Abstract
Realistic neural networks involve the co-existence of stiff, coupled, continuous differential equations arising from the integrations of individual neurons, with the discrete events with delays used for modeling synaptic connections. We present here an integration method, the local variable time-step method (lvardt) that utilizes separate variable step integrators for individual neurons in the network. Cells which are undergoing excitation tend to have small time-steps and cells which are at rest with little synaptic input tend to have large time-steps. A synaptic input to a cell causes re-initialization of only that cell’s integrator without affecting the integration of other cells. We illustrated the use of lvardt on three models: a worst-case synchronizing mutual-inhibition model, a best-case synfire chain model, and a more realistic thalamocortical network model.
Introduction
We have previously demonstrated some advantages of the global variable time-step integrators CVODE and CVODES (Cohen, 1994) over traditional fixed step methods such as Euler, Runge-Kutta or Crank-Nicholson for simulating single cells. (Hines & Carnevale, 2001) The major advantage was due to the fact that neuron activity features spikes, requiring short time-steps, followed by interspike intervals, which allow long time-steps. The associated speed-up in the single-cell integration realm does not extend to simulation of networks however. A major reason is that the global time-step is governed by the fastest changing state-variable. In an active network, some cell is usually firing, requiring a small time-step for the entire network. Another, related reason, is that synaptic events generally cause a discontinuity in a parameter or state-variable. This requires a re-initialization as the integrator must start again with a new initial-condition problem. In a network simulation, this means re-initialization of the entire network due to a single state variable change in one cell. With re-initialization, the integrator is working without any past history. Hence the first step can only be first-order accurate and must be very short.
We demonstrate here that the poor network performance of the global variable time-step method can be overcome by giving each neuron in the system an independent variable time-step integrator. Thus, a single cell’s individual integrator uses a large dt when a neuron is quiescent or changing slowly, even though activity in other neurons in the network may cause those other integrators to proceed forward with many steps (small dt). When a cell receives a synaptic event, only that cell’s integrator has to be re-initialized.
The critical problem in the implementation of the local time-step method (lvardt) is to ensure that when an event arrives at a cell at time te that all the state-variables for the receiving cell are also at time te. This requires coordinating individual integrators so that one cell does not get so far ahead that it cannot receive a synaptic signal from another cell.
To maximally challenge lvardt, we used the mutual-inhibition model, a model which fully synchronizes. In this case, the expected superiority of multiple integrators is expected to be negated by the fact that all integrators are doing the same thing at the same time. At the other extreme we show that synfire chains enjoy a dramatic performance improvement when using lvardt. We generalize the synfire chain in a simulation of multiple delay-line rings to help understand the computational complexity of lvardt.
Additionally, we demonstrate the performance improvement obtained using lvardt on a more biologically detailed thalamocortical network.
Methods
The techniques and simulations described here are implemented in the NEURON simulator (Neuron web site). The simulator provides several global time-step integration schemes. For global fixed step methods (Hines, 1984; Hines and Carnevale, 1997) one can select either a first order backward euler integration scheme that is numerically stable for all reasonable neural models or the second order Crank-Nicholson method. There are two global variable step methods, both part of the Livermore SUNDIALS package, CVODES and IDA. (SUNDIALS web site; Hindmarsh & Serban, 2002) The current implementation of lvardt uses CVODES which solves ordinary differential equations (ODEs) of the form
| (1) |
CVODES, like many other variable step integrators, has an important property for the present usage. It allows rapid interpolation within the interval of the just-executed time-step.
Using a fully implicit fixed step method, accuracy is proportional to dt. With the variable step method, an absolute error tolerance (atol) is used to bound the error. In Neuron, absolute error tolerance is used in preference to proportionate error in order to avoid infinitesimal error tolerance near zero for state-variables, notably voltage, that approach or pass zero.
We replicated a fully-connected homogeneous inhibitory interneuron network that shows rapid synchronization through mutual inhibition. (Wang & Buzsaki, 1996) As a shorthand, we will call this the mutual-inhibition model. Variations on this model have been widely studied.(Traub et al., 1999) The basic simulation used parameters identical to those in the original paper.(Wang & Buzsaki, 1996) Porting this simulation to lvardt required minor alterations detailed in the results. We replicated the single cell model from the network, demonstrating a current-frequency response curve identical to that reported in the paper.(Wang & Buzsaki, 1996) We then used both Neuron’s fixed step method and global variable time-step methods to demonstrate comparable activity in the network simulation (precise activity is dependent on randomized initial conditions).
Both the original simulation and the lvardt version are available in runnable open-source form at the ModelDB web site (Hines et al., 2004). We also present a synfire simulation available as an example in the Neuron simulation package and a thalamocortical simulation based on published sources.(Bazhenov et al., 1998)
Simulations were run on 2.40 and 2.80 GHz Intel CPUs under Linux and Solaris operating systems.
Results
The global variable time-step method has advantages in any simulation where periods of intense simulation activity alternate with periods where state-variables remain relatively constant for a period of time. In general this situation is more likely to occur during simulation of a single cell rather than a network. The larger the network simulation, the greater the likelihood that a neuron somewhere in the network is showing spike activity. Using a global variable time-step method, this activity slows the entire simulation to tend to that one neuron’s integration needs. Neurons that are not active will be integrated with an unnecessarily short time-step.
