Abstract
Background and Aims
Although high light (HL) and high temperature (HT) stresses have been extensively investigated, a global analysis of their combined effects on the transcriptome of any plant species has not yet been described. Sunflower is an agronomically important oil crop frequently subjected to these stress factors. Because results in model plants may not always translate well to crop plants, responses of sunflower (Helianthus annuus) to HL, HT and a combination of both stresses were analysed by profiling gene expression in leaves and immature seeds.
Methods
Plants were grown in HL (600 µE m−2 s−1), HT (35 °C) and a combination of HL and HT (HL + HT), and gene expression in leaves and immature seeds was profiled using cDNA microarrays containing more than 8000 putative unigenes.
Key Results
Using two-way analysis of variance, 105, 55 and 129 cDNA clones were identified showing significant changes in steady-state transcript levels, across the two tissues, in response to HL, HT and HL + HT, respectively. A significant number of these transcripts were found to be specific to each stress. Comparing gene expression profiles between leaves and immature seeds revealed that 89, 113 and 186 cDNA clones can be considered as differentially expressed in response to HL, HT and HL + HT, respectively. More than half of the cDNA clones showing significant differences between embryo and leaf tissues in response to HL + HT were specific to this stress. Significant differences between leaves and seeds shared by all three stress treatments were observed for only eight genes.
Conclusions
Taken together, these results indicate that vegetative and reproductive tissues employ different transcriptome responses to these stress treatments. Careful examination of the putative functions of these genes revealed novel and specific responses. The potential roles of many of the differentially expressed genes in stress tolerance are mentioned and discussed.
Key words: cDNA microarray, Helianthus annuus, high light, high temperature, heal-time RT–PCR, gene expression, environmental stress
INTRODUCTION
High temperature and extensive light are among the major environmental stresses that affect plant growth and crop productivity. High temperatures (HT) can modulate many plant metabolic and physiological processes, such as photosystem II activity (Havaux et al., 1991), pollen and seed development and structure (Cheikh and Jones, 1994; Wallwork et al., 1998; Pressman et al., 2002), leaf growth (Beator et al., 1992), carbohydrate partitioning (Lafta and Lorenzen, 1995), reactive oxygen metabolism (Larkindale et al., 2005), auxin-mediated processes such as hypocotyl elongation (Gray et al., 1998), and membrane fluidity (Kim and Portis, 2005). Similarly, exposure of plants to excess light can lead to many harmful effects on various physiological process and cellular activities. Among the negative effects of high light (HL) are the inhibition of photosynthesis activity and the production of reactive oxygen species (ROS), which are toxic for many of cellular processes (Niyogi, 1999).
Despite the various effects of HT-stress on different aspects of plant physiology and development, only a limited number of factors have been defined that contribute to heat tolerance. Well-characterized and essential factors include heat shock proteins (HSPs). Plant HSPs can be classified into five evolutionarily conserved protein families, each with a distinct mechanism of action (Parsell and Lindquist, 1993). Preventing of denaturation or refolding of denatured proteins resulting from heat stress appears to be the principle function of HSPs (Parsell and Lindquist, 1993).
Recently, 76 HL and heat-shock stress-inducible genes, including a putative heat-shock transcription factor (HsfA2), have been isolated from Arabidopsis (Nishizawa et al., 2006). Transgenic Arabidopsis plants over-expressing HsfA2 showed increased tolerance to a combination of HL + HT stress (Nishizawa et al., 2006). Although heat-shock transcription factors have been identified as a key regulator in the induction of the defence response under several types of environmental stresses (Nishizawa et al., 2006), knowledge on the molecular mechanisms involved in HT signal transduction in plants remains limited. Global gene expression analyses provide insight into the transcriptional changes triggered by a specific stimulus and can shed light onto the mechanism(s) by which plants obtain thermotolerance. Although heat-stress responses in plants have received increasing attention in recent years, global transcriptional response to HT has been reported in few cases (Rizhsky et al., 2002, 2004; Rensink et al., 2005).
Under field conditions, plants are subjected to multiple stress conditions simultaneously. Different abiotic stress conditions such as cold, drought and salinity can result in the activation of similar responses (Chen et al., 2002; Kreps et al., 2002; Kimura et al., 2003). Nevertheless, the response of plants subjected to a combination of stress factors can be different from the response to each factor when applied independently. It was demonstrated that the response of tobacco and Arabidopsis plants to heat stress or drought is different from the response of plants subjected to a combination of these stresses (Rizhsky et al., 2002, 2004). These results indicate the complexity of plant signal transduction pathways that sense changes in environmental conditions and trigger appropriate counteractive responses. However, although HL and heat stress are among the environmental factors that are generally combined, no studies have addressed the combined effects of HL and HT together on the transcriptome of any plant species. The ability to dissect heat stress response, especially in combination with HL, may be of great importance in agricultural productivity.
In this study, we present transcriptome profiles of leaves and immature seeds in sunflower plants subjected to HT and HL, as well as to a combination of HL and HT stress. cDNA microarrays containing more than 8000 unigenes were used to identify genes whose expression is regulated by these stress factors.
MATERIALS AND METHODS
Plant material
Sunflower (Helianthus annuus L., Asteracea) genotype Ha300B, was grown under standard conditions in a greenhouse (25 °C, 14 h photoperiod, 150 µE m−2 s−1 light intensity and 70 % relative humidity, RH). Three days after the onset of flowering, plants were transferred to growth chambers and subjected to the stress treatments for 2 weeks. High light (HL) treatment was performed by subjecting plants to a light intensity of 600 µE m−2 s−1 with a 14 h photoperiod at 25 °C and 70 % RH. High temperature (HT) stress was applied by subjecting plants to a continuous temperature of 35 °C, 150 µE m−2 s−1 light intensity and 70 % RH. A combination of HL and HT (HL + HT) was imposed by subjecting plants to a continuous temperature of 35 °C and light intensity of 600 µE m−2 s−1 with a 14 h photoperiod and 70 % RH. Control plants were also transferred to a growth chamber and grown under standard conditions of 150 µE m−2 s−1 light intensity with a 14 h photoperiod, 70 % RH and a temperature of 25 °C. To avoid drought stress induced by HL and/or HT all treated plants were frequently watered if needed. The experiment was performed in triplicate with each replicate containing at least six plants. Leaf samples were of the youngest mature leaves (i.e. the third leaf from the top of the plant) and seed samples included embryos at the early cotyledonary stage. For all treatments, leaf and immature seed samples were collected at the same time during the day, at 2000 h after 13 h of the photoperiod.
Construction of microarrays and hybridization of probes
A total of 21 807 estimated sequence tags (ESTs) derived from different cDNA libraries, including embryos at different developmental stages, leaves, stems and apex, as well as from previously described libraries (Tamborindeguy et al., 2004; Ben et al., 2005) were clustered in 8025 contigs using Phragment Assembly Program (PHRAP, University of Washington Genome Center) at a strict assembly criteria of >95 % identity in a 40-bp overlap. For 1219 contigs, two non-overlapping ESTs at both 5′ and 3′ ends were selected. For the remaining 6806 contigs only one EST at the 3′ end was selected. Thus, a total of 9244 ESTs representing 8025 putative unigenes were selected and amplified successfully. The size and quality of all PCR products were tested by agarose gel electrophoresis. PCR samples showing double bands were removed or replaced by another cDNA clone belonging to the same contig. The full list of the selected clones, as well as the positive and negative controls, can be found in the Supplementary Information (Supplementary Information table S1, available online). The PCR products were concentrated in 40 µL water to obtain a concentration of approximately 300–400 ng μL−1 and were arrayed with five touches on a Hybond N+ (Amersham) nylon membrane using the MicroGrid II (Biorobotics LTD, Cambridge, UK) with 64 microarraying pins in a 13 × 13 gridding pattern and a distance of 0·325 µm between spots; this process was carried out at the Centre de resources-Genotypage, Sequencage in Toulouse, France. To increase the reliability of signal, each PCR sample was arrayed twice in unadjusted spots to yield a total of 21 632 data points including negative and positive controls. After spotting, the nylon membranes were placed face up onto Whatman paper moistened with denaturation solution (1·5 m NaCl and 0·5 m NaOH) followed by neutralization solution (1·5 m NaCl and 1 m Tris–HCl, pH 7·4) for 20 min each. The treated membranes were then dried at 80 °C for 2 h, followed by UV cross-linking with a UV Stratalinker 1800 (Stratagene). The quality of spotting was tested after oligonucleotide hybridization. RNA preparation, probes radio-labelling, hybridization and signal quantification were performed as described by Hewezi et al. (2006). To ascertain the reproducibility of the changes in gene expression patterns, three separate hybridization experiments were performed with three separately prepared radio-labelled probes from three biologically independent RNA samples.
