Abstract
The mechanisms of tyrosine nitration by peroxynitrous acid or nitrosoperoxycarbonate were investigated with the CBS-QB3 method. Either the protonation of peroxynitrite, or a reaction with carbon dioxide gives a reactive peroxide intermediate. Peroxynitrous acid mediated nitration of phenol occurs via the unimolecular decomposition to give nitrogen dioxide and hydroxyl radicals. Nitrosoperoxycarbonate also undergoes unimolecular decomposition to give carbonate and nitrogen dioxide radicals. The reactions of tyrosine with the hydroxyl or carbonate radicals give a phenoxy radical intermediate. The reaction of the nitrogen dioxide with this radical intermediate followed by tautomerization gives nitrated tyrosine in both cases. According to CBS-QB3 calculations, the rate-limiting step for the nitration of phenol is the decomposition of peroxynitrous acid or of nitrosoperoxycarbonate.
INTRODUCTION
Nitric oxide (NO) is an important small molecule that is involved in various biochemical pathways1 such as neurotransmission,2 dilation of blood vessels3 and the immune response.4 Under normal conditions, NO is produced by nitric oxide synthase at low concentrations.5 However, the production of excess amounts of NO can have detrimental effects via the formation of reactive nitrogen species (RNS) such as dinitrogen trioxide (N2O3) and peroxynitrite (ONOO−).5 The formation of these RNS has been considered to be the basis for NO related cytotoxicity in biology.5 The formation of ONOO− is particularly important because it can cause both oxidative and nitrosative stress under physiological conditions.
Peroxynitrite is produced via the diffusion-limited reaction of NO with the superoxide radical;6 a reaction that has been studied extensively, the formation of peroxide is maximal when equal amounts of NO and superoxide are present.7,8 During the inflammatory processes, cells produce high levels of superoxide and NO;9 consequently, peroxynitrite formation is expected to be facile during the inflammatory processes. For example, increased levels of proteins containing 3-nitrotyrosine were identified when transplanted organs are rejected.9
Peroxynitrite, with a pKa of 6.8, is readily protonated under physiological conditions.10,11 The protonation of peroxynitrite generates peroxynitrous acid (ONOOH). Peroxynitrite can also react with CO2 to form a reactive CO2 adduct of peroxynitrite, nitrosoperoxycarbonate, ONOOCO2 −.12 We have studied the mechanism of tyrosine nitration by peroxynitous acid and nitrosoperoxycarbonate using the high-accuracy CBSQB3 method. The nitration of tyrosine by peroxynitrous acid was found to take place via a radical mechanism. The rate-determining step in this mechanism is the decomposition peroxynitrous acid to nitrogen dioxide and hydroxyl radical. The reaction of CO2 with peroxynitrite is very favorable, and nitrosoperoxycarbonate decomposes to carbonate radical and nitrogen dioxide. These radicals react with tyrosine to give 3-nitrotyrosine.
Biology of Tyrosine Nitration
Peroxynitrous acid can nitrate free or protein bound tyrosine.13 The nitration of tyrosine at the 3-position is considered to be a biomarker for the formation of peroxynitrite in vivo.5 The nitration of protein bound tyrosine may alter the structure and/or diminish the function of proteins. For example, tyrosine nitration can alter the structure of actin,14 neuroflament-L15 and tyrosine kinase.16,17 The nitration of protein bound tyrosine may also inhibit the activity of enzymes; for example the activities of cytochrome P450,18 α-thrombin,19 mitochondrial ATPase,20 tyrosine hydroxylase,21 tyrosine kinase16 and manganese and iron superoxide dismutase are known to be diminished in the presence of peroxynitrite.22 The inhibition of tyrosine hydroxylase and tyrosine kinase may cause dopamine deficiency and inhibit kinase dependent phosphorylation, respectively.16,21
Tyrosine nitration by peroxynitrite may also be dependent on the local protein environment. The presence of nearby acidic residues facilitates the nitration of certain tyrosine residues in proteins.23 In addition, the abundance of tyrosine residues in proteins does not correlate with the yields of tyrosine nitration in proteins.13 For example, β-casein, with a relatively low tyrosine content (1.9 % tyrosine), is more susceptible to tyrosine nitration by peroxynitrite than phospholipase A2, which contains 7.9 % tyrosine.13
Mechanism of Tyrosine Nitration
Two different mechanisms have been proposed for the nitration of tyrosine. They involve the formation of (i) the nitronium ion24 or (ii) the nitrogen dioxide radical.25 The formation of nitronium ion from peroxynitrite is favored over the formation of nitrogen dioxide radical in the presence of protein bound or free metal ions.24 It was proposed that the redox reaction of the metal ions with the peroxynitrite can generate the nitronium ion and metal oxide.22 The nitronium ion reacts rapidly with water to give nitric acid, but can also attack tyrosine in proteins.