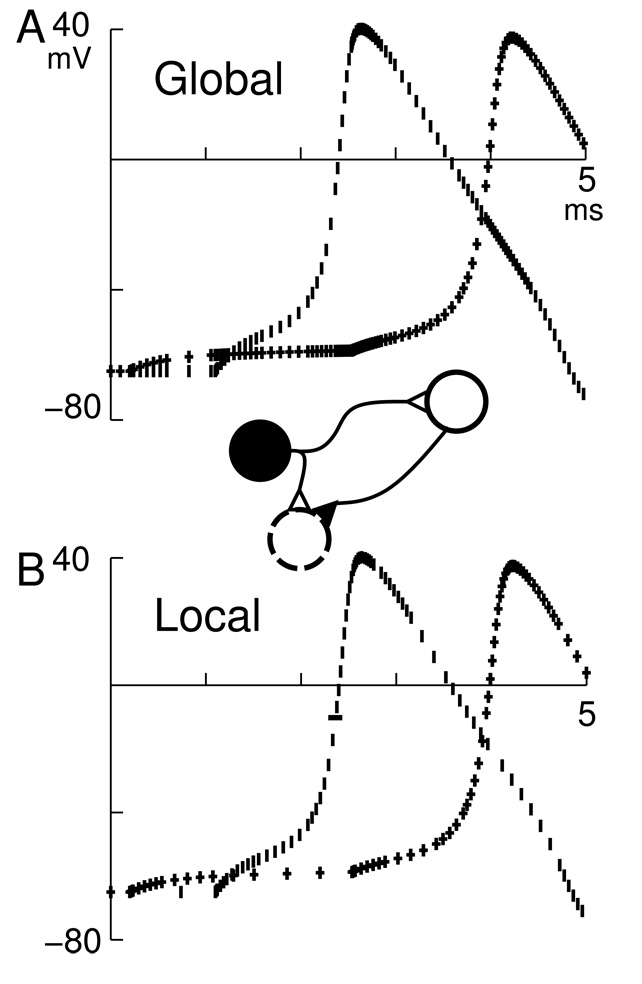
These considerations suggested the development of the local variable time-step method (lvardt) to integrate a network piece-meal, providing short time-step integration for active neurons and long time-step integration for inactive neurons, while maintaining consistent integration accuracy throughout. Neurons that fire at different times get their state-variables calculated at different times and, more importantly, different intervals (Fig. 1). Using the global method (top graph), the two cells have their trajectories calculated at the same times. With lvardt (bottom graph), integration points are independent. This is most obvious at the beginning of the simulation, where the cell that fires first (at right on schematic; vertical lines on graph) has only 2 integration points while the other cell (bottom; crosses) has 12 integration points. Where the trajectories cross, the first-spike cell integrator is called frequently while the second-spike cell integrator is using longer dt. At the peak of the first spike, both cells are being updated frequently since the second-spike cell is coincidentally approaching threshold. At the peak of the second spike, however, the first-spike cell is on the falling phase of its spike and has far fewer integration points.
Fig. 1.
Voltage trajectories for two cells are shown for the global variable step method (A) and lvardt(B). dt (interval between marks) varies together at top and separately at bottom. Network is shown in schematic: stimulator cell (filled circle) fires at time 0 and drives bottom cell weakly with a delay of 0.1 ms (+’s on graphs) and the right cell strongly with a delay of 1 ms (vertical lines on graphs). The right cell drives the bottom cell moderately with a delay of 0.1 ms. The two cells have Hodgkin-Huxley dynamics. The right cell spike threshold is −10 mV and that second-order correct threshold event is marked with a “-” on the bottom panel. The threshold event does not necessarily lie on an integration time-step boundary.
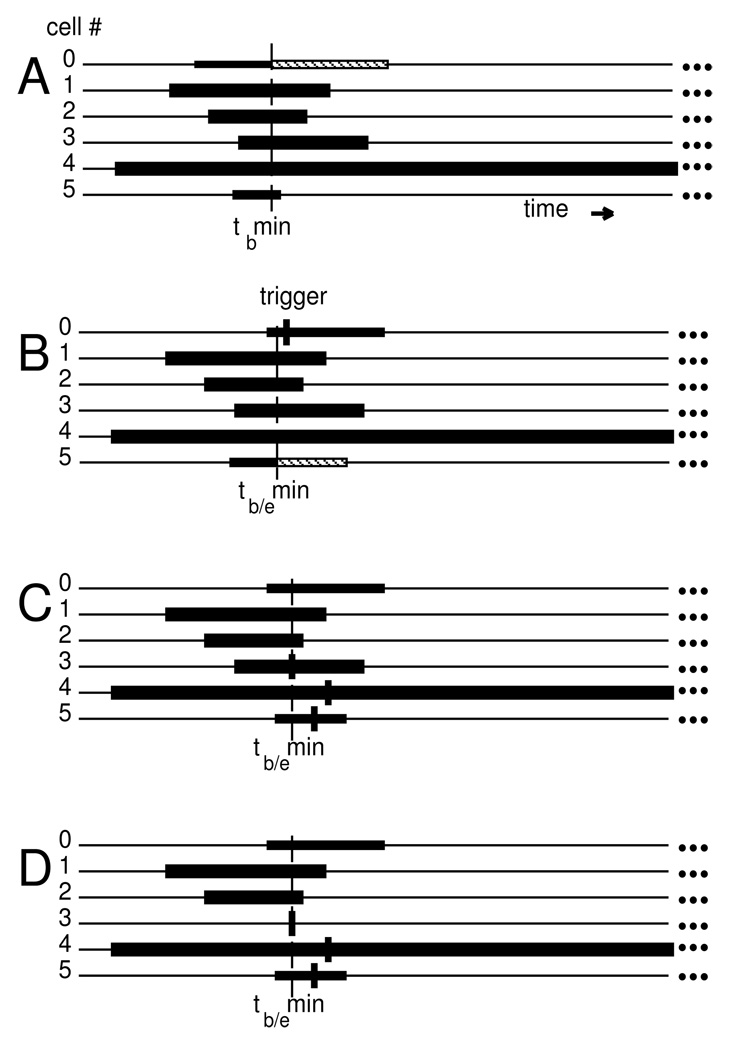
Typical sequence of local integration steps in a 6 cell example. The ta to tb intervals of possible interpolation are shown as black rectangles. The tb/emin is shown as a vertical dashed line.
