations (Lande, 1988; Frankham et al., 2002).
This research reported on a novel minisatellite in P. kwangtungensis and the population structure in this pine species. It showed that Quaternary climate cycles had the most important role in shaping its genetic architecture, which may have significant implications for its conservation, especially in the context of current global warming. It is expected that continuing work should explore the evolutionary history of P. kwangtungensis in more detail, such as identifying ancestral haplotypes and the exact locations of refugia, inferring migratory routes, divergence times and historical demography.
SUPPLEMENTARY INFORMATION
The following Supplementary Information is available online at http://aob.oxfordjournals.org/. Table S1: Universal primers used for screening sequence variation in the Pinus kwangtungensis plastid genome. Table S2: Designed primers according to P. koraiensis plastid genome sequence. Fig. S1: Distribution of fourteen plastid haplotypes within and among populations of Pinus kwangtungensis according to all polymorphic sites. Fig. S2: Distribution of four plastid haplotypes within and among populations of Pinus kwangtungensis according to the SNPs. Fig. S3: Neighbour-joining tree of populations of Pinus kwangtungensis based on pairwise FST of all polymorphic sites.
ACKNOWLEDGEMENTS
We are indebted to Qing-Jun Yuan and Lin-Bin Zhang, for help with the molecular technique, to Hong-Wei Wang, Xin-Wei XU and Liang Tang, for help with data analyses, and to Xin-Sheng Qin for help in collecting experimental materials. Many thanks are given to Dr Ding-Rong Wu for his assistance in using the ArcGIS program. We are grateful to Dr Robert L. Conner of Agriculture and Agri-Food Canada for polishing the language in this manuscript. We are greatly indebted to three anonymous reviewers whose comments were of great help for improving the quality of this paper. This work is supported by the National Science Foundation of China (NSFC 30460020).
LITERATURE CITED
- Ann W, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Molecular Biology and Evolution. 2007;24:90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- Avise JC. Molecular markers, natural history and evolution. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Bates JM. The genetic effects of forest fragmentation on five species of Amazonian birds. Journal of Avian Biology. 2002;33:276–294. [Google Scholar]
- Birky CW., Jr The inheritance of genes in mitochondria and plastids: laws, mechanisms, and models. Annual Review of Genetics. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- Buard J, Shone AC, Jeffreys AJ. Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90) The American Journal of Human Genetics. 2000;67:333–344. doi: 10.1086/303015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YP, Hwang SY, Lin TP. Potential refugia in Taiwan revealed by the phylogeographical study of Castanopsis carlesii Hayata (Fagaceae) Molecular Ecology. 2005;14:2075–2085. doi: 10.1111/j.1365-294X.2005.02567.x. [DOI] [PubMed] [Google Scholar]
- Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A. Molecular evolution of a plastid tandem repeat locus in an orchid lineage. Journal of Molecular Evolution. 2003;a 57:41–49. doi: 10.1007/s00239-003-0006-3. [DOI] [PubMed] [Google Scholar]
- Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A. Fine-scale phylogeographical analysis of Mediterranean Anacamptis palustris (Orchidaceae) populations based on plastid minisatellite and microsatellite variation. Molecular Ecology. 2003;b 12:2783–2792. doi: 10.1046/j.1365-294x.2003.01958.x. [DOI] [PubMed] [Google Scholar]
- Dechaine EG, Martin AP. Historic cycles of fragmentation and expansion in Parnassius smintheus (Papilionidae) inferred using mitochondrial DNA. Evolution. 2004;58:113–127. doi: 10.1111/j.0014-3820.2004.tb01578.x. [DOI] [PubMed] [Google Scholar]
- Dechaine EG, Martin AP. Marked genetic divergence among sky island populations of Sedum lanceolatum (Crassulaceae) in the Rocky mountains. American Journal of Botany. 2005;92:477–486. doi: 10.3732/ajb.92.3.477. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin of the Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Edwards A, Hammond H, Jin L, Caskey C, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genemics. 1992;12:241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Estoup A, Angers B. Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In: Carvalho G, editor. Advances in molecular ecology. Amsterdam: NATO Press; 1998. pp. 55–86. [Google Scholar]
- Faber JE, Stepien CA. Tandemly repeated sequences in the mitochondrial DNA control region and phylogeography of the pike-perches Stizostedion. Molecular Phylogenetics and Evolution. 1998;10:310–322. doi: 10.1006/mpev.1998.0530. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Fu LG, Chen TQ, Lang KY, Hong T. Higher plants of China. Qingdao: Qingdao Publishing House; 2000. [Google Scholar]