Data normalization and ANOVA analysis
Analysis of variance (ANOVA) models were used to identify the differentially expressed genes and to estimate other sources of variation in the microarray data, as described by Wolfinger et al. (2001) and Kerr et al. (2000). After background correction, raw signal intensity values were transformed (log10) and analysed using two interconnected ANOVA models. A normalization ANOVA model of the form
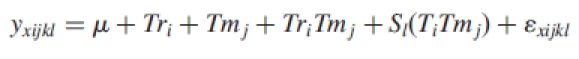 |
was fitted, where yxijkl is the expression level of gene x subjected to stress treatment i in the tissue j for the spot k on array l. The parameter μ is the overall mean of the normalized values for that gene. Tri is the treatment effect (HL, HT, HL + HT and control, i.e. i = 1, 2, 3, 4), Tmj is the tissue effect (leaves and immature seeds, i.e. j = 1, 2), Sl is the membrane effect within the combination of the factors and εxijkl is the stochastic error. The residuals from this model are referred as ‘normalized expression levels’. Genes with normalized expression values higher than the maximum of the empirical distribution of normalized expression levels of the negative controls in any treatment were considered as expressed genes and used in gene-specific models to identify the differentially expressed genes. The gene-specific models were of the form
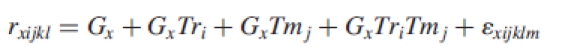 |
where rxijkl denotes the normalized expression levels of gene x. Gx represents the effect of gene x. GxTri corresponds to the interaction between the gene x and stress treatments, whilst GxTmj corresponds to the interaction between the gene x and the tissues. The GxTriTmj interaction effect describes gene expression levels in the two tissues as a function of the stress treatments. A test for heterogeneous variances for the normalized expression levels among treatments was done using the Levene test for each gene model. The Bonferroni method was used to conservatively reduce errors due to multiple tests. Computations were made on a PC running GNU/Linux (Suse 9·2, http://en.opensuse.org) and the R 2·0·0 (http://www.r-project.org) statistical system.
Gene annotation
The putative function of the differentially expressed genes was assigned by scanning the entire GenBank EST database (version May 2007) using the TBLASTX program, and E < 0·00001 was set as the level of significance. Genes with assigned putative functions were manually grouped into different functional categories according to annotation from the Functional Catalogue of the Munich Information Center for Protein Sequences.
Real-Time RT−PCR
The transcript abundance of 14 differentially expressed cDNA clones was analysed by quantitative real-time RT−PCR to confirm the microarray results. Gene-specific primers were designed using the Primer Express software, version 2·0 (Applied Biosystems, Courtaboeuf, France). Oligonucleotide primer sequences are shown in Table 1. First strand cDNA was reverse transcribed from 5 µg of DNase-treated RNA, as described by Hewezi et al. (2006). The reaction was performed in a 20-μL volume containing 10 µL 2× Sybr Green Mastermix (Applied Biosystems), 300 nm of each primer and 1 µL of 5-fold-diluted RT products. The PCR reactions were run in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using the following program: 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Following PCR amplification, the reactions were subjected to a temperature ramp to create the dissociation curve, measured as changes in fluorescence measurements as a function of temperature, by which the non-specific products can be detected. The dissociation program was 95 °C for 15 s, 60 °C for 15 s followed by 20 min of slow ramp from 60 °C to 95 °C. Three replicates of each reaction were performed and β-Actin (accession number AF282624), as a constitutively expressed gene, was used as an internal control to normalize gene expression levels. Quantifying the relative changes in gene expression was performed using the 2−ΔΔCT method as described by Livak and Schmittgen (2001). The construction of the cDNA microarray, the hybridization protocol, the statistical analysis and the validation of results meet the MIAME criteria as described by Brazma et al. (2001).
Table 1.
Oligonucleotide primers used for quantitative real-time RT−PCR
| Accession number | Forward Primer 5′ − 3′ | Reverse Primer 5′ − 3′ |
|---|---|---|
| CD848486 | CCTTGCCTTCAGAATCAATCTTG | AGTTCCTGCTGCAAAGACTCTTG |
| CD848534 | CGAGCTCCATGTAGTTGCAAGT | GATGGTCTTTATGGGCCAACTC |
| CX947086 | CCAAGACATGGGCGTTACCTAA | TTCCTTGGTGCTCATCTTACCA |
| CD847616 | CTCCACCCTTCTCTTCGTCTTC | GCTGTCAACTTGGCGAAAGC |
| CD847838 | TCAGGCTTTAGCGTGGACAA | TTGAAGGTTTCTTGCACCGTAA |
| CD845834 | ACCATGGTATCCATCCCACAA | GGGTGAGGACGGGTACTACAAG |
| CD845613 | CATAACCAGGATGCGGATCTATG | TTGGTTCCTCTTAGTGCTTCCAA |
| CD849757 | CCCCCAGTTTCGTGACATTT | GCATGCCGTTGCTATCAAAG |
| CD852690 | CATTGTGGTGGCCCTGCTAT | CCGCATGTACAGCAGACCAA |
| CD850746 | AGTTTGGCAGTGGGAATAGCAA | TGGATTCGGAAACTCTCCTTCA |
| CD849228 | GCAGTCGTGCTCAGACTTTCC | GACCATGATGGCCACCTACAA |
| CD855563 | TGTTCTCCCGCACGATCAT | ATTTCGCATCGCCTTTGGT |
| CX943668 | GAGCGCGAAAGATATGGAAAA | TTCCTGGTGCAAGAAAAGCA |
| CD846383 | TGTCCGTTAGAGTCTCCTGCAA | TCATGATCTTATGGCGGAAGAA |
| AF282624 | TCAATGTTCCCGCCATGTAT | GACCACTGGCATAGAGGGAAAG |
RESULTS
In order to study the global expression profiles specific to HL (600 µE m−2 s−1), HT (35 °C) and a combination of HL and HT, mature sunflower plants at the flowering stage were subjected to these stresses and compared with non-stressed plants 2 weeks after stress application. Subjecting sunflower plants to continuous HT of 40 °C for 2 weeks gave rise to sterile plants. Combination of HT of 35 °C with HL intensity of 900 µE m−2 s−1 dramatically affected seed set and only a few filled seeds were recovered. Thus, the HL (600 µE m−2 s−1) and HT (35 °C) treatments were selected in this experiment in order to study their combined effect whilst avoiding harmful effects on embryo development. The choice of a 2-week stress period to study global gene expression was an attempt to discover acclimation responses that are stress-specific, and to eliminate possible coinciding general stress responses. These stress treatments were designed to mimic stress conditions that can be encountered in sunflower plants during the seed-filling stage. These comparative analyses were also designed to reveal shared and specific expression patterns triggered by these three stress conditions in vegetative and reproductive tissues. The normalization of gene expression values was performed using an ANOVA normalization model, according to treatment effects (HL, HT, HL + HT and controls) and tissue effects (leaves and immature seeds) that were found to be highly significant (data not shown); this revealed the importance of the normalization steps in the analysis of microarray data. Over the range of normalized expression values, the correlation between the two spots for each clone regardless of the stress treatment was found to be very high (r2 = 0·996). A total of 1219 genes are represented by two ESTs at both 5′ and 3′ ends and the correlation between the standardized expression values of both ESTs was very high (r2 = 0·95). Finally, the correlation between the biological replicates on independent membranes was also found to be very significant (r2 ranging from 0·85 to 0·90). Taken together, these data indicated that measurements of gene expression values were reproducible and that there were no significant differences due to sample heterogeneity. The residuals from the normalization model, which represent the normalized expression values, were analysed statistically using a two-way analysis of variance method as described in the Material and Methods. This method enabled us to identify genes that displayed similar responses in both tissues as a function of treatment effects (main effect), as well as genes that exhibited significant differences in transcriptional activity between the two tissues in response to the stress treatments (interaction effect). To confirm the results obtained from microarray experiments, the transcript abundance of 14 differentially expressed ESTs were tested using quantitative real-time RT−PCR. These 14 genes were selected to represent different expression levels and different functional categories. Total RNA was extracted from stress treatments in a repeat of the experiment, subjected to first-strand cDNA synthesis and used in triplicate in SYBR Green quantitative real-time RT−PCR assays. Estimated expression levels by DNA microarray and real-time RT−PCR are compared in Table 2. In general, the fold-change values estimated by real-time RT−PCR are in good agreement with the microarray data (r2 value of 0·79). In addition, we tested the expression levels of these genes under stress treatments other than those that were found to affect their expression patterns. Only non-significant changes ranging between 1·05-fold down-regulation and 1·09-fold up-regulation were detected (data not shown), confirming the microarray data. Differences between the microarray data and some of the real-time RT−PCR results can be explained by the biological variability between the two independent experiments.