A. Integration-coordinator requests integration for lagging cell (# 0 with minimum tb). Integrator advances by dt (length of hashed rectangle). In that step, we suppose cell 0 crosses threshold and a threshold event is generated — labeled “trigger” in panel B. This is an event whose time is tentative since unprocessed synaptic events could still influence this cell. B. Cell 5 has minimum tb and integrates forward. C. The trigger event is at tb/emin. The handling of this trigger creates 3 events to be delivered to cells 3,4,5 at varying delays (short vertical lines). We suppose that its delay places the cell-3 input event earlier than any tb. D. Event in cell 3 is now tb/emin. Cell 3 back-interpolates, the event is handled and cell 3 re-initializes, giving . Cell 3 will be the next cell to integrate forward.
State-variables change quickly near threshold and at the peak of the action potential. At these times, the integrators use short time-steps to accurately follow the trajectory through state space. At other times, much larger dt’s can be used to achieve the same accuracy for the more slowly varying state-variables. Using lvardt, a neuron that is inactive does not waste CPU time. Overall performance evaluation for this simple simulation demonstrates that the global method integrates its 8 state-variables (the 4 Hodgkin-Huxley variables m, h, n, V for each cell) 177 times for a total of 1416 state-variable integrations. On the other hand the integrators in the lvardt example integrate 4 state-variables 138 times (first-spike cell) and 115 times (second-spike cell) for a total of 4 · (138 + 115) = 1012 state-variable integrations. Thsi suggests the possibility of a 40% speed-up.
The lvardt method creates a separate CVODES integrator for each of Nc cells in the network. Although there are many more integrators, each integrator is more compact since it only has to handle the state-variables belonging to its particular neuron. Whether using one or Nc integrators, the total number of state-variables remains the same. Although the expected relative performance gain with lvardt by function-call statistics in Fig. 1 is 40%, there is constant overhead for each step associated with each integrator (total overhead proportional to Nc) and overhead required to determine which cell is to be integrated next, proportional to log(Nc) for each step (total overhead proportional to Nc · log(Nc)). We discuss queue overhead in the section, “Estimating simulation complexity,” but, except for very large numbers of cells, it reduces performance only slightly.
Because the system now is being calculated forward in time by multiple, independent integrators, an integration-coordinator is used to maintain the overall coherence of the integration. If the various neurons in the network are not connected, as in the case of testing parameter variation over a set of neurons, such coordination is not needed. However, in a network, the integration-coordinator is vital to permit synaptic signals to be communicated at appropriate times.
Handling events
Handling events with lvardt requires that when an event arrives at a cell all of the state-variables for that cell are at their appropriate values for that time. This is accomplished with 3 standard variable-step-integrator operations: single step integration, interpolation, and re-initialization. Using these operations, we ensure that a) incoming events are always within reach of the receiving cell’s integrator. b) individual integrators do not move too far beyond the network as a whole.
The individual integrators maintain state and derivative information on the interval of the most recent time-step. That is, each neuron’s integrator can access states over an interval between the beginning ta and the end tb of a time-step: for the ith neuron. This gives each individual integrator the ability to provide fast, high-order interpolation to a state at any time within the interpolation-range defined by the two bounding times. The integration-coordinator ensures that there is always overlap in these interpolation-ranges: . We define tb/emin as the time of the earliest event te or least-advanced integration bound tb. To guarantee interpolation-range overlap, the integration-coordinator either handles the least-time event or single-steps the least advanced integrator, whichever is earlier. In this way, no integrator’s tb, and no event, ever falls behind any ta.
Fig. 1 illustrates most of these operations in the context of a sample integration for 6 cells. Note that an input event normally requires a three step sequence of: 1. interpolation to the event time, 2. handling the event (or all the outstanding events to the cell with that delivery time), and 3. re-initializing the integrator. This full three-step sequence is only required for events that alter the course of the integration. By contrast, an event which only records the value of a state requires the interpolation step, since the recorded cell’s integration can then continue from tb rather than from the interpolation point. Similarly, threshold detection does not affect the integrator. However, it must be noted that threshold detection remains tentative until the threshold time is reached by tb/emin, because an event received by the cell in the interim may re-initialize the cell to a time prior to the tentatively calculated threshold time.
Porting the mutual-inhibition model to lvardt
The mutual-inhibition model is an all-inhibitory network with full-connectivity. Each neuron is a single compartment (point neuron) with spike-generating sodium and potassium voltage-dependent currents of the Hodgkin-Huxley type. The dynamics of the mutual-inhibition model permits initially asynchronous firing to coalesce into synchronous firing within a few spikes. In the original versions of the mutual-inhibition model,(Wang & Buzsaki, 1996) the entire network is implemented as one large continuous set of linked ODEs. This is done by making the opening-rate (kC→O in /ms) of the postsynaptic conductance a continuous and continuously differentiable Boltzmann function of presynaptic voltage:
| (2) |
This continuous-activation synapse model has the advantage of making the entire simulation somewhat more tractable analytically. However, the continuous-activation model can be criticized as being non-biophysical, since it represents postsynaptic conductance as being activated by somatic presynaptic voltage at all voltage levels.(Destexhe et al., 1994a; Destexhe et al., 1994b) At rest, this activation is generally infinitesimal and will have no effect on the simulation. A more important disadvantage of the continuous-activation synapse model is that it does not allow explicit definition of axonal and synaptic delays.