- Ge XJ, Chiang YC, Chou CH, Chiang TY. Nested clade analysis of Dunnia sinensis (Rubiaceae), a monotypic genus from China based on organelle DNA sequences. Conservation Genetics. 2002;3:351–362. [Google Scholar]
- Haber JE, Louis EJ. Minisatellite origins in yeast and humans. Genomics. 1998;48:132–135. doi: 10.1006/geno.1997.5153. [DOI] [PubMed] [Google Scholar]
- Hewitt G. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hong YP, Hipkins VD, Strauss SH. Plastid DNA diversity among trees, populations and species in the California closedcone pines Pinus radiata, Pinus muricata and Pinus attenuata. Genetics. 1993;135:1187–1196. doi: 10.1093/genetics/135.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarne P, Lagoda PJL. Microsatellites from molecules to populations and back. Trends in Ecology and Evolution. 1996;1110:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314:767–773. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jorgensen SM, Hamrick JL. Biogeography and population genetics of whitebark pine Pinus albicaulis. Canadian Journal of Forestry Research. 1997;27:1574–1585. [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M, McCauley DW. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Ledig FT. In: Genetic variation in Pinus Ecology and biogeography of Pinus. Richardson DM, editor. Cambridge: Cambridge University Press; 1998. pp. 251–280. [Google Scholar]
- Liepelt S, Bialozyt R, Ziegenhagen B. Wind-dispersed pollen mediates postglacial gene flow among refugia. Proceedings of National Academy of Sciences of the USA. 2002;99:14590–14594. doi: 10.1073/pnas.212285399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SY, Peng CI, Cheng YP, Hong KH, Chiang TY. Plastid DNA phylogeography of Cunninghamia konishii (Cupressaceae), an endemic conifer of Taiwan. Genome. 2001;44:797–807. [PubMed] [Google Scholar]
- Nakamura Y, Leppert M, Connell PO, Wolff R, Holm T, Culver M, et al. Variable number of tandem repeat VNTR markers for human genetic mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, et al. Complete nucleotide sequence of liverwort Marchantia polymorpha plastid DNA. Plant Molecular Biology Reporter. 1986;4:148–175. [Google Scholar]
- Otha T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genetical Research. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of plastid, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet J-M. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends in Ecology and Evolution. 2001;16:142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. Phylogeographic studies in plants, problems and prospects. Molecular Ecology. 1998;7:465–474. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin, Version 2·0: a software for population genetic data analysis. Geneva: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Shen L, Chen XY, Zhang X, Li YY, Fu CX, Qiu YX. Genetic variation of Ginkgo bilobaL. (Ginkgoaceae) based on cpDNA PCR-RFLPs: inference of glacial refugia. Heredity. 2005;94:396–401. doi: 10.1038/sj.hdy.6800616. [DOI] [PubMed] [Google Scholar]
- Shi YF, Cui ZJ, Li JJ. Quaternay glacials and environmental issues of east China. Beijing: Science Press; 1989. [Google Scholar]
- Song BH, Wang XQ, Wang XR, Ding KY, Hong DY. Cytoplasmic composition in Pinus densata and population establishment of the diploid hybrid pine. Molecular Ecology. 2003;12:2995–3001. doi: 10.1046/j.1365-294x.2003.01962.x. [DOI] [PubMed] [Google Scholar]
- Tautz D, Schlotterer C. Simple sequences. Current Opinion in Genetics and Development. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ, McCauley DE. Extinction and recolonization: their effects on the genetic differentiation of local populations. Evolution. 1990;42:995–1005. doi: 10.1111/j.1558-5646.1988.tb02518.x. [DOI] [PubMed] [Google Scholar]
- Wang HW, Ge S. Phylogeography of the endangered Cathaya argyrophylla (Pinaceae) inferred from sequence variation of mitochondrial and nuclear DNA. Molecular Ecology. 2006;15:4109–4122. doi: 10.1111/j.1365-294X.2006.03086.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Xie Y. China species Red List. Beijing: Higher Education Press; 2004. [Google Scholar]
- Weber J, May P. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. American Journal of Human Genetics. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, plastid, and nuclear DNAs. Proceedings of National Academy of Sciences of the USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell UG. An allozyme analysis of whitebark pine. Reno: Master's Thesis, University of Nevada; 1992. [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G, Clegg MT, Brown AHD. The nature of nucleotide sequence divergence between barley and maize plastid DNA. Genetics. 1984;106:735–749. doi: 10.1093/genetics/106.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]