In the absence of metal ions, the nitration of tyrosine is likely to take place through the formation of nitrogen dioxide. Prutz et al. demonstrated that the reaction of tyrosine with the nitrogen dioxide radical gives phenoxyl radicals in the aqueous phase; further reaction of phenoxyl radical with nitrogen dioxide gives nitrotyrosine. 25 Peroxynitrous acid may react with tyrosine (Scheme 1) or undergo peroxide bond homolysis to give hydroxyl radical and nitrogen dioxide. The reaction of the hydroxyl radical and nitrogen dioxide with tyrosine can also give nitrated tyrosine (Scheme 2). The measured pseudo first-order rates for nitration of 4-hydroxyphenylacetic acid by peroxynitrite at pH 7.5 (0.36 s−1) and the first-order rate of spontaneous decomposition of peroxynitrite under physiological conditions (0.34 s−1) are similar.24 Peroxynitrite is readily protonated under these conditions.10,11 Therefore, it is possible that both tyrosine nitration and peroxynitrite decomposition proceed via the same mechanism. On the other hand, peroxynitrite can react with CO2 to form nitrosoperoxycarbonate,12 ONOOCO2− which can react with tyrosine to give nitro-tyrosine and bicarbonate (Scheme 3) or decompose into nitrogen dioxide and carbonate radicals. The reaction of tyrosine with the nitrogen dioxide and carbonate radicals can also give the nitro-tyrosine (Scheme 4).26–31
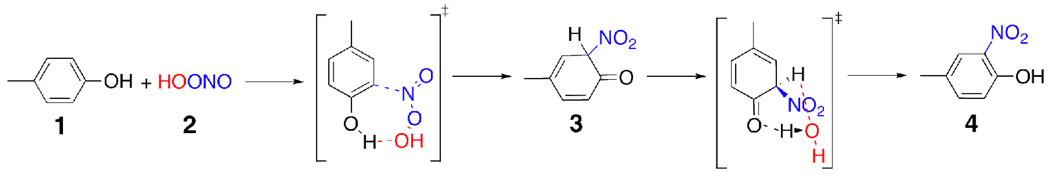
Scheme 1.
Reaction of ONOOH with p-methylphenol via a concerted pathway.
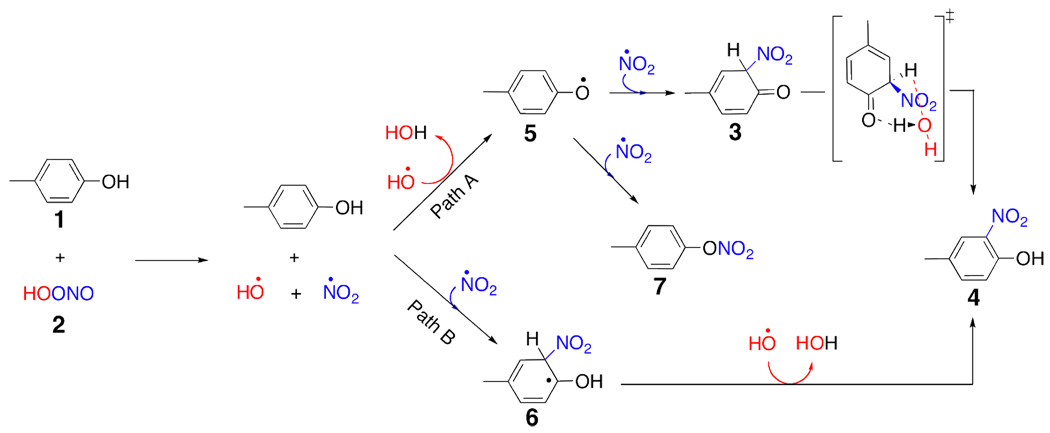
Scheme 2.
Reaction of ONOOH with p-methylphenol via radical pathways.
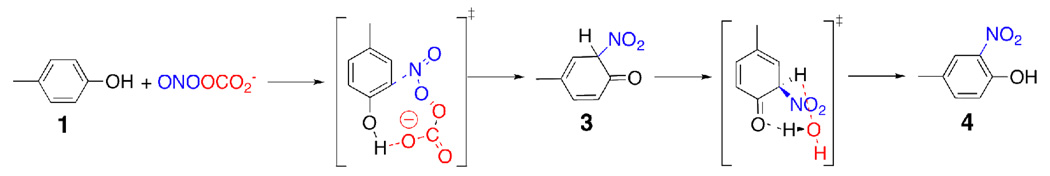
Scheme 3.