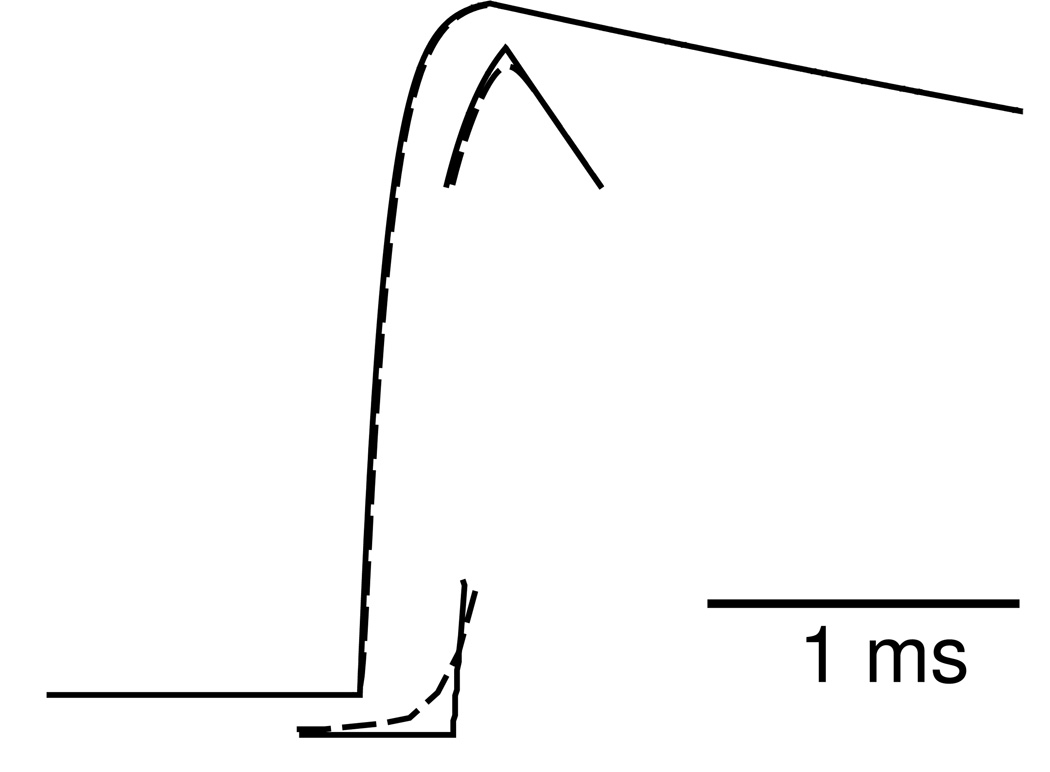
In order to implement the mutual-inhibition model using lvardt, we needed to translate the continuous-activation synapses to event-driven synapses. Equation 2 for the continuous-activation synapse gives a steeply rising sigmoid. Thus the transmitter release is significant only in the period in which the presynaptic action potential is above some threshold around 0 mV. Furthermore, since the action potential trajectory in this region is relatively insensitive to changing synaptic inputs, the transmitter release is well approximated by a threshold triggered stereotypical pulse of transmitter of duration, Cdur. This latter synapse model has been extensively used.(Destexhe et al., 1994; Lytton, 1996) Adjusting the event threshold and pulse duration parameters to least squares best fit the continuous-activation synapse conductance, Fig. 2, gives synaptic conductance trajectories so similar that simulations with the two kinds of synapses produce graphically identical results.
Fig. 2.
Comparison between event-triggered (solid) and continuously activated (dashed) synaptic conductance elicited by a presynaptic action potential. Curves superimpose except for slight deviations at initiation and peak, demonstrated by 50 fold blow-ups at these locations. Threshold = −5.7 mV, Cdur = 0.41 ms
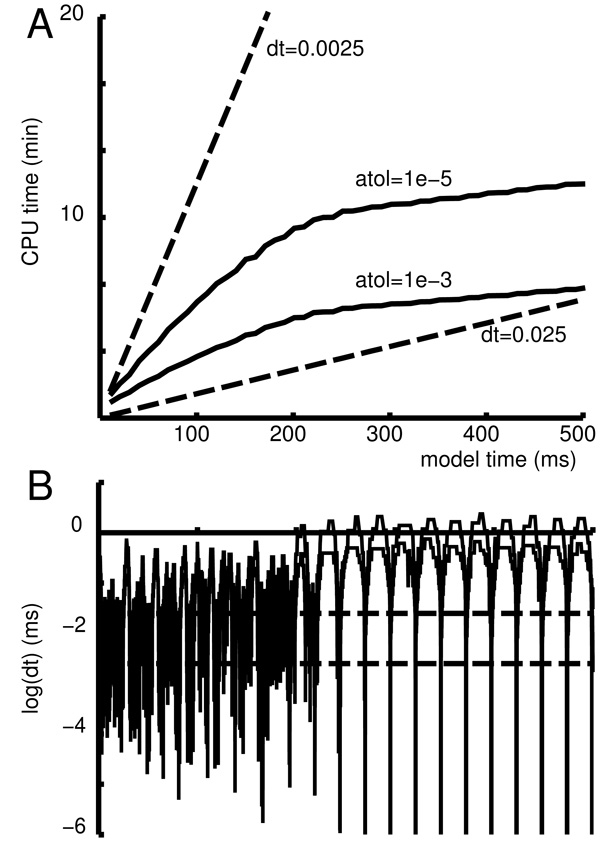
Spike time deviations between the event-driven simulation and the original continuous simulation were minimal: 108± 64 µs (mean ± standard deviation), well below the duration of an action potential. Improving the fit by, for example, adjusting the maximum kC→O was unnecessary. For the mutual-inhibition model, event-driven simulations run a bit faster than the equivalent continuous simulations. This is due to the use of a single generalized synapse for each cell that accepts all of the connecting input event streams and discontinuously changes just two state variables when an event arrives. This is orders of magnitude more efficient than the continuous model where there are about Nc synapses per cell, each with an ODE that requires an evaluation of equation 2 every time-step. Given that fixed time-step methods remain the simulation standard, we compared the fixed time-step performance with lvardt for the mutual-inhibition model (Fig. 3). We found that a fixed time-steps of 0.0025 ms gave results closely comparable to those of lvardt with absolute error tolerance (atol) of 1 · 10−3 or 1 · 10−5. In this simulation, firing of all cells is powerfully drawn into the synchronizing attractor, making the result qualitatively similar for any integration method or tolerance that produced reasonable spike trajectories for the individual cells. Fig. 3A demonstrates that the lvardt simulations are relatively slow in the early phase of the simulation where irregular firing generates high frequency input events in all cells but then become far more efficient once synchrony sets in. Fig. 3B explains this by showing the time-steps used during the simulations. The fixed dt methods are represented here by horizontal lines. The lvardt methods produces time-steps that jump around during the initial presynchrony phase of the simulation and then settle down to an alternation between large dt (>1 ms with atol=1 · 10−3) in the long intervals separating the population spikes and extremely short dt during the population spike itself. This is readily understood by noting the need to calculate not only individual cell spiking during the period of the population spike but also handle the instantaneous (zero-delay) exchange of spikes and synaptic responses to these spikes during this same brief period. Profiling of these simulations demonstrates that lvardt consumes about 12% of its total CPU time performing these event deliveries and the associated interpolations. This relatively high figure is due to the fact that spikes that are tentatively triggered in a particular cell may then need to be taken back as other inputs into that cell arrive and alter the spike time.
