Abstract
The T-cell death-associated gene 8 (TDAG8) is a pH-sensing GPCR with a reported immune-specific expression profile. Here, we demonstrate pH-induced activation of TDAG8 receptor cloned from rodent brain (rTDAG8). Cloned rTDAG8 transcript showed 88–95% homology with human and mouse transcripts of lymphoid origin. RT-PCR revealed high expression of TDAG8 in forebrain limbic regions. Extracellular acidification induced significantly elevated intracellular cyclic AMP, and phosphorylated CREB in TDAG8 expressing cells. Acidification-induced LDH release was significantly attenuated in cells expressing TDAG8, suggesting neuroprotective potential against acidosis-related cell injury. Our results open up new areas of investigation into the relevance of TDAG8 in pH homeostasis and pathological states associated with acid-base dys-regulation in the brain such as ischemia and panic disorder.
Keywords: acid-sensing, pH-sensing, cloning, T cell death-associated gene 8, TDAG8, cAMP, pCREB, forebrain limbic
Introduction
The T-cell death-associated gene 8, TDAG8 (also known as GPR65), is a G-protein coupled receptor (GPCR) belonging to the G2A/OGR1 subfamily of lysophospholipid receptors [1]. TDAG8 was first identified by differential mRNA display during mouse thymocyte apoptosis induced by T-cell receptor engagement [2]. Human and mouse homologs of TDAG8 indicated 90% similarity [3]. Pharmacological studies report acidification induced signaling by TDAG8, [4,5] identifying it as a proton-sensing GPCR.
Initial publications reported selective expression of TDAG8 mRNA in lymphoid tissues, including peripheral blood leukocytes, spleen, thymus and lymph nodes [2, 6]. TDAG8 transcription is up-regulated during glucocorticoid-induced apoptotic cell death of thymocytes [2]. Studies using TDAG8 knockout mice did not present a clear immune phenotype suggesting a permissive rather than obligatory role for TDAG8 in immune development and function [7].
Evidence on TDAG8 receptor expression and characterization in other tissues is limited. Recently, expression of TDAG8 was shown in nociceptors of dorsal root ganglion [8], supporting TDAG8 expression in neuronal tissues.
Since pH homeostasis is relevant to central nervous system (CNS) function and physiology, we investigated the expression of TDAG8 in rodent brain. Here, we report the cloning and acid-sensing capability of TDAG8 receptor predominantly expressed in rat forebrain regions. Pharmacological studies reveal that brain TDAG8 receptor acts as an acid-sensing receptor coupled to specific cell signaling pathways. Our observations open new areas of investigation studying contributions of brain TDAG8 to cerebral pH homeostasis and perhaps pathological states associated with central pH malfunction.
Materials and Methods
Animals
PCR template was generated from adult male Sprague-Dawley rats (Charles River; 250–300 g). Animals were maintained in constant temperature/humidity vivarium with standardized lighting and free access to chow and water. Procedures were approved by the institutional animal care and use committee (IACUC).
Cloning TDAG8 from brain
For cloning of brain TDAG8, total RNA was prepared from amygdala, a region with significant TDAG8 mRNA expression as identified by RT-PCR. Following cervical dislocation and brain isolationn, amydala was rapidly dissected from 5 mm thick coronal brain slices at the level of the hypothalamus. Our dissection procedure does not completely exclude potential inclusion of RNA from surrounding areas such as the piriform cortex. However, we expect a predominance of amygdalar RNA in our preparation. Total RNA was isolated by single step guanidine thiocyanate-phenol extraction using TRI-REAGENT (Molecular Research Center, Cincinnati, OH) following the manufacturer’s instructions. Concentration and purity of RNA samples were determined by spectrophotometric measurements at 260 and 280 nm. First strand cDNA was synthesized from total RNA using oligo(dT) primers (Invitrogen, Carlsbad, CA). Reverse transcription-polymerase chain reaction (RT-PCR) using primers specific to the predicted rat GPR65 mRNA sequence (NCBI Accession number XM_234367) and amygdalar cDNA template was used to obtain the full-length TDAG8 sequence. Oligonucleotides primers corresponded to bp 1–18 and 1016–1032 and included EcorRI restriction enzyme cleavage sites. PCR product of expected size was gel purified, digested with EcoR1 and subsequently cloned into vector pIRESneo3 (Clontech Lab., Mountain View, CA). Sequencing at the Cincinnati Children’s Hospital Medical Center DNA Core sequencing facility confirmed the identity of the plasmid insert.