Reaction of ONOOCO2− with p-methylphenol via concerted pathway.
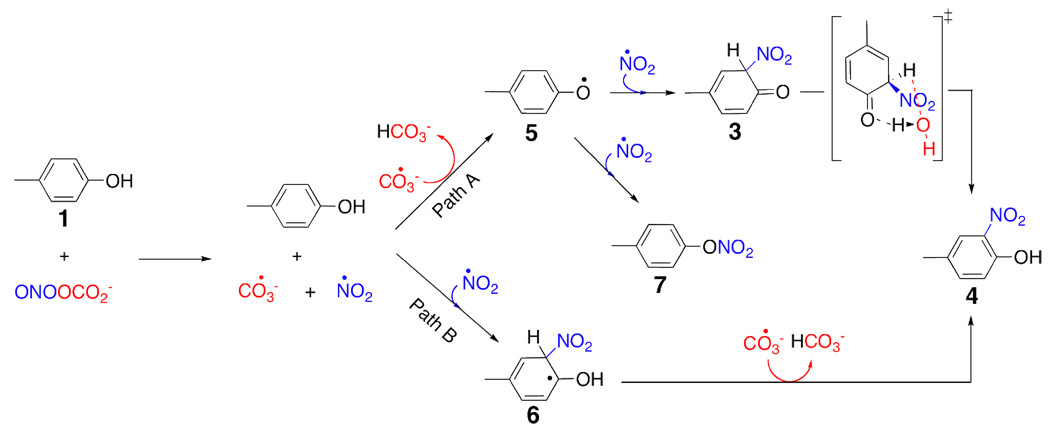
Scheme 4.
Reaction of ONOOCO2− with p-methylphenol via radical pathways.
The formation of dityrosine is also observed in the presence of free tyrosine and peroxynitrous acid, nitrogen dioxide radical or nitrosoperoxycarbonate.31 The amount of dityrosine formed exceeds that of nitrotyrosine in the presence of hydroxyl or carbonate radical scavengers or at low peroxynitrite concentrations.31 Under such conditions, nitrogen dioxide radical reacts with the tyrosine to give a phenoxy radical intermediate and HONO. The formation of nitrotyrosine requires the reaction of nitrogen dioxide with the phenoxy radical intermediate; therefore, the formation of nitrotyrosine occurs via a second-order rate constant with respect to nitrogen dioxide radical. However, the dimerization of phenoxy radicals gives the experimentally observed dityrosine. Therefore the formation of dityrosine is expected to be faster than the formation of nitrotyrosine under such conditions.31
We have investigated the mechanism and energetics of tyrosine nitration by using CBS-QB3 method.32,33 In our calculations, tyrosine was modeled with p-methylphenol (p-cresol). The reaction energetics were computed for the reaction of p-methylphenol with peroxynitrous acid and nitrosoperoxycarbonate for two competing reaction mechanisms: i) the direct reaction of ONOOH or ONOOCO2− with the p-methylphenol (Scheme 1 & Scheme 3) and ii) the reaction of p-methylphenol with the peroxynitrous acid or nitrosoperoxycarbonate decomposition products, hydroxyl or carbonate radical and nitrogen dioxide (Scheme 2 & Scheme 4). When hydroxyl or carbonate radical and nitrogen dioxide are generated, p-methylphenol can be converted to 2-nitro-4-methyl phenol (4) via two competing pathways. The initial reaction of p-methylphenol with nitrogen dioxide gives the radical intermediate (6) (Path B, Scheme 2 & Scheme 4). The reaction of hydroxyl radical with this intermediate forms the final product (4). Alternatively, the initial reaction of p-methylphenol with the hydroxyl (carbonate) radical gives the radical the intermediate (5) (Path A, Scheme 2 & Scheme 4). This radical intermediate can react with the nitrogen dioxide to give intermediate (3). In this pathway, the re-aromatization of the intermediate (3) gives the final product (4).
RESULTS AND DISCUSSION
The reaction of ONOOH with p-methylphenol
The concerted formation of intermediate (3) from p-methylphenol and peroxynitrous acid is illustrated in Scheme 1. We were unable to locate the hypothetical transition state that is shown in Scheme 1 for the direct reaction of peroxynitrous acid with p-methylphenol at the UB3LYP/6-311G(2d, d, p) level of theory. Attempts to locate such a concerted transition state led to a transition state for the decomposition of peroxynitrous to hydroxyl radical and nitrogen dioxide in the presence of one water molecule and one molecule of p-methylphenol in the gas phase.