Table 2.
Comparison of gene expression levels of 14 selected differentially expressed genes estimated from quantitative real-time RT−PCR and microarray analysis
| (A) Treatment × control | |||||
|---|---|---|---|---|---|
| Accession number | Functional annotation | Quantitative RT−PCR fold change |
Microarray fold change |
||
| HL vs. Control | |||||
| CD848486 | Putative ribosomal protein S12 (Oryza sativa) | 1·3 | 1·84 | ||
| CD848534 | Lipid transfer protein (A. thaliana) | 2·01 | 3 | ||
| HL + HT vs. Control | |||||
| CX947086 | Beta-tubulin (Zinnia elegans) | 1·49 | 2·04 | ||
| CD847616 | Aquaporin (Helianthus annuus) | 2·69 | 2·48 | ||
| CD847838 | Oxidoreductase NAD-binding domain-containing protein (A. thaliana) | 1·63 | 2·73 | ||
| (B) Treatment × Organ | |||||
| Quantitative RT–PCR fold change | Microarray fold change | ||||
| Accession number | Functional annotation | Leaves | Immature seeds | Leaves | Immature seeds |
| HL × Organ | |||||
| CD845834 | Cysteine protease (Aster tripolium) | 1·66 | 1 | 3·02 | −1·83 |
| CD845613 | ss-1,3-glucanase (Cichorium intybus × Cichorium endivia) | 2·91 | −1·18 | 4·44 | −1·62 |
| CD849757 | Expansin 3 (Zinnia elegans) | −2·48 | 1·34 | −1·33 | 2·08 |
| HT × Organ | |||||
| CD852690 | Cytochrome P450 family protein (A. thaliana) | −1·67 | 2·14 | −1·90 | 1·78 |
| CD850746 | Pectin acetylesterase (EC 3·1·1.-) precursor − (Vigna radiata) | 4·81 | −1·4 | 2·57 | −2·00 |
| CD849228 | Ferredoxin (Helianthus annuus) | 2·85 | 1·17 | 5·17 | −1·20 |
| HL + HT × Organ | |||||
| CD855563 | Unknown protein (A. thaliana) | −1·74 | 1·16 | −1·41 | 2·32 |
| CX943668 | Esterase/lipase/thioesterase family protein (A. thaliana) | 1·48 | −2·27 | 3·01 | −1·11 |
| CD846383 | Homeobox-leucine zipper protein (HD-ZIP protein 5) (A. thaliana) | 1·4 | 1·74 | 3·61 | 1·01 |
Fourteen cDNA clones exhibiting different expression patterns and belonging to different functional categories were subjected to real-time RT−PCR analysis to confirm the microarray results. The expression levels of the target genes derived from RT−PCR were normalized using β-actin as an internal control and the fold-change values were calculated using the 2−ΔΔCT method and represent changes of mRNA abundance between the compared treatments. Fold-change values derived from the microarray analysis were computed as the difference between normalized expression values of the compared treatments. Positive values denote up-regulation and negative values denote down-regulation.
Main effects of stress treatment
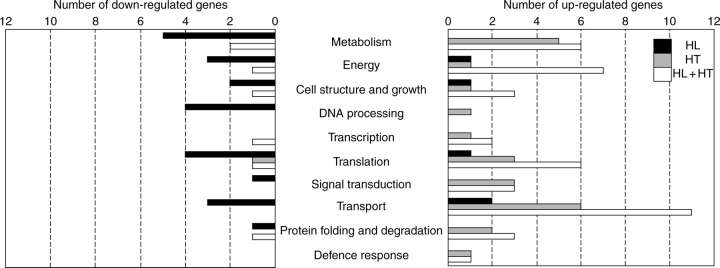
At a P-value cut off of 0·001, we identified 105, 55 and 129 cDNA clones showing significant changes in steady-state transcript levels across the two tissues in response to HL, HT and HL + HT, respectively. The complete lists of these differentially expressed genes as well as their putative functions are provided in the Supplementary Information (Supplementary Information table S2, available online). In order to explore the biological processes in which the differentially expressed genes are involved, we have classified the regulated genes with assigned putative functions into different functional categories. As shown in Fig. 1, many biological processes have been found to display down-regulation in response to HL. Most of the genes with potential roles in primary metabolism, energy, DNA processing and translation have been shown to be repressed. In contrast to HL-responses, most of HT-regulated genes with predicted functions have been induced. The functional categories with the highest number of genes were those involved in metabolism and transport activities (Fig. 1). Several biological processes have been shown to display induction or repression in response to the combined effect of HL and HT. Genes potentially involved in metabolism, energy, cell structure, translation, and protein folding and degradation have appeared to be more induced than repressed. In addition, all genes related to signal transduction and transport activities have been induced (Fig. 1).
Fig. 1.
Functional classification of differentially expressed genes in response to HL, HT and HL + HT across leaf and seed tissues. Genes with putative functions were classified according to annotation from the Functional Catalogue of the Munich Information Center for Protein Sequences. Genes annotated as ‘no hits’ or unknown are not shown.
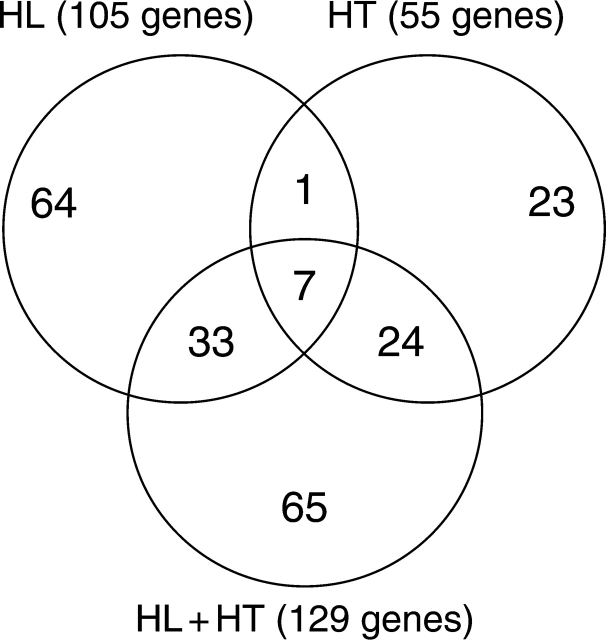
In order to provide information on the shared and stress-specific responses, we illustrate the differentially expressed genes in Venn diagrams (Fig. 2). A significant number of the differentially expressed genes were found to be specific to each stress. As shown in Fig. 2, 64, 23 and 65 genes were exclusively regulated by HL, HT and HL + HT, respectively. The putative functions of the differentially expressed genes that are exclusively regulated during HL + HT treatment are provided in Table 3. Careful examination of this gene list indicates that the transcriptional response of mature sunflower exposed to a combination of HL and HT stress was different from that of plants exposed to these stresses individually. Venn diagram analysis also revealed an overlap of 33 transcripts between HL and HL + HT. Similarly, an overlap of 24 transcripts was observed between HT and HL + HT. However, an overlap of only one gene (AJ540169) with unknown function was found uniquely between HL and HT. Finally, seven genes were found to be regulated by all of these stresses.
Fig. 2.
Venn diagram showing the number of differentially expressed genes in sunflower plants in response to HL, HT and HL + HT across leaf and seed tissues.
The number in the overlapping areas indicate the number of cDNA clones that exhibited shared responses to either two or three stress treatments compared to non-stressed controls.
Table 3.