Fig. 3.
Comparison of fixed and variable time-step methods for the mutual-inhibition model. A. CPU time increases linearly with simulation time for fixed step method (dashed lines for dt=2.5 µs – upper curve; dt=25 µs – lower) There is a reduction in CPU load at onset of synchrony using the variable step method (solid lines with absolute error tolerance 1 · 10−5 – upper; 1 · 10−3 – lower). B. Time-step size as a function of time. Log(dt) is shown for fixed dt (horizontal dashed lines) and variable dt in one neuron (solid lines).
When using the global variable time-step method, this species of thrashing behavior sometimes resulted in severe inefficiencies. Due to the all-to-all connectivity, the near-simultaneous spiking of 100 neurons places 9900 (n2 − n since no self-connectivity) near-simultaneous events on the event queue after the occurrence of spikes in all of the cells. Using lvardt these events are generally handled in sequential order with only occasional need to recalculate threshold time (Fig. 1C). However, the global variable time-step method attempts to reconcile the mutual influence of all of these competing events using the single integrator. This led to large computer time increases (up to 60-fold) in some simulations. It is possible to provide efficient handling of such massive event influx under the global method by artificially defining a resolution interval within which events would be considered to be simultaneous. However, such an ad hoc event-handling approach would not be desirable for other types of simulations. Instead, we regard the lvardt as the natural implementation for the event-driven form of the mutual-inhibition model. We further note that the all-to-all connectivity and near-perfect synchrony of the mutual-inhibition model represents an extreme simulation situation.
Generalization of mutual-inhibition model using lvardt
The use of the event-driven simulation for mutual-inhibition model provides the desirable side effect of allowing arbitrary delays to be introduced into the simulation. We have begun exploring the effects of delays primarily in order to ensure that these would be handled readily without introducing unexpected errors or inefficiencies. We found that CPU times using lvardt decreased slightly with the introduction of 2 ms delay (from 1.43 min to 1.07 min). CPU times were also similar with introduction of inhomogeneity in the cells’ intrinsic frequencies (1.33 min), introduction of variability in the delays (1.33 min), or variability in both intrinsic frequency and delay times (1.57 min).
Introduction of brief delay shifted but did not otherwise interfere with synchronization in the homogeneous case. Introduction of a range of delays (1.9–2.1 ms; uniform distribution) also did not interfere with synchronization. Further increasing the delay range to 2–5 ms produced slightly broader population spikes. However activity still synchronized within 4–5 cycles as before. These manipulations increased lvardt integration efficiency by 10–20%.
With an inhomogeneous population of neurons having different natural firing frequencies, the population spike broadened considerably, again without interfering significantly with the number of cycles required to achieve synchrony. Here again there was only a mild reduction of integration speed, comparable to that seen with randomization of delays.
Use of lvardt with a synfire chain
The synfire chain, introduced by Abeles,(Abeles, 1991; Aviel et al., 2003) is optimal for application of the lvardt method. In this classical simulation, sets of cells are fired sequentially due to synaptic connectivity density that is greatest from each set to a follower set. At any one time, activity is restricted to a small set of cells that are carrying the signal forward, while other cells in the simulation are quiescent or are firing at lower background rates. Our evaluation of simulations consisting of 100 single compartment HH cells showed a 20-fold speed-up using lvardt as compared to fixed dt method with similar accuracies. In general, synfire simulations can be expected to show speed-ups of one to two orders of magnitude depending on the size of the chain, background firing rates and forward vs. lateral connectivity densities. Profiling of these simulations demonstrated that event handling overhead was insignificant.
Use of lvardt in thalamocortical simulation
The highly structured, stereotyped simulations described above were meant to highlight situations in which lvardt would be particularly useful (synfire chain) or would be likely to encounter problems (mutual-inhibition model). We also benchmarked lvardt with a more complex thalamocortical simulation more closely related to activity in the nervous system. This simulation features four cell types, cortical pyramidal neurons and interneurons, thalamocortical cells and thalamic reticular neurons.(Bazhenov et al., 1998) The two thalamic cell types produced bursts with prolonged interburst intervals, a situation particularly advantageous for the use of the lvardt algorithm.
To preserve accurate spike times out to 150 ms of simulation time, we had to use high accuracy simulations: an error tolerance (atol) of 1 · 10−6 for the lvardt method and a dt of 1 · 10−4 for the fixed time-step method. (The concept of accuracy is somewhat problematic when considering these complex network simulations, as will be discussed further below.) The results were striking: 10 hour 20 minute simulation time for fixed dt and 6 minutes 13 seconds for lvardt, a 100-fold speed-up. Less dramatic results were obtained when comparing to a more typical fixed dt of 1 · 10−2, comparable in this simulation to the lvardt method with error tolerance of 1 · 10−3. In this case, the simulations took 2 minutes for lvardt and 5 minutes 53 seconds for fixed dt, a 3-fold speed-up.