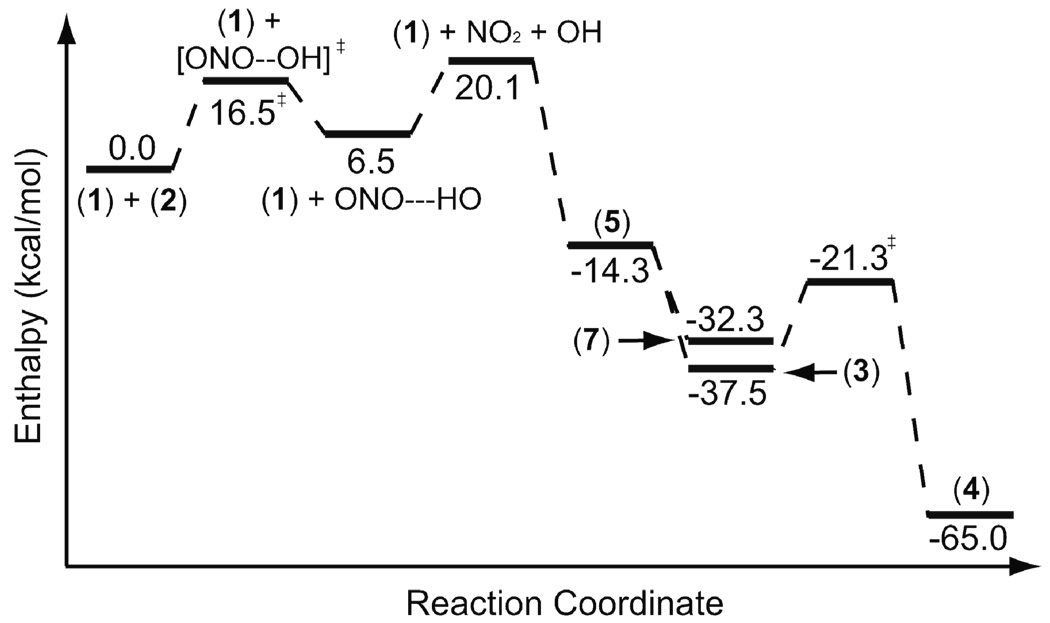
The transition state located for the decomposition of peroxynitrous acid in the presence of p-methylphenol suggests that ONOOH decomposition transition state is lower in energy that the hypothetical transition state shown in Scheme 1. The mechanism and dynamics of the decay of peroxynitrous acid to nitrate in water have recently been studied with molecular dynamics simulations.34 Peroxynitrous acid decays to nitrate via the formation of nitrogen dioxide and hydroxyl radicals.34 The fluctuations of the water molecules solvating the peroxynitrous acid and the dynamics of the nitrogen dioxide and hydroxyl radicals within the water cage determine the ratio of the formation of the solvent separated radical pair versus nitrate in water.34 Once the peroxynitrous acid decomposition takes place, the nitration of p-methylphenol can proceed via two competing radical pathways described in Scheme 2. We have computed the reaction energy profile for these two competing mechanisms separately in order to find which one of these competing pathways is favored. Relative enthalpies of the transition states and intermediates for the lowest energy pathway with respect to infinitely separated peroxynitrous acid and p-methylphenol are given in Figure 1.
Figure 1.
Energy diagram for the reaction of the p-methylphenol with peroxynitrous acid via the hydroxyl radical and nitrogen dioxide. (3) is converted to (4) via a water assisted TS.
The stepwise mechanism proposed here requires the initial decomposition of peroxynitrous acid. According to our calculations, the energy of infinitely separated hydroxyl radical and nitrogen dioxide is 20.1 kcal/mol higher than that of peroxynitrous acid in the gas phase. The separated products are higher in energy than the energy of the transition state in the gas phase. This is because hydroxyl radical and nitrogen dioxide form a complex (6.5 kcal/mol higher in energy than reactants) in the gas phase. The mechanism of peroxynitrous acid decomposition in the gas phase has been discussed in detail elsewhere.35 The experimentally measured activation energy for the aqueous phase decomposition of peroxynitrous acid is 16 kcal/mol.36 The CBS-QB3 method computed activation enthalpy (16.5 kcal/mol) is consistent with the experimental value. Once the peroxynitrous acid decomposition takes place, the hydroxyl radical and nitrogen dioxide can diffuse away from each other in water.34 The reaction of nitrogen dioxide with the pmethylphenol gives the intermediate (6). This step is endothermic, and the infinitely separated species are 30.4 kcal/mol above peroxynitrous acid and p-methylphenol in the gas phase. The reaction of intermediate (6) with the hydroxyl radical can spontaneously form the nitrated product 2-nitro-4-methyl phenol (Scheme 2). The nitration of the p-methylphenol via this mechanism requires an overall activation enthalpy of 31.6 kcal/mol.