Putative functions of differentially expressed genes exclusively regulated during the HL + HT treatment
| Sequence name | Accession number | P-value | Putative function | E value | Fold-change |
|---|---|---|---|---|---|
| DH0AB66ZA04RM1 | CD847838 | 1·18E–09 | Oxidoreductase NAD-binding domain-containing protein (A. thaliana) | 1·00E–50 | 2·73 |
| DH0AB67ZD03RM1 | CD847935 | 3·18E–06 | Putative ribosomal protein L11 (A. thaliana) | 2·00E–72 | 2·72 |
| DH0AC010ZF02FM1 | CD848727 | 4·27E–06 | Putative ribosomal protein S11 (A. thaliana) | 5·00E–68 | 2·25 |
| DH0AB56ZF04RM1 | CX943605 | 7·34E–06 | Chlorophyll a/b-binding protein type I (Asarina barclaiana) | 1·00E–51 | 2·40 |
| DH0AB43ZD08RM1 | CD846206 | 9·78E–06 | Ribosomal protein L44 isoform b (Gossypium hirsutum) | 1·00E–57 | 2·49 |
| DH0AB61ZG06RM1 | CD847616 | 1·57E–05 | Aquaporin (Helianthus annuus) | 5·00E–51 | 2·48 |
| DH0AMM2ZB04ZZM1 | CD854391 | 1·72E–05 | 60S ribosomal protein L37a (Capsicum chinense) | 1·00E–46 | 2·16 |
| DH0AGB14ZH01RM1 | CX944089 | 3·35E–05 | Chloroplast oxygen-evolving enhancer protein (Manihot esculenta) | 1·00E–11 | 2·20 |
| DH0AGB15ZC04RM1 | CX944112 | 4·63E–05 | Probable cleavage and polyadenylation specificity factor (A. thaliana) | 7·00E–16 | 1·97 |
| DH0AB009ZH08RM1 | CD845977 | 6·74E–05 | Triose-phosphate isomerase (EC 5·3·1·1) precursor, chloroplast (Spinacia oleracea) | 3·00E–61 | 2·40 |
| DH0AC014ZG10FM1 | CD849044 | 1·12E–04 | Protein phosphatase 2C (A. thaliana) | 5·00E–83 | 2·05 |
| DH0AB42ZG11RM1 | CD846162 | 1·20E–04 | Photosystem I subunit PSI-E (Nicotiana sylvestris) | 6·00E–32 | 2·17 |
| DH0AGB10ZH02RM1 | CX943929 | 1·22E–04 | NADH-plastoquinone oxidoreductase chain 1 (Medicago truncatula) | 3·00E–24 | 1·90 |
| DH0AB44ZH02RM1 | CD846320 | 1·33E–04 | Delta-aminolevulinic acid dehydratase, chloroplast precursor (Porphobilinogen synthase) | 9·00E–57 | 2·30 |
| DH0AG15ZE04RM1 | CD858367 | 1·34E − 04 | Putative peptide transport protein (A. thaliana) | 3·00E–40 | 1·92 |
| DH0AB008ZF02RM1 | CD845931 | 1·47E–04 | Calmodulin (A. thaliana) | 5·00E–49 | 2·07 |
| DH0AB48ZC09RM1 | CD846655 | 1·59E–04 | RubiscO (SSU) (Helianthus annuus) | 2·00E–94 | 1·85 |
| DH0AMM32ZC04ZZM1 | CD854627 | 2·37E–04 | Ribosomal protein S18, putative (A. thaliana) | 6·00E–69 | 1·67 |
| DH0AQA1ZH02RM1 | CX947086 | 2·41E–04 | Beta-tubulin (Zinnia elegans) | 6·00E–107 | 2·04 |
| HaCotR003G01 | AJ828911 | 2·97E–04 | Acetyl-CoA carboxylase beta subunit (Nicotiana tabacum) | 2·00E–73 | 2·03 |
| DH0AB55ZE03RM1 | CD847156 | 3·01E–04 | Peptidase M48 family protein (A. thaliana) | 1·00E–98 | 2·32 |
| DH0AB49ZG06RM1 | CD846762 | 3·04E–04 | DAD1 Defender against cell death 1 (DAD-1) (Solanum lycopersicum) | 3·00E–62 | 2·24 |
| DH0AB52ZD01RM1 | CD846920 | 3·66E–04 | Chlorophyll a/b-binding protein type III precursor (Solanum lycopersicum) | 4·00E–61 | 1·89 |
| DH0AG14ZC04RM1 | CD858263 | 4·79E–04 | Plasma membrane H + -ATPase (Daucus carota) | 6·00E–21 | 1·92 |
| DH0AC027ZG12FM1 | CD850008 | 5·00E–04 | F1 ATPase; adenosinetriphosphatase (Helianthus annuus) | 3·00E–44 | 2·16 |
| DH0AG16ZD08RM1 | CD858445 | 5·13E–04 | Aquaporin (Helianthus annuus) | 9·00E–41 | 1·81 |
| DH0AC42ZD06RM1 | CD850620 | 5·79E–04 | Putative esterase D (A. thaliana) | 8·00E–55 | 2·10 |
| DH0AB008ZF10RM1 | CD845937 | 7·20E–04 | Transport protein subunit-like (A. thaliana) | 1·00E–11 | 1·94 |
| DH0AB45ZD12RM1 | CD846365 | 7·31E–04 | Thylakoid lumenal protein-like (Oryza sativa) | 2·00E–70 | 1·77 |
| DH0AB008ZG05RM1 | CD845941 | 8·18E–04 | Putative 16 kDa membrane protein (Nicotiana tabacum) | 4·00E–12 | 1·92 |
| DH0AB58ZG06RM1 | CD847382 | 9·11E–04 | 33 kDa subunit of oxygen evolving system of photosystem II (Solanum tuberosum) | 1·00E–107 | 1·70 |
| DH0ALL29ZE09ZZM1 | CD852547 | 9·41E–04 | L-galactose-1-phosphate phosphatase (Actinidia deliciosa) | 1·00E–115 | −1·68 |
Only differentially expressed genes with assigned putative functions are presented: unknown genes are not included. The full list of differentially expressed genes that were exclusively regulated by HL + HT treatment is provided in the Supplementary Information (table S2, available online). The fold-change values represent changes of mRNA abundance in HL + HT-treated plants vs. controls across the two tissues, and are computed as the difference between normalized expression values. Positive values denote up-regulation and negative values denote down-regulation.
Interaction effects
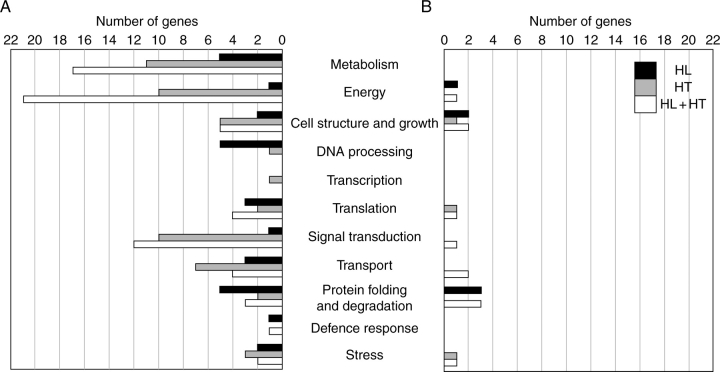
Organ-specific gene expression in response to environmental stress is needed to define the stress-response characteristics of each organ and to identify the mechanisms involved. In order to investigate the effects of stress treatments on sink and source tissues, ANOVA interaction analysis was used as a tool to identify gene expression differences between leaves and immature seeds. Because not all changes in transcriptional profiles between leaf and seed tissues are expected to be a direct consequence of stress treatments, we identified and excluded all genes that exhibited significant variation in expression levels between both tissues under control conditions. This allows the comparison between both tissues in response to the stress treatments (interaction effects) to be potentially free of the tissues' main effect. At a P-value cut-off of 0·001, we identified 89, 113 and 186 cDNA clones showing significant differences between the two tissues in response to HL, HT and HL + HT respectively (see Supplementary Information table S3, available online). The differentially expressed genes, in which the putative function can be assigned, are distributed throughout different functional groups and are illustrated in Fig. 3. Careful comparison of the putative functions of the differentially expressed genes between leaves and immature seeds in response to HL highlighted that all transcripts encoding proteins potentially involved in metabolism, DNA processing, translation, transport and stress response were found to be significantly more highly expressed in leaves compared with immature seeds. Similarly, all genes classified as being involved in metabolism, energy, signal transduction, transport functions and protein degradation were more highly transcribed in leaves compared with seeds in response to HT. However, genes belonging to cell structure and growth, translation, signal transduction, transport activity, and protein folding and degradation functional categories were found to display enhanced expression levels in both leaves and in seeds in response to HL + HT, with a larger proportion identified as being highly expressed in leaves compared with immature seeds (Fig. 3). Based on Venn diagram analysis, we identified shared and specific interaction responses of gene expression among HL, HT and HL + HT treatments. As shown in Fig. 4, out of the 89 ESTs showing a HL × tissue interaction, 53 were unique to this interaction. Similarly, 33 cDNA clones were unique to the HT × tissue interaction (Fig. 4). Interestingly, 102 cDNA clones were specific to the HL + HT × tissue interaction. Out of these 102 clones, the putative function of 43 was assigned (Table 4). An overlap of 12 transcripts between the HL × tissue interaction and the HT × tissue interaction was detected. Similarly, overlaps of 16 and 60 transcripts were observed between the HL × tissue and the HL + HT × tissue interactions, and HT × tissue and HL + HT × tissue interactions, respectively. Finally, an overlap of only eight genes was found between all three interactions, representing genes that are tissue-specific and shared by all three stress conditions.