As in the original Bazhenov et al. model, several cell parameters were randomized to introduce variability into the model. In general, added variability would be expected to be advantageous for lvardt, increasing the likelihood that the integrators would require different time-steps at a give point in the simulation. In practice, repetitive drive in this simulation dominated dynamics so that changing the degree of cellular variability made little difference. Similarly, addition of noisy inputs had little effect in this simulation. In general, addition of strong, high-frequency noisy inputs would be expected to remove lvardt advantages, requiring interrupts and re-initialization at the input frequencies. Such an extreme case would also adversely affect performance of the global variable time-step method. Although the fixed dt method would be unaffected, it would be inaccurate: all events would be rounded off to the nearest dt, an aliasing producing false input synchrony. Addressing this by reduction to an appropriately small fixed dt would again leave the variable time-steps with an advantage.
Estimating simulation complexity
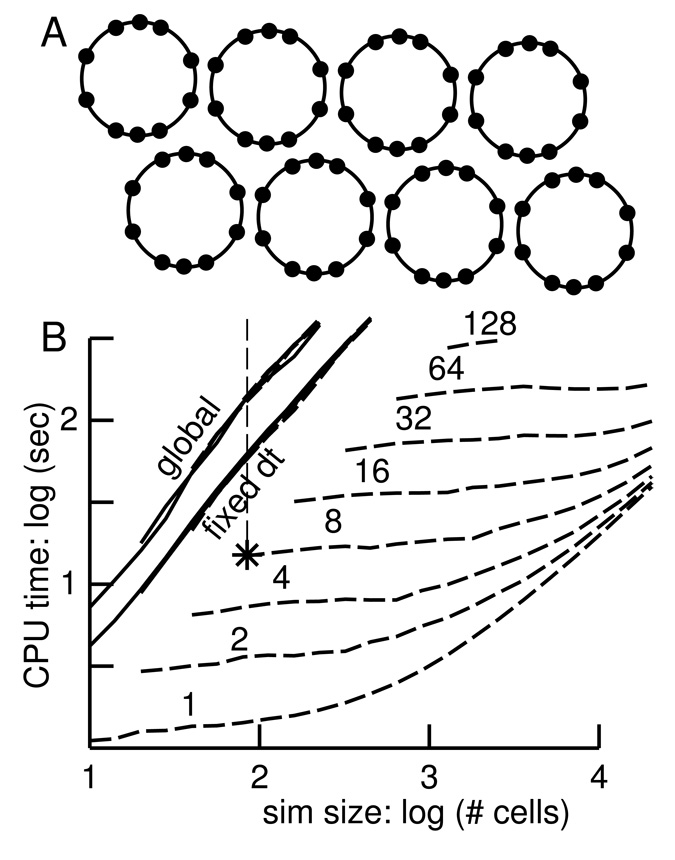
In order to perform detailed benchmarking and complexity evaluation, we needed a hybrid simulation that would allow us to scale simulation size without altering the simulation pattern. This was not readily done with the thalamocortical simulation, where scaling to greater numbers of neurons produced activity spread that depended critically on boundary conditions and parameter-scaling choices, despite preservation of qualitative activity pattern. We therefore went back to the synfire chain, reconfiguring it as a set of rings (Fig. 4A) to make it 1. scale well and 2. produce continuous activity. Each neuron in this rings simulation has 10 active compartments with Hodgkin-Huxley sodium, potassium and leak conductances, i.e., 40 states. Increasing the number of neurons in a ring increases the size of the simulation without increasing the amount of parallel activity: each ring is a delay line in which only one neuron is active at a given time. An increase in the number of rings increases the amount of activity occurring simultaneously.
Fig. 4.
A. Schematic of a rings simulation using 8 rings of ten neurons. The 10 tightly-coupled compartments of each neuron have standard Hodgkin-Huxley channels. Activity is passed around each ring independently. B. Log-log plot of simulation time vs. simulation size. Simulations range in size from 10 to 20480 neurons (total of 400 to 819200 state variables) in 1 to 128 rings. With global and fixed dt, results for different number of rings overlap. With lvardt (dashed lines) simulation time increases with increased number of rings (number above each line). Asterisk shows the simulation represented in A with with dashed vertical line indicating run time with fixed and global methods. Times are on a Pentium CPU running at 3 GHz.
As expected, simulation time for the global and fixed methods depends only on the number of cells and not on how they are divided into rings. Therefore the “global” and “fixed dt” curves for different number of rings all overlap in Fig. 4B. The relative location of these curves reflects the usual choice of time-step=0.025 ms for the fixed method and atol=1 · 10−3 for the global variable step method.
The lvardt curves indicate an enormous speed advantage when run with a small number of rings (lower set of dashed curves labeled 1, 2, 4) where only 1, 2 respectively 4 cells will be active at any given time. As we increase the number of rings, hence increasing the number of cells simultaneously active, lvardt takes more computation time to simulate a given number of cells. As the number of simultaneously active cells approaches the total number of cells, lvardt will be placed at a disadvantage compared to the other integration methods as it wastes time maintaining the integrator-queue and because of the Nc–fold increase in constant integrator overhead.
At the left side of each lvardt curve (small number of cells per ring), doubling the number of rings doubles the number of active cells and approximately doubles simulation time. As the number of cells per ring rises, simulation time increases only very slightly at first and then rises more steeply causing the curves to converge somewhat as the number of cells per ring gets very large. With these very large rings, the time spent integrating the cell that is firing is swamped by the time doing maximum time-steps in the large number of quiescent cells.