Primers corresponding to bp 7–29 and bp 977–1000 of the TDAG8 sequence were used for TDAG8 detection in various brain regions. PCR products were separated by electrophoresis on 1.2 % agarose gel and stained with ethidium bromide. (Primer sequences and other details are available in the online supplement.)
Cell culture and Stable expression of TDAG8
CHO-K1 cells (ATCC, Manassas, VA) were cultured in Ham’s F12 medium (Kaighn’s modification) containing 10% bovine serum albumin (Invitrogen Corp.). Cells were transfected with either the pIRES-neo3-rTDAG8 vector or the empty vector using Lipofectamine 2000 reagent (Invitrogen Corp.). After 24 hours, transient expression and functional activation of TDAG8 was confirmed by RT-PCR and cAMP assay, respectively (data not shown). Stably transformed clonal populations were selected by successive passages in G418 (0.4 mg/ml) and tested for functional activity. The T6 line exhibited optimal cAMP response to pH, therefore all pharmacological data were generated using these cells unless indicated otherwise. In initial experiments untransfected CHO cells and vector-transfected cells were used as controls. No significant differences in pH-induced responses were observed between untransfected and vector-transfected cells. Therefore, all data presented here show vector-transfected cells as controls.
Accumulation of intracellular cyclic AMP
T6 cells or vector-transfected cells were plated in twelve well plates at 0.2×106 cells/well. 24 hours before experimentation the media was changed to fresh serum-free F12-Ham’s containing 0.1% bovine serum albumin to reduce serum-induced basal activation of cells. For the experiment, cell monolayers were rinsed once with medium and pre-incubated at 37 °C for 10 min in assay buffer (pH 7.6). After pre-incubation, stimulation was initiated by replacing the neutral buffer with assay buffer of various pHs for 30 min. To cover a wide pH range, the assay medium was buffered with HEPES/EPPS/MES (HEM; 8mM each). Following incubation, cells were washed twice with assay buffer and exposed to 1 ml of lysis buffer (Amersham cAMP kit). Intracellular cAMP concentration was measured by an ELISA kit (Amersham Pharmacia Biotech). (see online supplement).
Measurement of intracellular pCREB
The PathDetect CREB Signal Transduction pathway trans-reporting systems (Stratagene) was used as described [9]. CHO cells were co-transfected with pCREB and luciferase reporter plasmids and either TDAG8 or vector plasmids. 24 hours following transfection, cells were incubated with physiological or acidic medium for 30 min, washed and incubated for another 4 hr. Luciferase assays were performed with 50 μL of cell extract and 100 μL luciferase substrate (Promega) using a Tropix TR717 Microplate Luminometer, Applied Biosystems. (see online supplement)
Measurement of Lactate Dehydrogenase (LDH) release
Acidosis induced cell injury was assessed quantitatively from LDH release using a cytotoxicity detection kit (Roche Diagnostics, Indianapolis, IN). Cells were exposed to media of varying pH for one hour, followed by regular growth medium for another 16 hr. Cells were then incubated with kit reagents for 15 min. Absorbance measurements were read at 490 nm using a 96 well microplate reader (SoftmaxPro ELISA plate reader, Molecular Devices Corp.). LDH units per sample were calculated and expressed as a ratio of LDH released/LDH total (LDH released + LDH extracted post lysis).