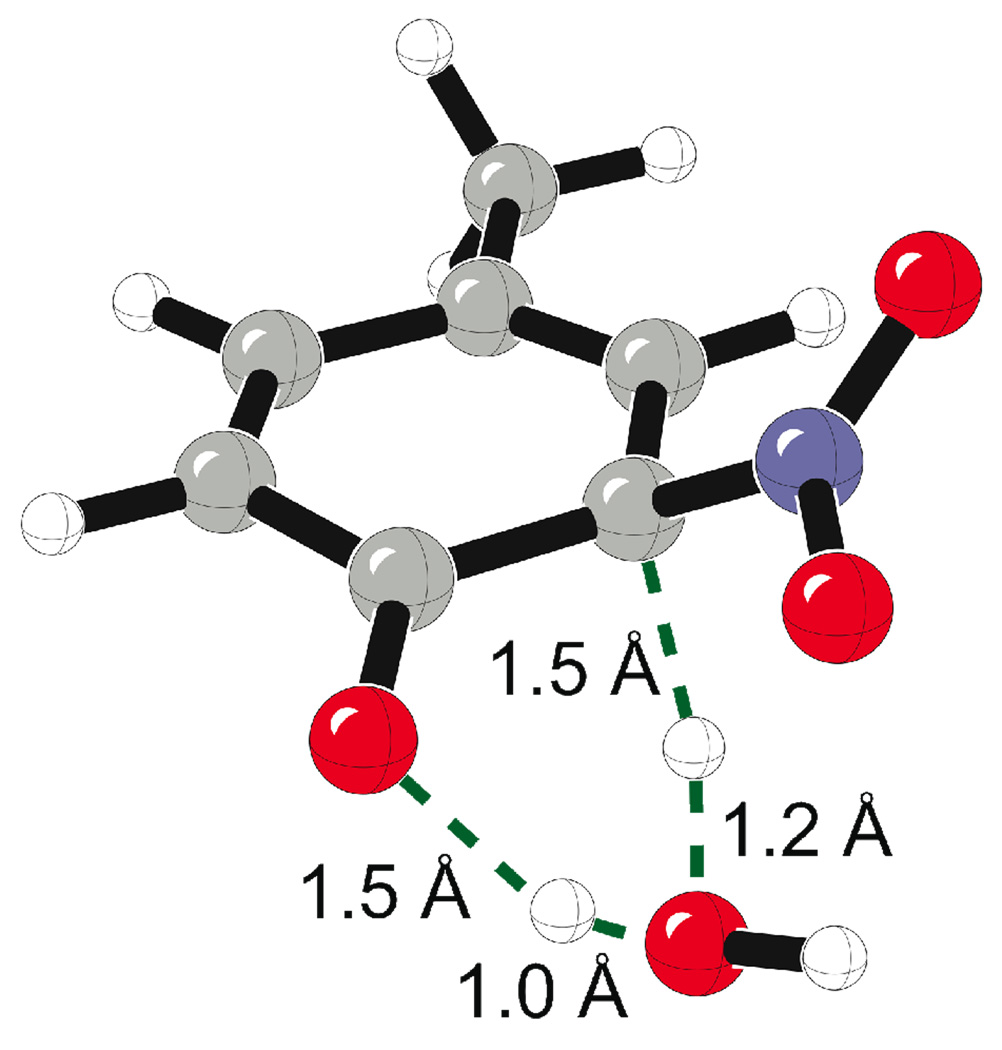
In the competing pathway, p-methylphenol reacts with the hydroxyl radical; the enthalpy profile for this pathway is given in Figure 1. This reaction gives intermediate (5). This step is barrierless in the gas phase and exothermic by 14.3 kcal/mol with respect to the infinitely separated peroxnitrous acid and p-methylphenol. Intermediate (5) reacts with the nitrogen dioxide to give p-tolyl nitrate in a barrierless process that is exothermic by 32.3 kcal/mol with respect to the infinitely separated starting materials. The reaction of the intermediate (5) with the nitrogen dioxide can also form the intermediate (3) without a barrier. Intermediate (3) is thermodynamically favored over intermediate (7) by 5.2 kcal/mol (Figure 1). The re-aromatization of the intermediate (3) gives the nitrated product (4). The activation enthalpy for this re-aromatization step is 16.2 kcal/mol in the gas phase for the water-assisted mechanism (Scheme 1 and Scheme 2). The transition state for this re-aromatization step is given in Figure 2. Traces of acid or base would presumably give a lower-energy process for this re-aromatization step.