Evaluation of the lvardt integration in the ring simulation allowed us to develop an empiric weighting of simulation time which indicates the complexity of the lvardt method:
| (3) |
Total simulation time (Trun) for the lvardt method will be proportional to number of cells (Nc) and total model time (tstop). The characteristic inverse dependence on time-step (Δtsmall) must here be weighted by a θ factor indicating the proportion of neuron-time spent crawling through spikes at small dt. The proportion of is also included since inactive cells will be expected to have a characteristic time-step related to subthreshold synaptic input interval in the interspike interval. For the fixed step method, Δtbig = Δtsmall. For the global variable step method, θ is dependent on the proportion of active to inactive in the network as a whole. For all methods, simulation time will be dependent on the number of state variables s per neuron and the time Ts required to integrate a single state variable. lvardt has 2 more dependencies: general overhead time To for handling each integrator, as well as queue handling overhead for determining which integrator goes next. This latter term scales as Nc · log(Nc) rather than Nc, reflecting the scaling for a queue sorting algorithm. Profiling demonstrates that integrator-queue effect on simulation time is minimal (coefficient Tq is relatively small). For 128 rings of size 160 each, management of the integrator-queue is under 3% of the simulation time while integration is 94%. When using 1-compartment instead of 10-compartment cells (4 instead of 40 state variables) with this size network, state integration still dominates the calculation with queue time increasing to only 6% of the simulation time.
In Fig. 1 we depict times for events and integration boundaries as they would appear on a single queue. In the Neuron implementation, events are maintained on an event-queue while integrators are maintained on a separate integrator-queue. Only the latter is considered in the Tq term of Eqn. 3. We did not consider the time for the event-queue in this equation since it is the same regardless of which of the 3 integration methods is being used.
Discussion
The opposing demands of the continuous and event-triggered aspects of neural simulation suggest that the problem can be split up. Individual neurons are computationally demanding. By contrast, connectivity does not require much CPU time, although its representation may be demanding of memory (up to n2 of neuron number). Thus it is natural to separate the simulation problem into calculation of the continuous neural potentials and calculation of events and their effects. Splitting the problem thusly, it is immediately recognized that the computational demands of each neuron will differ among themselves at any given time. This suggests the use of the local time-step method.
Event driven simulation
Prior work has demonstrated a variety of methods for efficient handling of event-driven neural network simulations.(Makino, 2003; Mattia & Del Giudice, 2000; Watts, 1994) However, these networks have been restricted to use with artificial cells which permit analytic solution or approximation of cell states based on values at an arbitrary prior time. In such a network, cell states are calculated at the time of event receipt based on values determined at the prior event. In addition to external events (te), the event-queue for an artificial network may also contain self-events. For example, an artificial cell may use an event to alert itself to the end of its refractory period, permitting resetting of an internal state flag. Such self-events are typically of fixed period and can be added effortlessly into the queue.
In an artificial cell network, the simulator maintains a queue of scheduled events that are then evaluated in order. To handle an event, the simulator updates the states of follower cells and places on the queue any new events generated by these followers. Since many practical simulations involve only a small set of event delay times, the need for O(logNc) queue sorting is avoided and the event-queue can make use of efficient algorithms such as the O(1) algorithm presented by Mattia and Del Giudice (2000).
Similar to the above, a network in Neuron can be constructed entirely of event-driven artificial cells. In an artificial cell network without realistic cells, there is no need for integration. In this setting, lvardt creates no integrators and does not need an integrator-queue, only making use of the event queue. States for artificial cells are computed analytically upon the arrival of events and output events are then added to the event queue.
By contrast with the event-queue, the integrator-queue always contains Nc ordered tb events, where Nc is the number of cells. The tb times are uncorrelated with each other and with any te times. This variability in event order means that the integrator-queue requires a fully general algorithm with characteristic log(Nc) complexity per time-step. In Neuron, both the event- and integrator-queues are based on Jones’s (1986) implementation of the splay-tree algorithm of Sleator and Tarjan (1983). As we have shown, integrator-queue execution time is negligible even for simulations on the order of 10000 cells.
Contrasting example simulations
In order to demonstrate the general usefulness of the method, we explored its performance in several examples. The mutual-inhibition model is relatively unsuited to the use of any variable time-step method, proceeding to full synchronization within the round-off error for the double precision representation available. With regard to lvardt, such synchronization is a worst-case scenario from a performance perspective, since perfect synchrony of identical neurons means that all integrators are redundantly performing the same calculations at the same time. In addition, the mutual-inhibition model places increasing demands on event accounting, as the event-delivery algorithm attempts to reconcile the mutual effects of the n2 − n events arriving nearly simultaneously. Performance on the mutual-inhibition model simulation can be greatly improved by providing a window wherein arriving events are considered simultaneous and can be handled in the order received. We have explored this implementation but did not pursue it since it is an ad hoc solution to a peculiar simulation with pathologically synchronized behavior, and since the lvardt method performed adequately despite this handicap.
Moving beyond this artificially handicapped situation, it can readily be seen that the best use of lvardt will be in larger simulations involving heterogeneous neural populations where activity bounces from one area to another or spreads across the network. The synfire chain simulation represents the best-case extreme with only a small subset of neurons being active at a given time. lvardt can integrate these few, devoting no resources to the many that are done or await their moment. The ring simulation generalizes the synfire chain to allow the simulation size and the extent of simultaneous activity to be independently scaled. With a large enough number of rings, lvardt will be expected to lose its advantage as integrator switching between simultaneously active cells will dominate.
The more complex thalamocortical simulation was also assessed to demonstrate that the new method has real virtual-world application. While performance of lvardt on this simulation produced excellent results, comparison of simulations run using different integration methods with different degrees of precision raised questions of the appropriate standard of accuracy for simulations. In general, network simulations will not converge to a single, correct result since long runs will invariably yield a near-threshold event which will produce spikes at slightly different times or not at all, ultimately dependent on round-offs, an example of sensitivity to initial conditions. From this point onward, the divergence of activity in the single neuron may spread to alter firing patterns throughout the network. In the case of a synchronizing network such as the mutual-inhibition model, the strength of the synchronizing attractor will resolve such deviations. In the general case however, these deviations result in entirely different spike trains after a certain point, regardless of the degree of accuracy requested. This invariable variability requires some metric other than strict spike occurrence times to identify firing pattern similarities and the adequacy of an integration method. (Victor and Purpura, 1996)
Parallel computation
The use of parallel integrators for different neurons naturally raises the issue of porting this simulation method to a parallel computer. The NEOSIM project (NEOSIM web site) has developed a “Parallel Discrete Event” sample implementation for the delayed delivery of spike events from a source cell on one cpu to a target cell on another. This implementation coexists with Neuron’s lvardt method. High performance requires that there be a significant minimum spike delay time , between source cell i and target cell j. In this case, our strict integration assertion property is relaxed to . The Neuron portion of the NEOSIM + Neuron implementation merely accepts a request from NEOSIM to integrate a specific cell or group of cells to a specified stop time consistent with the relaxed assertion. When a Neuron cell fires at time t it notifies NEOSIM. At this time, no cell has a . Note that in our zero-delay mutual-inhibition model model, this parallel method would be useless. However, most network models have a significant minimum delay between different cells and can realize significant performance gains with the NEOSIM Parallel Discrete Event delivery technique.