Statistical Analysis
Data were analyzed by a two-way analysis of variance (ANOVA) using transfection and pH as variables and the Bonferroni’s test for post hoc analysis (GraphPad Prism, GraphPad Inc. San Diego, CA)
Results
Cloning and expression of TDAG8 receptor in rat brain
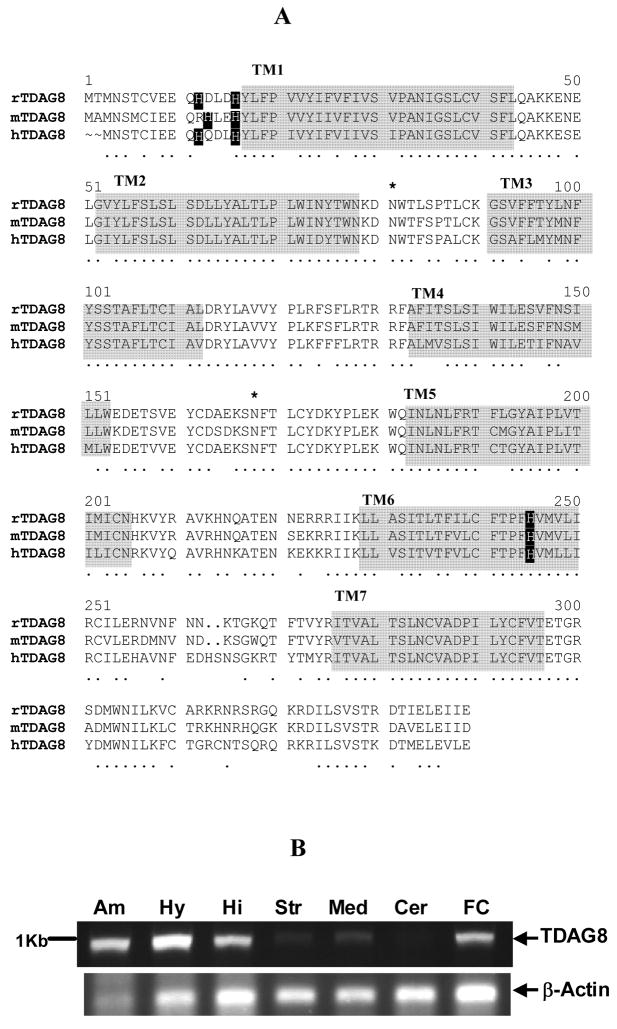
A full-length rTDAG8 cDNA sequence was amplified from rat brain (GenBank accession number EF405799). Phylogenetic analysis revealed that rTDAG8 is closely related to the G2A/OGR1 family of pH-sensing G-protein coupled receptors, and a member of the TDAG8 sub-family. The deduced amino acid sequence revealed a 337 amino acid protein (Fig IA) homologous to mouse and human TDAG8 sequences (approximately 95% and 88% identity, respectively). Primary sequence of rTDAG8 contains potential N-linked glycosylation sites (residues 4, 81 and 168) similar to mouse and human TDAG8 receptors. Importantly, rTDAG8 sequence contains residues specific to the G2A receptor family. Histidine residues (H12 and H16) assumed to be critical for proton sensing exist in the N terminal region of rTDAG8 in alignment with mouse and human TDAG8 (Fig IA). Additionally, a histidine residue in TM6 conserved in all proton-sensing G2A family GPCRs is also present in the rTDAG8 sequence.
Figure I.
(A) Sequence alignment of rTDAG8 cloned from brain with mouse (mTDAG8) and human (hTDAG8) sequences previously cloned from lymphocytes. Alignments were created using multiple sequence alignment software using Clustal W algorithm. Conserved residues between sequences are denoted by a (·). Residues within transmembrane regions (TM1-TM7) are shaded. Potential N-glycosylation sites are marked (*). Black boxes show conserved histidine residues important for proton sensing.
(B) Distribution of rTDAG8 mRNA expressed in rat brain: Amygdala (Am), Hypothalamus (Hy), Hippocampus (Hi), striatum (Str), Medulla (Med), cerebellum (Cer) and frontal cortex (FC).
rTDAG8 mRNA is expressed in forebrain regions
RT-PCR revealed a single amplification product of the predicted size of TDAG8 ~ 1Kb (Fig IB). TDAG8 is expressed in multiple brain regions including hypothalamus, amygdala, hippocampus, frontal cortex, striatum and medulla (Fig IB). Amplification of rTDAG8 transcript from DNAse treated samples confirmed that amplicons were generated from RNA and not from contaminating genomic DNA. High expression was apparent in forebrain and diencephalic limbic regions. Negligible expression of rTDAG8 mRNA was observed in cerebellum, pons, spinal cord and midbrain (data not shown).