Figure 2.
Transition state for the water assisted re-aromatization of intermediate (3).
The reaction of ONOOCO2− with p-methylphenol
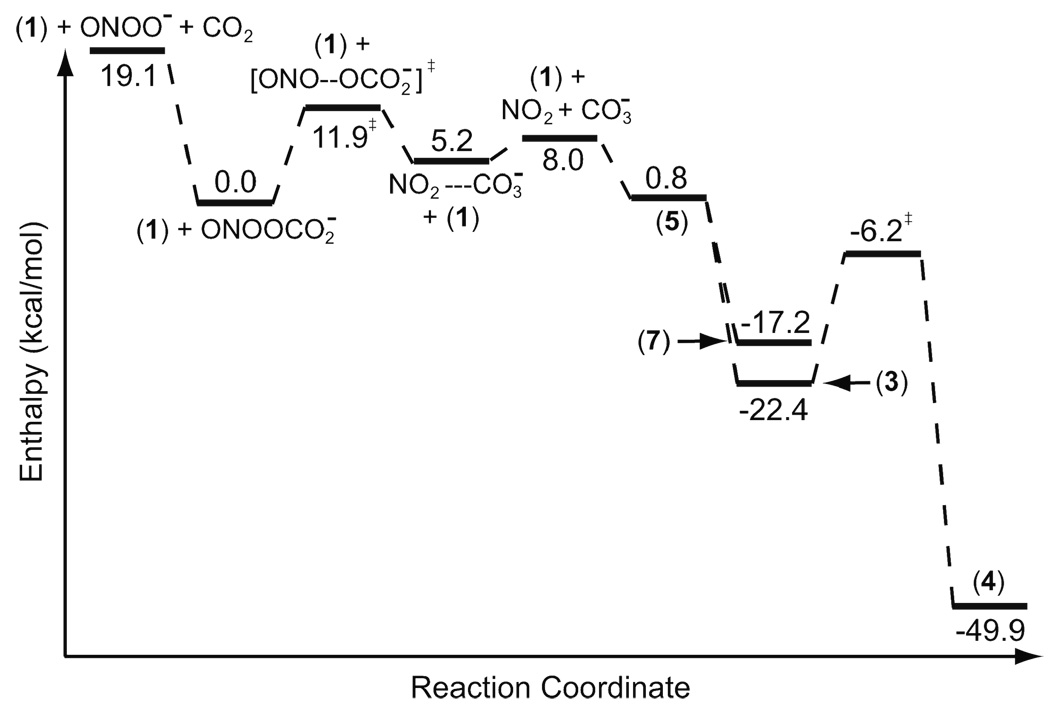
The reaction of peroxynitrite with carbon dioxide is exothermic by 19.1 kcal/mol and is expected to take place whenever peroxynitrate is exposed to carbon dioxide (Figure 3). This reaction gives nitrosoperoxycarbonate. The concerted reaction of (1) and ONOOCO2− gives intermediate (3) (Scheme 3). The activation enthalpy for this concerted transition state is 17.2 kcal/mol with respect to the substrate complex at the UB3LYP/6-311G(2d, d, p) level of theory. However, the unimolecular decomposition of nitrosoperoxycarbonate into nitrogen dioxide and carbonate radicals requires only 11.9 kcal/mol. The reaction of (1) with the nitrogen dioxide and carbonate radicals can also give (4) (Scheme 4). Just like the reaction of peroxynitrous acid with (1), the rate-limiting step for the reaction of (1) with ONOOCO2− is the unimolecular decomposition step; the activation enthalpy for this step is 11.9 kcal/mol, 4.6 kcal/mol lower in energy than the rate-limiting step in the peroxynitrous acid mediated pathway (Figure 2 & Figure 3). In this mechanism, the reaction of the carbonate radical with (1) generates intermediate (5). The reaction proceeds via the same mechanism described for the reaction of ONOOH with (1) after the generation of intermediate (5), and the energies are given in Figure (3).
Figure 3.