Summary
In summary, lvardt offers substantial speed ups for simulations of networks of realistic ion-channel-based neurons. The advantages will be greatest in situations where some neurons are quiescent during periods when other portions of the network are active. This would be the case for simulations involving serial activation of areas, as for example in hippocampus, or involving cell types with very different firing properties, as for example in a thalamocortical or basal ganglia simulation. lvardt’s mixture of event-driven and differential equation simulation also make it ideal for implementation of hybrid networks where artificial cells with analytically-soluble states are combined with realistic neurons requiring full ODE integration.
As with any computational method, the suitability of lvardt is dependent on the exact problem to be solved. The individual user will want to benchmark a particular simulation across methods before deciding on which one to use. A general assessment of suitability can be performed by considering the factors laid out in equation 3. Our heuristic conclusion is that for medium size networks in which average synaptic input intervals to a single cell are much greater than a fixed step dt, the lvardt method will have better performance than the fixed step method.
Acknowledgements
Supported by NINDS grants NS11613 and NS32187. We thank the reviewers for many helpful suggestions
References
- Abeles M. Corticonics : Neural Circuits of the Cerebral Cortex. New York: Cambridge University Press; 1991. [Google Scholar]
- Aviel Y, Mehring C, Abeles M, Horn D. On embedding synfire chains in a balanced network. Neural Computation. 2003;15:1321–1340. doi: 10.1162/089976603321780290. [DOI] [PubMed] [Google Scholar]
- Bazhenov M, Timofeev I, Steriade M, Sejnowski T. Computational models of thalamocortical augmenting responses. Journal of Neuroscience. 1998;18:6444–6465. doi: 10.1523/JNEUROSCI.18-16-06444.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Hindmarsh A. Cvode user guide. Livermore, CA: Lawrence Livermore National Laboratory; Tech. rep. 1994
- Destexhe A, Mainen Z, Sejnowski T. An efficient method for computing synaptic conductances based on a kinetic model of receptor binding. Neural Computation. 1994a;6:14–18. [Google Scholar]
- Destexhe A, Mainen Z, Sejnowski T. Synthesis of models for excitable membranes, synaptic transmission and neuromodulation using a common kinetic formalism. J Comput Neurosci. 1994b;1:195–230. doi: 10.1007/BF00961734. [DOI] [PubMed] [Google Scholar]
- Hindmarsh A, Serban R. User documentation for cvodes, an ode solver with sensitivity analysis capabilities. Lawrence Livermore National Laboratory; Tech. rep. 2002
- Hines M, Carnevale N. Neuron: a tool for neuroscientists. The Neuroscientist. 2001;7:123–135. doi: 10.1177/107385840100700207. [DOI] [PubMed] [Google Scholar]
- Hines M, Morse T, Migliore M, Carnevale N, Shepherd G. Modeldb: a database to support computational neuroscience. J Comput Neurosci. 2004;17:73–77. doi: 10.1023/B:JCNS.0000023869.22017.2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. An empirical comparison of priority-queue and event-set implementations. Comm. ACM. 1986;4:300–311. [Google Scholar]
- Lytton W. Optimizing synaptic conductance calculation for network simulations. Neural Computation. 1996;8:501–510. doi: 10.1162/neco.1996.8.3.501. [DOI] [PubMed] [Google Scholar]
- Makino T. A discrete-event neural network simulator for general neuron models. Neural Computing & Applications. 2003;11:210–223. [Google Scholar]
- Mattia M, Del Giudice P. Efficient event-driven simulation of large networks of spiking neurons and dynamical synapses. Neural Computation. 2000;12:2305–2329. doi: 10.1162/089976600300014953. [DOI] [PubMed] [Google Scholar]
- Sleator D, Tarjan R. Self adjusting binary trees. Proc. ACM SIGACT Symposium on Theory of Computing. 1983:235–245. [Google Scholar]
- Traub R, Jefferys J, Whittington M. Fast Oscillations in Cortical Circuits. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- Victor J, Purpura K. Nature and precision of temporal coding in visual cortex: a metric-space analysis. J Neurophysiol. 1996;76:1310–1326. doi: 10.1152/jn.1996.76.2.1310. [DOI] [PubMed] [Google Scholar]
- Wang X, Buzsaki G. Gamma oscillation by synaptic inhibition in a hippocampal interneuronal network model. J Neurosci. 1996;16:6402–6413. doi: 10.1523/JNEUROSCI.16-20-06402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts L. Event-driven simulation of networks of spiking neurons. In: Cowan J, Tesauro G, Alspector J, editors. Advances in neural information processing systems. vol. 6. Morgan Kaufmann Publishers; 1994. pp. 927–934. [Google Scholar]
- ModelDB. Web site retrieved Apr 26, 2004. http://senselab.med.yale.edu/senselab/ModelDB.
- NEOSIM - Neural Open Simulation. Web site retrieved Apr 26, 2004. http://www.neosim.org.
- NEURON for empirically-based simulations of neurons and networks of neurons. Web site retrieved Apr 26, 2004. http://www.neuron.yale.edu.
- SUite of Nonlinear and DIfferential/ALgebraic equation Solvers. Web site retrieved Apr 26, 2004. http://www.llnl.gov/CASC/sundials/