Extracellular acidification increases intracellular cyclic AMP and phosphoCREB concentrations in TDAG8-expressing cells
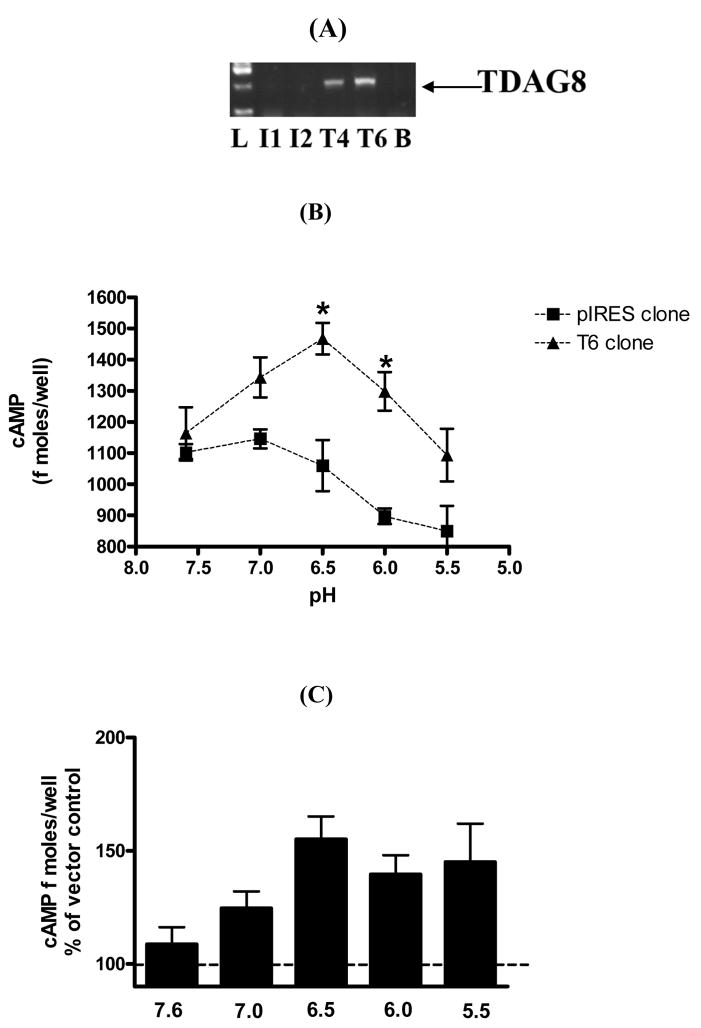
We generated CHO cell clones stably expressing TDAG8 (T4 and T6, Fig II A). The T6 clone elicited higher acid-induced activation of cAMP despite similar rTDAG8 expression between clones (data not shown). No expression of rTDAG8 was observed in CHO cells expressing empty pIRES vector (I1 and I2, Fig II A); these cells served as control. Significant accumulation of intracellular cAMP was observed in T6 cells on exposure to pH 6.5 and 6.0 (Fig II B) [pH, F(4,19) = 7.045; p<0.0012]. A significant effect of transfection was observed since vector transfected cells did not elicit elevated cAMP levels at acidic pH [transfection, F(1,19) = 41.28; p<0.0001]. Post hoc analysis revealed significant effects at pH 6.5 and 6.0, but not at other pH values, indicating that TDAG8 activation may occur across a selective range of acidification. Collective data from independent experiments confirmed optimal activation at pH 6.5 (Fig II C).
Figure II.
Extracellular acidification increases intracellular cyclic AMP concentration in rTDAG8 expressing Chinese Hamster Ovary cells. (A) Generation of stable clones T4 and T6 (expressing TDAG8) and I1 and I2 (empty vector). All data was generated with the T6 line since functional response was greater in these cells. (B) pH dependence of intracellular cAMP induction. TDAG8-transfected T6 cells ( ▴ ) and vector transfected CHO-K cells ( ▪ ) were incubated in assay buffer adjusted to pH 5.5–7.6, supplemented with 5 mM theophylline (see online supplement). After 30 minutes cells were solubilized and cAMP concentrations measured in lysates. Plot of cAMP concentration versus pH values are shown. Data are representative of three independent experiments with similar results. (C) Optimal activation of TDAG8 at pH 6.5. Data represent mean of three independent experiments with each sample run in triplicate.