Fig. 3.
Functional classification of differentially expressed genes between leaves and immature seeds in response to HL, HT and HL + HT. (A) Genes with significantly higher expression in leaves than in seeds. (B) Genes with significantly higher expression in seeds than in leaves. Genes with putative functions were classified according to annotation from the Functional Catalogue of the Munich Information Center for Protein Sequences. Genes annotated as ‘no hits’ or unknown are not shown.
Fig. 4.
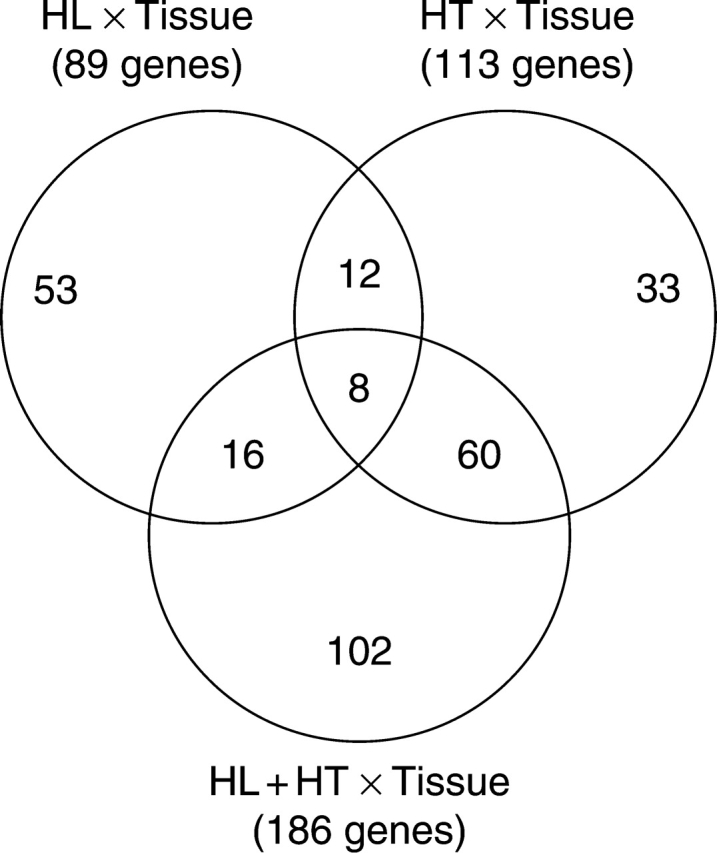
Venn diagram showing the number of genes having significantly different expression levels in leaves and immature seeds in response to HL, HT and HL + HT. The numbers in the overlapping areas indicate the number of cDNA clones that exhibited shared interaction responses to either two or three stress treatments compared with non-stressed controls.
Table 4.
Putative function of tissue-specific differentially expressed genes exclusively regulated during the HL + HT treatment
| Sequence name | Accession number | P-value | Putative function | E-value | Fold-change |
|
|---|---|---|---|---|---|---|
| Seeds | Leaves | |||||
| DH0AGB13ZB06RM1 | CX944002 | 1·19E–07 | Putative photosystem II protein (Tectona grandis) | 7·00E–79 | −2·2791 | 7·8012 |
| DH0AB50ZD07RM1 | CD846797 | 1·17E–06 | Peptidyl-prolyl cis-trans isomerase / cyclophilin (A. thaliana) | 2·00E–37 | −1·4349 | 3·9635 |
| DH0AB53ZA07RM1 | CD846970 | 1·22E–06 | Short-chain dehydrogenase/reductase (SDR) family protein (A. thaliana) | 4·00E–86 | −1·2841 | 2·7377 |
| DH0AB41ZB12RM1 | CD846049 | 1·79E–06 | Putative RNA-binding protein (A. thaliana) | 3·00E–54 | −1·2016 | 3·6500 |
| DH0AGB20ZD11RM1 | CX944380 | 9·18E–06 | Photosystem II protein D1 (Bassia scoparia) | 1·00E–32 | −1·7503 | 4·8678 |
| DH0AG16ZC11RM1 | CD858436 | 1·08E–05 | Ribulose-bisphosphate carboxylase (EC 4·1·1·39) small chain precursor (Helianthus annuus) | 2·00E–24 | −1·2968 | 4·4031 |
| DH0AB58ZE09RM1 | CD847366 | 1·09E–05 | Malate dehydrogenase (Glycine max) | 5·00E–75 | −1·3735 | 3·2679 |
| DH0AB008ZF03RM1 | CX943561 | 1·10E–05 | Ribulose-bisphosphate carboxylase (EC 4·1·1·39) small chain precursor (Helianthus annuus) | 6·00E–13 | −1·1247 | 3·5740 |
| DH0AC007ZA11FM1 | CD848487 | 1·47E–05 | Plastocyanin (Lactuca sativa) | 3·00E–27 | −1·0806 | 6·1530 |
| DH0AB58ZH07RM1 | CD847394 | 3·05E–05 | O-methyltransferase (Mesembryanthemum crystallinum) | 3·00E–65 | −1·1098 | 3·1417 |
| DH0AB002ZC03RM1 | CD845669 | 3·33E–05 | Chlorophyll a/b-binding protein (cab-11) (Solanum lycopersicum) | 7·00E–27 | −1·8892 | 3·2064 |
| DH0AB63ZE05RM1 | CD847711 | 4·49E–05 | Cytosolic fructose-1,6-bisphosphate (Lactuca sativa) | 6·00E–20 | −1·2578 | 3·7438 |
| DH0AB49ZG04RM1 | CD846760 | 4·88E–05 | L-allo-threonine aldolase homolog, (A. thaliana) | 5·00E–90 | −1·4100 | 3·5358 |
| DH0AB43ZG02RM1 | CD846229 | 5·12E–05 | Thylakoid membrane phosphoprotein 14 kDa, chloroplast precursor (A. thaliana) | 7·00E–38 | −1·0155 | 4·2720 |
| DH0AB47ZA04RM1 | CD846554 | 5·35E–05 | Carbonic anhydrase (Flaveria bidentis) | 3·00E–28 | −1·4100 | 4·1296 |
| DH0AGB9ZG01RM1 | CX944985 | 7·53E–05 | ATP synthase beta subunit (Echinops bannaticus) | 1·00E–29 | −2·6869 | 1·5697 |
| DH0AB51ZC12RM1 | CD846849 | 9·06E–05 | Ribosomal protein S7 (Brassica napus) | 5·00E–49 | −1·0224 | 5·4261 |
| DH0AB53ZB03RM1 | CD846976 | 9·24E–05 | Nucleoside diphosphate kinase (Helianthus annuus) | 1·00E–68 | −1·2182 | 3·6211 |
| DH0AC011ZH08FM1 | CD848816 | 9·42E–05 | Ribosomal protein L2 (Nicotiana tabacum) | 1·00E–14 | −1·4961 | 6·5873 |
| DH0AB45ZG03RM1 | CD846383 | 1·07E–04 | HD-ZIP protein 5 (A. thaliana) | 6·00E–33 | −1·0086 | 3·6098 |
| HaCotR003B07 | AJ828437 | 1·49E–04 | Hydroxyproline-rich glycoprotein-like (Oryza sativa) | 9·00E–33 | 2·2750 | −1·6942 |
| DH0AB57ZG05RM1 | CD847313 | 1·58E–04 | Putative isoleucyl-tRNA synthetase (Oryza sativa) | 2·00E–61 | −1·6844 | 2·0572 |
| DH0AB59ZB01RM1 | CD847408 | 1·87E–04 | Zinc finger (AN1-like) family protein (A. thaliana) | 1·00E–31 | −1·2595 | 3·6436 |
| DH0AB007ZD09FM1 | CD845873 | 1·96E–04 | Ribulose-bisphosphate carboxylase (EC 4·1·1·39) small chain precursor (Helianthus annuus) | 1·00E–19 | −1·1373 | 4·0489 |
| DH0AMM27ZB11ZZM1 | CD854183 | 2·16E–04 | Synaptobrevin-like protein (A. thaliana) | 7·00E–85 | 2·4706 | −1·3509 |
| DH0AB67ZD08RM1 | CD847940 | 2·63E–04 | Putative palmitoyl-protein thioesterase (Tropaeolum majus) | 2·00E–101 | −2·3623 | 1·3585 |
| DH0AB008ZH04RM1 | CD845946 | 2·95E–04 | Aldo/keto reductase family protein (A. thaliana) | 5·00E–05 | 1·6695 | −1·4788 |
| DH0AB41ZH06RM1 | CD846095 | 2·96E–04 | Photosystem II reaction center W protein, (Spinacia oleracea) | 1·00E–30 | 1·2541 | 4·3425 |
| DH0ALL17ZD12ZZM1 | CD851683 | 3·09E–04 | Cytoplasmic ribosomal protein S13 (Solanum demissum) | 2·00E–69 | −1·0052 | 3·2242 |
| DH0AB67ZF04RM1 | CD847955 | 3·22E–04 | Translation elongation factor EF-Tu precursor, chloroplast (Glycine max) | 1·00E–58 | −1·1660 | 3·2571 |
| DH0AB010ZB11RM1 | CD845988 | 3·54E–04 | Light-regulated chloroplast-localized protein (Solanum tuberosum) | 1·00E–10 | 1·9388 | −1·8164 |
| DH0AB001ZH09RM1 | CX943532 | 4·23E–04 | Photosystem II 10K protein precursor–(Solanum tuberosum) | 3·00E–34 | −1·0242 | 3·6878 |
| DH0AB42ZH05RM1 | CD846167 | 4·46E–04 | 14-3-3 protein (Solanum tuberosum) | 1·00E–47 | −1·0250 | 3·0156 |
| DH0AB66ZG08RM1 | CD847899 | 5·31E–04 | PSI type III chlorophyll a/b-binding protein (Brassica juncea) | 1·00E–39 | 1·1505 | 5·6922 |
| DH0AC002ZB04FM1 | CX943668 | 5·69E–04 | Esterase/lipase/thioesterase family protein (A. thaliana) | 3·00E–27 | −1·1133 | 3·0077 |
| DH0AB011ZC10RM1 | CD846022 | 5·79E–04 | Endochitinase (Nicotiana sylvestris) | 6·00E–59 | −1·3580 | 4·0356 |
| DH0AB59ZC05RM1 | CD847419 | 5·98E–04 | Oxygen-evolving enhancer protein 3 precursor-like protein (A. thaliana) | 1·00E–36 | −1·0465 | 3·9955 |
| HaCotR003A08 | AJ828340 | 6·22E–04 | Kinase interacting family protein (A. thaliana) | 3·00E–26 | 2·0190 | −1·6361 |
| DH0AG7ZC07RM1 | CD857998 | 6·73E–04 | Chlorophyll a/b-binding protein (Solanum lycopersicum) | 1·00E–62 | −1·0702 | 2·8950 |
| DH0AB47ZA12RM1 | CD846561 | 7·41E–04 | Putative triosephosphate isomerase (A. thaliana) | 2·00E–61 | 1·1662 | 4·1200 |