CBS-QB3 enthalpy diagram for the reaction of the p-methylphenol with the carbonate radical and nitrogen dioxide formed from nitrosoperoxycarbonate. (3) is converted to (4) via a water assisted TS.
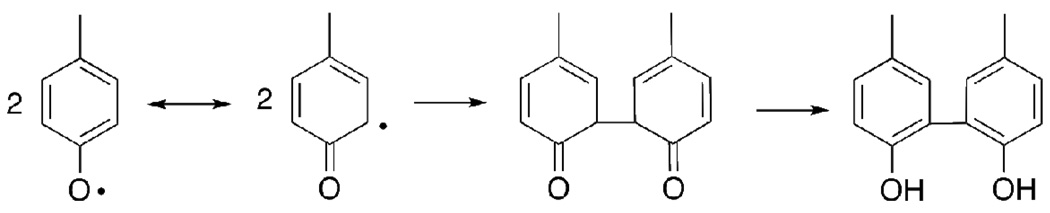
In the experiments, the formation of dityrosine has also been observed. Dityrosine can be formed from intermediate (5), which has appreciable spin density at the ortho carbon. The reaction of nitrogen dioxide with (5) at the ortho carbon is barrierless (Figure 2 & Figure 3). Radical (5) can react with itself to form the experimentally observed dityrosine. According to UB3LYP/6-311G(2d, d, p) calculations, the formation of dimer in Scheme 5 is exothermic by 50.4 kcal/mol whereas the reaction of (5) with nitrogen dioxide is exothermic by 42.5 kcal/mol.
Scheme 5.
The formation of dityrosine from ortho radicals.
CONCLUSIONS
In conclusion, according to CBS-QB3 calculations, the nitration of p-methylphenol proceeds via radical mechanism. Hydroxyl or carbonate radicals and nitrogen dioxide are generated via the unimolecular decomposition of peroxynitrous acid or nitrosoperoxycarbonate. The diffusion-limited reactions of p-methylphenol with the hydroxyl or carbonate radicals generate the radical intermediate (5). The reaction of (5) with nitrogen dioxide is diffusion limited and gives intermediate (3). The rearomatization of intermediate (3) gives the final product 2-nitro-4-methyl phenol. In this mechanism, the rate-determining step is the generation of the hydroxyl or carbonate radical plus nitrogen dioxide. According to CBS-QB3 calculations, the reaction of nitrosoperoxycarbonate with (1) is faster than the reaction of peroxynitrous acid with (1). This proposed reaction mechanism is also supported by experimental kinetic data; the rate of peroxynitrite decomposition in the absence of 4-hydroxyphenylacetic acid and the rate of 4-hydroxyphenylacetic acid nitration by peroxynitrite are similar, and the presence of carbon dioxide speeds up the nitration process.24,27 The observation of the phenoxyl by Prutz et al. also supports the stepwise radical mechanism.25
Supplementary Material
Coordinates for all CBS-QB3 optimized geometries. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENTS
We are thankful to the NIGMS-NIH (Grant GM059446) for the financial support of this research. The computations were performed on the UCLA CNSI and HOFFMAN2 computer clusters.
REFERENCES
- 1.Ignarro LJ. Nitric Oxide: Biology and Pathobiology. San Diego: Academic Press; 2000. [Google Scholar]
- 2.Bult H, Boeckxstaens GE, Pelckmans PA, Jordaens FH, Vanmaercke YM, Herman AG. Nature. 1990;345:346–347. doi: 10.1038/345346a0. [DOI] [PubMed] [Google Scholar]
- 3.Pawloski JR, Hess DT, Stamler JS. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan C. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 5.Liaudet L, Soriano FG, Szabo C. Crit Care Med. 2000;28:N169–N169. doi: 10.1097/00003246-200004001-00005. [DOI] [PubMed] [Google Scholar]
- 6.Huie RE, Padmaja S. Free Radical Res Com. 1993;18:195–199. doi: 10.3109/10715769309145868. [DOI] [PubMed] [Google Scholar]
- 7.Grisham MB, Granger DN, Lefer DJ. Free Radical Bio Med. 1998;25:404–433. doi: 10.1016/s0891-5849(98)00094-x. [DOI] [PubMed] [Google Scholar]
- 8.Miles AM, Bohle DS, Glassbrenner PA, Hansert B, Wink DA, Grisham MB. J Biol Chem. 1996;271:40–47. doi: 10.1074/jbc.271.1.40. [DOI] [PubMed] [Google Scholar]