* p<0.05 versus vector transfected cells.
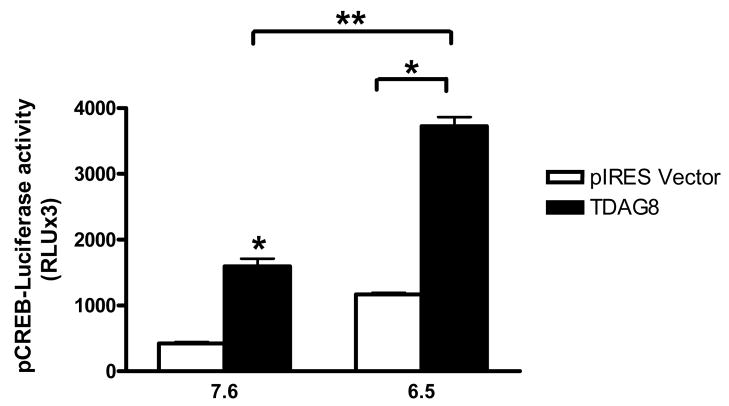
Downstream effectors associated with acid-evoked signaling have not been investigated. Exposure of TDAG8-expressing CHO cells to pH 6.5 resulted in a significant increase in pCREB over basal levels (Fig III). Two-way ANOVA revealed significant effects [transfection F(1,4)=904.82; p<0.0001] and [pH F(1,4)=151.96; p<0.0002]. In vector-transfected CHO cells pCREB expression did not rise significantly above basal levels upon acidification. Baseline pCREB expression was higher in TDAG8 cells as compared to controls, possibly due to the constitutive activity of TDAG8 receptor.
Figure III.
Elevation in intracellular phosphorylated cAMP response element binding protein (pCREB) concentration in rTDAG8 and vector expressing CHO cells in response to neutral (7.6) and acidic (6.5) pH. pCREB was assessed using the PathDetect CREB Signal Transduction pathway trans-reporting system. Relative luminescence units were measured to assess pCREB-luciferase activity. Data are representative of three experiments with each sample run in triplicate. * p<0.05 versus vector transfected cells; ** p<0.05 versus pH 7.6
Attenuation of acidosis-induced toxicity in cells expressing TDAG8
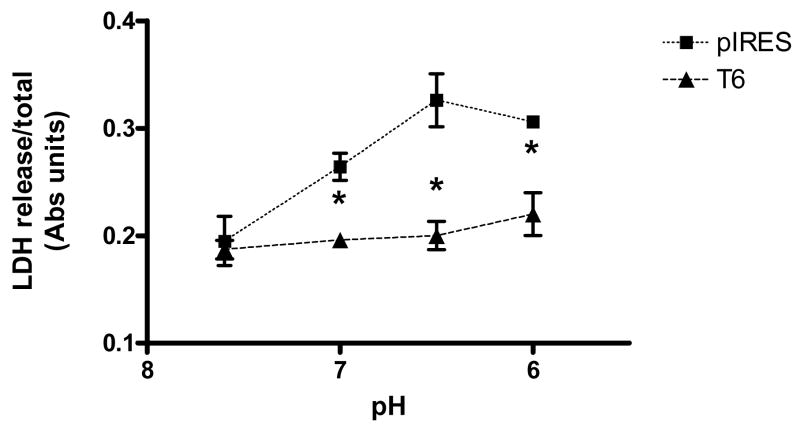
Prolonged extracellular acidification may contribute to cellular toxicity. Vector and TDAG8-expressing cells were exposed to acidified medium and lactate dehydrogenase (LDH) release was measured to assess cell damage. Relative to controls, T6 cells released significantly less LDH following exposure to acidic pH (Fig IV). Two-way ANOVA revealed significant effects [transfection F(1, 20)=34.31; p<0.0001] and [pH F(4, 20)=120.0; p<0.0001]. Thus, TDAG8 mediated resistance to pH- induced toxicity at acidic pH is consistent with cAMP response and pCREB activation at this pH range.