| DH0ALL13ZG05ZZM1 | CD851449 | 7·55E–04 | Sterol C-24 reductase (Zinnia elegans) | 7·00E–112 | 1·7798 | −1·9677 |
| DH0AG8ZC07RM1 | CD858089 | 7·98E–04 | Putative alanine aminotransferase (A. thaliana) | 3·00E–79 | −1·1320 | 3·7155 |
| DH0AL5ZG01ZM1 | CD851057 | 9·06E–04 | Tubulin Alpha-6 Chain (A. thaliana) | 3·00E–79 | −1·4728 | 2·3562 |
Only differentially expressed genes with assigned putative functions are presented, while unknown genes are not included. The full list of tissue-specific differentially expressed genes that were exclusively regulated by the HL + HT treatment is provided in the Supplementary Information (table S3, available online). The fold-change values represent changes of mRNA abundance in leaf and immature seed tissues in the HL + HT-treated plants compared with non-stressed controls, and computed as the difference between normalized expression values. Positive values denote up-regulation and negative values denote down-regulation.
DISCUSSION
In this study, we investigated sunflower responses to high light (HL), high temperature (HT) and a combination of HL and HT by carrying out a global analysis of gene expression in leaf and immature seed tissues. A set of 217 genes were identified that displayed significant changes in expression in response to stress treatments across the leaf and seed tissues (main effects). In addition, 284 genes exhibiting significant differential expression between leaves and seeds in response to the stress treatment (interaction effects) were identified.
Stress treatment main effects
HL-specific responses
Data analysis revealed that a major response of sunflower to HL involves down-regulation of gene expression, since 58 out of the 64 HL-specific genes were down-regulated. An extensive down-regulation of gene expression was also reported in Arabidopsis subjected to HL stress (Rossel et al., 2002). These HL down-regulated genes are distributed throughout different functional categories with the largest category being ‘no hits’ in the GenBank database; these sequences could be sunflower-specific. Among the HL-induced genes, two lipid transfer proteins (LTP) were found. Several studies have reported the induction of LTPs under a wide range of biotic and abiotic stresses including HL (Rossel et al., 2002; Kimura et al., 2003; Hewezi et al., 2006). The HL down-regulation of fructose-bisphosphate aldolase (EC: 4·1·2·13), S-adenosyl-L-methionine synthetase (EC: 2·5·1·6) and GTP cyclohydrolase II (EC: 3·5·4·25), which are key enzymes involved in the metabolism of glycolysis, methionine and riboflavin, respectively, suggests that these pathways may be affected by HL. Among the specific HL-regulated genes, two subunits of PSI were found with opposite expression patterns: PSI-V was induced whereas PSI-D was repressed. Transcripts coding for subunits of PSI were also found to exhibit opposite expression patterns in sunflower plants in response to water-deficit stress under field conditions (Roche et al., 2007). This contrasting expression pattern suggests different regulatory roles of these subunits during plant responses to HL. A translational control of the HL response in sunflower is reflected by regulation of many cDNA clones coding for elongation factors and ribosomal proteins (see Supplementary Information table S2, available online). Genes involved in translation machinery were also highly represented in the differentially expressed gene list in response to HL in Arabidopsis (Vandenabeele et al., 2004).
HT-specific responses
In contrast to HL-responses, which mainly involved down-regulation of gene expression, the majority of HT-regulated genes were induced. A putative inositol-3-phosphate synthase (EC: 5·5·1·4) is among the HT up-regulated genes and converts D-glucose 6-phosphate to 1D-myo-inositol 3-phosphate, the rate-limiting step of de novo inositol biosynthesis. Inositol-3-phosphate synthase mRNA was also induced in Arabidopsis plants during heat acclimation (Lim et al., 2006), suggesting a role of this gene during the response of plants to heat stress. Heat stress stimulates H2O2 generation and hence oxidative stress in plants (Dat et al., 1998). Among the HT-induced genes, genes were identified that can act as antioxidants and/or detoxificants. These include glyceraldehyde-3-phosphate dehydrogenase and glutathione transporter. Transcripts encoding ROS-detoxifying enzymes were also found to be induced under heat stress in tobacco (Rizhsky et al., 2002). The induction of these genes may contribute to HT tolerance.
HL + HT-specific responses
In contrast to the treatments where HL or HT was applied individually, a combination of HL and HT resulted in the induction of different stress response (Table 3). It was found that all genes involved in protein synthesis and energy metabolism were up-regulated in the HL + HT treatment, indicating that the combination of the two environmental stresses affects cellular processes differently than when the stresses are applied individually. The up-regulation of energy-related genes may be required for plant cells to preserve the increased expression of genes encoding proteins involved in the adaptive or protective responses. The induction of many ribosomal proteins may provide an insight into an important role of these proteins in the direction of synthesis machinery to produce proteins important or critical for cell maintenance. It is of interest that a significant number of transport-related genes, including two aquaporins, displayed up-regulation; the induction of aquaporin-encoding genes may be an adaptive response to maintain cellular homeostasis. Up-regulation was also observed for genes encoding potential signal transduction components, including protein phosphatase 2C and calmodulin. Calmodulin is known as a mediator of calcium signal transduction and controls target gene expression. This finding suggests that a calcium sensing and regulatory network could be involved in the long-term adaptation to HL + HT stress. In accordance with this suggestion, calcium and calcium-activated calmodulin have been reported to be involved in heat shock signal transduction in wheat (Liu et al., 2003).