- 9.MacMillanCrow LA, Crow JP, Kerby JD, Beckman JS, Thompson JA. P Natl Acad Sci USA. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pryor WA, Jin X, Squadrito GL. P Natl Acad Sci USA. 1994;91:11173–11177. doi: 10.1073/pnas.91.23.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kissner R, Nauser T, Bugnon P, Lye PG, Koppenol WH. Chem Res Toxicol. 1997;10:1285–1292. doi: 10.1021/tx970160x. [DOI] [PubMed] [Google Scholar]
- 12.Lymar SV, Hurst JK. Journal of the American Chemical Society. 1995;117:8867–8868. [Google Scholar]
- 13.Ischiropoulos H. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 14.Boota A, Zar H, Kim YM, Johnson B, Pitt B, Davies P. Am J Physiol-Lung C. 1996;15:L932–L938. doi: 10.1152/ajplung.1996.271.6.L932. [DOI] [PubMed] [Google Scholar]
- 15.Crow JP, Ye YZ, Strong M, Kirk M, Barnes S, Beckman JS. J Neurochem. 1997;69:1945–1953. doi: 10.1046/j.1471-4159.1997.69051945.x. [DOI] [PubMed] [Google Scholar]
- 16.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Febs Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 17.Kong SK, Yim MB, Stadtman ER, Chock PB. P Natl Acad Sci USA. 1996;93:3377–3382. doi: 10.1073/pnas.93.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janig GR, Kraft R, Blanck J, Ristau O, Rabe H, Ruckpaul K. Biochim Biophys Acta. 1987;916:512–523. doi: 10.1016/0167-4838(87)90198-1. [DOI] [PubMed] [Google Scholar]
- 19.Lundblad RL, Noyes CM, Featherstone GL, Harrison JH, Jenzano JW. J Biol Chem. 1988;263:3729–3734. [PubMed] [Google Scholar]
- 20.Guerrieri F, Yagi A, Yagi T, Papa S. J Bioenerg Biomembr. 1984;16:251–262. doi: 10.1007/BF00744279. [DOI] [PubMed] [Google Scholar]
- 21.Ara J, Przedborski S, Naini AB, Jackson-Lewis V, Trifiletti RR, Horwitz J, Ischiropoulos H. P Natl Acad Sci USA. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 23.Beckman JS, Carson M, Smith CD, Koppenol WH. Nature. 1993;364:584–584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 24.Beckman JS, Ischiropoulos H, Zhu L, Vanderwoerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Arch Biochem Biophys. 1992;298:438–445. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 25.Prutz WA, Monig H, Butler J, Land EJ. Arch Biochem Biophys. 1985;243:125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- 26.Gow A, Duran D, Thom SR, Ischiropoulos H. Arch Biochem Biophys. 1996;333:42–48. doi: 10.1006/abbi.1996.0362. [DOI] [PubMed] [Google Scholar]
- 27.Lymar SV, Jiang Q, Hurst JK. Biochemistry-Us. 1996;35:7855–7861. doi: 10.1021/bi960331h. [DOI] [PubMed] [Google Scholar]
- 28.Vandervliet A, Oneill CA, Halliwell B, Cross CE, Kaur H. Febs Lett. 1994;339:89–92. doi: 10.1016/0014-5793(94)80391-9. [DOI] [PubMed] [Google Scholar]
- 29.Denicola A, Freeman BA, Trujillo M, Radi R. Arch Biochem Biophys. 1996;333:49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 30.Lymar SV, Hurst JK. Inorg Chem. 1998;37:294–301. [Google Scholar]
- 31.Pfeiffer S, Schmidt K, Mayer B. J Biol Chem. 2000;275:6346–6352. doi: 10.1074/jbc.275.9.6346. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery JA, Frisch MJ, Ochterski JW, Petersson GA. J Chem Phys. 1999;110:2822–2827. [Google Scholar]
- 33.Montgomery JA, Frisch MJ, Ochterski JW, Petersson GA. J Chem Phys. 2000;112:6532–6542. [Google Scholar]
- 34.Gunaydin H, Houk KN. Journal of the American Chemical Society. 2008;130:10036–+. doi: 10.1021/ja711365e. [DOI] [PubMed] [Google Scholar]
- 35.Zhao YL, Houk KN, Olson LP. J Phys Chem A. 2004;108:5864–5871. [Google Scholar]
- 36.Goldstein S, Czapski G, Lind J, Merenyi G. Chem Res Toxicol. 2001;14:657–660. doi: 10.1021/tx010066n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coordinates for all CBS-QB3 optimized geometries. This material is available free of charge via the Internet at http://pubs.acs.org.