Figure IV.
Acidosis induced LDH release from rTDAG8 and pIRES expressing cell lines. For effect of pH acidification on LDH release, cells were exposed to specified pH solutions for 1 hr and LDH release was assessed 16 hr later. Data show means from a representative experiment using triplicate wells and repeated thrice. p<0.05 versus pIRES vector expressing cells.
Discussion
To our knowledge this is the first report of expression and signaling by acid-sensing receptor TDAG8 in the brain, a tissue where fluctuations in pH may have significant implications. Our observations are supported by recent work demonstrating expression of TDAG8 in the peripheral and central nervous system using similar techniques [8]. It is likely that the RT-PCR protocol is more sensitive than northern blot analysis used in earlier studies atleast for TDAG8 detection. The presence of rTDAG8 in brain is not surprising. Expression of Ovarian G protein coupled receptor-1 (OGR1), another member of the G2A group, has been detected in the CNS [10], although acidification induced functional responses have not been reported to date.
Rat brain TDAG8: a member of the G2A family of proton-sensing receptors
High homology (88–95%) between rTDAG8 and previously cloned sequences from mouse and human lymphoid tissue [2,3] suggests that rTDAG8 belongs to this subfamily within the G2A family. Phylogenetic analysis further confirmed this classification. Structural evidence strongly supports the potential of rTDAG8 as a pH-sensing receptor. Functionally relevant conserved histidine residues (H10, H14 or H243) are present at analogous positions in the rTDAG8 sequence as in human TDAG8 sequence. These residues have been shown to be relevant for proton-dependent cAMP accumulation [5].
rTDAG8 is widely expressed in forebrain limbic regions
Profiling of rTDAG8 distribution revealed expression in multiple brain regions. Our results suggest a predominance of TDAG8 in forebrain limbic regions such as hypothalamus, amygdala, hippocampus, frontal cortex and striatum. Expression was also detected in the medulla, a region rich in chemosensory and acid-sensing mechanisms [12, 13]. Parallels can be drawn between rTDAG8 and central expression of the acid sensing ion channel ASIC1 [14]. The ASIC1 also shows predominant expression in the hippocampus and amygdala, suggesting that proton sensing may be important in physiological processes and behaviors controlled by the limbic system.
Transduction of acid-induced signaling by central TDAG8
To date, well-characterized acid-sensing mechanisms in the brain have been ion-channels or transporters [15, 16, 17, 18]. Our results provide the first evidence of pH-induced signaling by a GPCR cloned from rat brain. Intracellular cAMP concentration increased significantly following acid-induced activation of the rTDAG8 receptor. This agrees with TDAG8-mediated signaling in the periphery [4, 5, 19]. Therefore, TDAG8 may be a central chemoreceptor that can detect interstitial fluid pH changes. The G2A family of proton-sensing GPCRs, including TDAG8, are fully activated at pH 6.4–6.8 in vitro, a physiologically attainable range. Extracellular acidosis also resulted in accumulation of downstream effector pCREB in TDAG8-expressing cells suggesting activation of the rTDAG8 receptor may couple to the cAMP-PKA-pCREB signaling pathway. Interestingly, acidosis mediated activation of the cAMP-pCREB pathway is reported in skeletal tissue via a proton sensing GPCR, suspected to be TDAG8 [20].
Our pharmacological data suggests that the TDAG8 receptor is capable of transducing extracellular pH changes into cellular responses at the transcriptional level involving factors like phosphoCREB.
TDAG8 promotes cell survival
Previous literature supports a protective role of cAMP and pCREB in cell survival [21, 22, 23]. Therefore, we questioned whether TDAG8 activation may lead to improved cell survival. Significantly reduced LDH release by TDAG8-expressing cells following acidosis suggests a mechanism whereby rTDAG8 activation may engage neuroprotective signaling cascades and promote cell survival. In contrast, pro-apoptotic responses have been reported for the TDAG8 receptor in the immune system [24]. Although downstream effector coupling has not been established for the peripheral receptor, it is possible that these pathways differ between central and peripheral TDAG8 receptors.