Non-specific main effects
Among the 129 genes showing HL + HT responses, only 33 and 24 genes were also regulated by HL and HT, respectively (see Supplementary Information table S2, available online). These results demonstrate that sunflower responses to a combination of stress factors are different from the response to each stress when applied independently. Therefore, the response of plants to multiple environmental stress conditions can not be deduced from studies that apply these stress factors individually.
Our analysis revealed that only seven genes were shared by all of the three stress conditions and exhibited down-regulation (see Supplementary Information table S2). This shared expression response indicates that different stress treatments can led to the repression of the same set of genes, probably via common signalling pathways. This gene down-regulation seems to be a common adaptive response that enables plants to cope with the new environmental conditions, possibly in order to conserve energy that can be used to activate tolerance responses.
Interaction effects
HL × tissue-specific responses
Among the 53 transcripts showing significant changes in transcriptional activity between immature seeds and leaves exclusively in response to HL stress (see Supplementary Information table S3, available online), we found a putative expansin with a preferential expression profile in embryonic tissues. Most expansin genes are thought to encode proteins that mediate cell expansion and tissue growth (Cho and Kende, 1997). Investigations of expansin gene expression have indicated that different expansin genes are expressed in a tissue-specific manner, and that they respond distinctively to light treatments (Cosgrove et al., 2002). In accordance with our results, analysis of transcript accumulation in tomato revealed higher levels of expansin gene expression in light-treated, slow-growing tissue than in dark-treated, rapidly elongating tissue (Caderas et al., 2000). Another cell wall-related gene with a preferential expression pattern in immature seeds was a transcript with high sequence similarity to soybean proline-rich protein (PRP). The high transcriptional activity of this gene in seeds may be needed to modify the structure of the wall in order to protect the cell during embryo development under HL stress. An increase in PRP mRNA in response to HL has also been reported (Rossel et al., 2002). Three genes encoding components involved in protein degradation were identified as being differentially expressed between leaf and seed tissues in response to HL stress – cysteine protease, ubiquitin and hexaubiquitin. This expression pattern suggests an important role of protein degradation during the plant response to stress, possibly in order to maintain appropriate levels of short-lived and regulatory proteins. Remarkably, we found two cDNA clones encoding putative 11S seed storage globulin among the highly down-regulated genes in immature seeds. Storage proteins are defined as proteins that accumulate during the grain-filling period and are used as nitrogen sources during seed germination (Shewry and Halford, 2002). The repression of these genes suggests that accumulation and/or synthesis of the major fraction of sunflower seed storage protein can be negatively affected by long-term HL exposure.
HT × tissue-specific responses
Careful examination of the putative functions of the genes that displayed HT × tissue-specific responses revealed novel and specific responses. Glyoxylate metabolism appears to play a role in sunflower under HT stress. Genes potentially encoding malate synthase and phosphoglycolate phosphatase, which are involved in glyoxylate cycle, were found to be preferentially expressed in leaves. Transcriptional activity of genes involved in the glyoxylate cycle has also been found to be affected by far-red light (Ma et al., 2001) and low-oxygen stress (Klok et al., 2002). Environmental factors can also affect the accumulation of intracellular free metal ions in plant cells and could be the reason for the specific induction of a copper chaperone in leaves under HT. In Arabidopsis, copper chaperone mRNA was found to be up-regulated in response to ozone treatment, suggesting a role in protection from oxidative stress (Himelblau et al., 1998). Another specific interaction response to HT was the induction in leaves of 3β-hydroxy steroid dehydrogenase (EC: 1·1·1·145), a key enzyme of steroid metabolism. A plant steroid has recently been reported to increase plant resistance to heat stress (Confraria, 2007).
HL + HT × tissue-specific responses
Response of sunflower to a combination of HL and HT was indicated by the up-regulation of genes involved in photochemical reactions (Table 4). Similarly, the steady-state levels of transcripts encoding potentially key enzymes of carbon fixation were also found to be up-regulated in leaves and unchanged significantly in seeds, including ribulose-bisphosphate carboxylase (EC: 4·1·1·39) and triosephosphate isomerase (EC: 5·3·1·1). The up-regulation of these two genes was accompanied by the induction of a cDNA clone encoding plasma membrane carbonic anhydrase, which facilitates CO2 acquisition (Fisher et al., 1996). In cyanobacteria and the unicellular alga Dunaliella salina, carbonic anhydrase was found to optimize CO2 uptake under salt stress (Fisher et al., 1996; Liska et al., 2004). This suggests that high assimilation of CO2 by Rubisco seems to be an adaptive response of sunflower plants to maintain high energy production under stressful conditions. The apparent accumulation of the metabolic intermediate pyruvate via alanine aminotransferase (EC: 2·6·1·2) and malate dehydrogenase in leaves appears to be an important metabolic response during the long-term adaptations to HL + HT. A stress-specific metabolic pathway, starting from pyruvate, can be suggested. Lipid degradation in response to HL + HT is reflected by an increase of transcript abundance of a putative lipase and palmitoyl-protein thioesterase (EC: 3·1·2·22). An interesting response of sunflower plants to HL + HT stress was the up-regulation in embryonic tissues of a cDNA coding for sterol C-24 reductase (EC: 1·3·1·71), a key enzyme in sterol biosynthesis. The crucial roles of sterols in embryo growth and development have been established (Schrick et al., 2002) and the functional role of C-24 reductase under stress needs to be determined further.
Signal transduction related-genes were found to be differentially expressed between leaves and seeds in sunflower plants subjected to HL + HT. This includes kinase interacting protein family member, 14-3-3 protein and two putative transcription factors, HD-ZIP and zinc finger proteins. These signal transduction components may be necessary for establishment of long-term HL + HT tolerance. Although transcription factors involved in plant responses to diverse environmental stress are well known, transcription factors controlling plant responses to a combined effect of HL and HT are still unknown. In this context, HD-ZIP and zinc finger family proteins can be considered as candidates for the transcriptional regulation of combined stress-responsive genes.
Non-specific interaction responses
Out of the 12 genes whose expression was found to differentiate significantly between seeds and leaves in response to HL and HT stress when applied individually, the putative functions of only four genes could be assigned (see Supplementary Information table S3, available online). They encode Skp1 (S-phase kinase-associated protein 1), endopeptidase inhibitor, dehydrin-cognate protein, and histone H2B1. Skp1 is an essential component of SCF ubiquitin ligases and mediates protein degradation by facilitating the ligation of ubiquitin to specific proteins (Kong et al., 2004). The expression pattern of the putative sunflower Skp1 sheds light on the possible regulatory roles of this protein in plant responses to environmental stress, probably through the selection of substrates for proteolysis.
Among the 16 genes whose expression was found to be significantly different between seeds and leaves in response to both HL and HL + HT (see Supplementary Information table S3, available online), we found DNAJ heat shock protein, 26S proteasome regulatory subunit S12 and proteasome maturation factor with enhanced expression in immature seeds. One of the hallmarks of the adaptive plant response to stress is the induction of a limited set of HSPs (Nishizawa et al., 2006). The interactive effects of light and temperature on heat-shock protein accumulation in Solidago altissima (goldenrod) have been reported both in the field and under controlled environments (Barua and Heckathorn, 2006). Additionally, elevated expression of HSPs has previously been reported in a thermotolerant sunflower hybrid (Senthil-Kumar et al., 2003). The expression in immature seeds suggests that the protective function of heat shock genes is not limited to photosynthetic tissues.
Among the 60 cDNA clones showing significant differences in expression patterns between seeds and leaves in response to HT and HL + HT (see Supplementary Information table S3, online), three transcripts were found encoding putative senescence-associated proteins. This finding suggests a role of senescence-related processes in sunflower response to these stress factors. Three genes encoding putative transcription factors belonging to MYB, RING zinc finger and B3 super families were induced in leaves and repressed in seeds by HT and HL + HT. These expression patterns suggest that these regulators have different roles in stress tolerance in the two tissues.
Only eight genes were identified as part of the interaction responses shared by all three stress treatments (see Supplementary Information table S3), and the expression profiles of these genes were similar under the three stress treatments (up-regulated in leaves and down-regulated in immature seeds). These genes represent only 2·3 % of all the interaction responses. This finding is consistent with the fact that plants switch from general stress responses to more specific responses progressively upon exposure to stress (Kreps et al., 2002). Although the putative function of these genes can not be assigned and they were annotated as ‘unknown function’, these genes could be of potential importance with regard to the elucidation of the mechanisms by which the plants develop multiple-stress tolerance.
CONCLUSIONS
Our microarray analysis confirmed the stress-responsive expression of a number of genes that previously have been reported to be stress-regulated in Arabidopsis and other plant species, such as glyceraldehyde-3-phosphate dehydrogenase, inositol-3-phosphate synthase, dehydrin, LEA protein, ferredoxin, aquaporin, cyclophilin, and DNAJ heat shock protein. We have shown that HL + HT-specific responses involve the up-regulation of genes related to energy metabolism, protein synthesis, cell wall activity and signal transduction components. Significant differences between vegetative and reproductive tissues, identified exclusively in response to HL + HT, were observed for genes involved in photochemical reactions, CO2 acquisition, pyruvate accumulation, lipid degradation, defence responses, protein folding and transcriptional control of gene expression. In addition, it was shown that a significant number of genes were found to be down-regulated in immature seed in response to HL + HT stress, indicating that down-regulation may be required in order to allow proper embryo formation and development under HL + HT stress. This is consistent with our observation that embryo development was not affected by this combined stress treatment in almost all cases. In this context, it may be important to note that there is obviously a source–sink relationship between the leaves and the developing embryos. By affecting the source leaves during the stress treatment this will invariably have effects on the embryo transcriptome, and these changes in gene expression may be due to a combination of a direct effect of the stress treatments on the embryo and indirect effects exerted from the source leaves. Additional application of large-scale transcriptional analysis of plant responses to various environmental stress combinations may help to elucidate multiple-stress sensing mechanisms and signalling.
SUPPLEMENTARY INFORMATION
Three sets of supplementary information are provided online at http://aob.oxfordjournals.org/ in the form of Excel files. Supplementary Information table S1 provides the full list of the spotted clones and their accession numbers; table S2 provides the complete list of the differentially expressed genes in response to HL, HT and HL + HT; and table S3 provides the complete list of the differentially expressed genes between leaves and immature seeds in response to HL, HT and HL + HT.
ACKNOWLEDGEMENTS
We are grateful to Professor Steve Rodermel and Dr Martijn van de Mortel (Iowa State University, USA) for their critical reading of the manuscript. We thank Annie Perrault for technical assistance, and Patrick Bermudes and Philippe Anson for care of the sunflower plants. The Génoplante program is acknowledged for authorizing the use of the cDNA clones for construction of the microarray.
LITERATURE CITED
- Barua D, Heckathorn SA. The interactive effects of light and temperature on heat-shock protein accumulation in Solidago altissima (Asteraceae) in the field and laboratory. American Journal of Botany. 2006;93:102–109. [Google Scholar]
- Beator J, Potter E, Kloppstech K. The effect of heat shock on morphogenesis in barley: coordinated circadian regulation of mRNA levels for light-regulated genes and of the capacity for accumulation of chlorophyll protein complexes. Plant Physiology. 1992;100:1780–1786. doi: 10.1104/pp.100.4.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben C, Hewezi T, Jardinaud MF, Bena F, Ladouce N, Moretti S, et al. Comparative analysis of early embryonic sunflower cDNA libraries. Plant Molecular Biology. 2005;57:255–270. doi: 10.1007/s11103-004-7532-2. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JKC, McQueen-Mason S, Kuhlemeier C. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiology. 2000;123:1399–1413. doi: 10.1104/pp.123.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheikh N, Jones RJ. Disruption of maize kernel growth and development by heat stress. Plant Physiology. 1994;106:45–51. doi: 10.1104/pp.106.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confraria A. Brassinosteroids protect plants against heat stress. Comparative Biochemistry and Physiology Part A: Physiology. 2007;146 [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant and Cell Physiology. 2002;43:1436–1444. doi: 10.1093/pcp/pcf180. [DOI] [PubMed] [Google Scholar]
- Dat JF, Lopez-Delgado H, Foyer CH, Scott IM. Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiology. 1998;116:1351–1357. doi: 10.1104/pp.116.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M, Gokhman I, Pick U, Zamir A. A salt-resistant plasma membrane carbonic anhydrase is induced by salt in Dunaliella salina. Journal of Biological Chemistry. 1996;271:17718–17723. doi: 10.1074/jbc.271.30.17718. [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Roman CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Greppin H, Strasser RJ. Functioning of photosystems I and II in pea leaves exposed to heat stress in the presence or absence of light. Planta. 1991;186:88–98. doi: 10.1007/BF00201502. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Léger M, El Kayal W, Gentzbittel L. Transcriptional profiling of sunflower plants growing under low temperatures reveals an extensive down-regulation of gene expression associated with chilling sensitivity. Journal of Experimental Botany. 2006;57:3109–3122. doi: 10.1093/jxb/erl080. [DOI] [PubMed] [Google Scholar]
- Himelblau E, Mira H, Lin SJ, Culotta VC, Peñarrubia L, Amasino RM. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiology. 1998;117:1227–1234. doi: 10.1104/pp.117.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. Journal of Computational Biology. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Kim K, Portis AR. Temperature dependence of photosynthesis in Arabidopsis plants with modifications in Rubisco activase and membrane fluidity. Plant and Cell Physiology. 2005;46:522–530. doi: 10.1093/pcp/pci052. [DOI] [PubMed] [Google Scholar]
- Kimura M, Yamamotoy Y, Seki M, Sakurai T, Sato M, Abe T, et al. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochemistry and Photobiology. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, et al. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H, Leebens-Mack J, Ni W, dePamphilis CW, Ma H. Highly heterogeneous rates of evolution in the SKP1 gene family in plants and animals: functional and evolutionary implications. Molecular Biology and Evolution. 2004;21:117–128. doi: 10.1093/molbev/msh001. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafta AM, Lorenzen JH. Effect of high temperature on plant growth and carbohydrate metabolism in potato. Plant Physiology. 1995;109:637–643. doi: 10.1104/pp.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiology. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Yang KA, Hong JK, Choi JS, Yun D-J, Hong JC, et al. Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells. Journal of Plant Research. 2006;119:373–383. doi: 10.1007/s10265-006-0285-z. [DOI] [PubMed] [Google Scholar]
- Liska AJ, Shevchenko A, Pick U, Katz A. Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella, as revealed by homology-based proteomics. Plant Physiology. 2004;136:2806–2817. doi: 10.1104/pp.104.039438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Li B, Shang ZL, Li XZ, Mu RL, Sun DY, Zhou RG. Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiology. 2003;132:1186–1195. doi: 10.1104/pp.102.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant Journal. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annual Review of Genetics. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Pressman E, Peet MM, Pharr DM. The effect of heat stress on tomato pollen characteristics is associated with changes in carbohydrate concentration in the developing anthers. Annals of Botany. 2002;90:631–636. doi: 10.1093/aob/mcf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Iobst S, Hart A, Stegalkina S, Liu J, Robin Buell CR. Gene expression profiling of potato responses to cold, heat and salt stress. Functional Integrative & Genomics. 2005;5:201–207. doi: 10.1007/s10142-005-0141-6. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiology. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology. 2004;134:1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche J, Hewezi T, Bouniols A, Gentzbittel L. Transcriptional profiles of primary metabolism and signal transduction-related genes in response to water stress in field-grown sunflower genotypes using a thematic cDNA microarray. Planta. 2007;226:601–617. doi: 10.1007/s00425-007-0508-0. [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiology. 2002;130:1109–1120. doi: 10.1104/pp.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jürgens G. Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant Journal. 2002;31:61–73. doi: 10.1046/j.1365-313x.2002.01333.x. [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Srikanthbabu V, Mohan Raju B, Ganeshkumar B, Shivaprakash N, Udayakumar M. Screening of inbred lines to develop a thermotolerant sunflower hybrid using the temperature induction response (TIR) technique: a novel approach by exploiting residual variability. Journal of Experimental Botany. 2003;54:2569–2578. doi: 10.1093/jxb/erg278. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG. Cereal seed storage proteins: structure, properties and role in grain utilization. Journal of Experimental Botany. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Tamborindeguy C, Ben C, Liboz T, Gentzbittel L. Sequence evaluation of four specific cDNA libraries for developmental genomics of sunflower. Molecular Genetics and Genomics. 2004;271:367–375. doi: 10.1007/s00438-004-0989-5. [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, et al. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant Journal. 2004;39:45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- Wallwork MAB, Jenner CF, Logue SJ, Sedgley M. Effect of high temperature during grain-filling on the structure of developing and malted barley grains. Annals of Botany. 1998;82:587–599. [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennet L, Hamade H, Bushel P, Afshira C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. Journal of Computational Biology. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]