Functional implications of rTDAG8 activity and relevance of a pH-sensing GPCR in brain
Currently, our knowledge of proton-sensing GPCRs, particularly TDAG8, is limited. Studies with TDAG8 knockout mice reported normal immune development and function [7] suggesting this receptor may not be critical for immunologic homeostasis. One can speculate the functional relevance of centrally expressed TDAG8; extracellular pH in the brain is a fundamental signal for regulating homeostatic arousal such as breathing and behavior. In the intact brain in vivo, the interstitial pH is generally ~7.1 to 7.25, but can fluctuate to 6.5, for example in response to hypoventilation [25]. Moreover, electrical stimulation can significantly lower extracellular pH [26, 27]. Synaptic pH can fall substantially during neurotransmitter release, as the intravesicular pH is ~5.2–5.7 [28]. These pH values are well within the range for rTDAG8 stimulation and signaling. TDAG8 may therefore contribute to synaptic physiology and homeostatic functions by responding to localized pH reductions in the synapse. Moreover, rTDAG8 activation by pH fluctuations has the capability to impact synaptic plasticity via pCREB signaling [23].
The profile of rTDAG8 expression in the brain suggests specialized physiological function(s) for this receptor. Most noteworthy is the preferential expression of rTDAG8 receptor in the forebrain limbic areas, particularly hypothalamus, amygdala, hippocampus, and cortex. Collectively, these regions are tied together in the control of stress homeostasis and integration. pH-sensing in the amygdala may be relevant to the physiology of panic disorder, as this condition involves a disturbance in acid-base homeostasis [29, 30]. Recent studies on the regulation of fear conditioning behaviors by acid-sensing ion channels suggest that acid-sensing mechanisms may be relevant in stress regulation and emotional behaviors [14, 32]. TDAG8-mediated effects may also be relevant in neuroprotection/injury, especially in pathologic states like cerebral ischemia where acid-base balance in brain is severely compromised. These speculations need to be tested in follow up studies. Studies with TDAG8 knockout mice may provide relevant information on function(s) associated with brain TDAG8 receptor.
The role of pH-sensing in the brain has gained significant interest in recent years. The field of proton-sensing GPCRs is relatively new. In contrast to ionotropic changes induced by H+-linked ion channels [32,33], a proton-sensing GPCR could propel the metabotropic activities of a variety of signaling molecules upon stimulation by extracellular acidosis. This opens up a new area of investigation with several pharmacotherapeutic possibilities.
Here we provide the first evidence on the functional activation of a pH-sensing GPCR, TDAG8 cloned from rat brain. Presence of a pH-sensing GPCR in the brain could have important and exciting implications for acidosis-related physiology and pathology.
Supplementary Material
Acknowledgments
This work was supported by NIH grants 1R21MH083213-01 (RS) and U01 HD37249 (FRS). Technical assistance of Ms. Somia Faroqui is greatly appreciated.
Footnotes
Disclosures
The authors report no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Im DS. Discovery of new G protein-coupled receptors for lipid mediators. J Lipid Res. 2004;45:410–418. doi: 10.1194/jlr.R300006-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Choi JW, Lee SY, Choi Y. Identification of a putative G protein-coupled receptor induced during activation-induced apoptosis of T cells. Cell Immunol. 1996;168:78–84. doi: 10.1006/cimm.1996.0051. [DOI] [PubMed] [Google Scholar]
- 3.Kyaw H, Zeng Z, Su K, Fan P, Shell BK, Carter KC, Li Y. Cloning, characterization and mapping of human homolog of mouse T-cell death-associated gene. DNA Cell Biol. 1998;17:493–500. doi: 10.1089/dna.1998.17.493. [DOI] [PubMed] [Google Scholar]
- 4.Ishii S, Kihara Y, Shimizu T. Identification of T Cell Death-associated Gene 8 (TDAG8) as a novel acid sensing G-protein-coupled receptor. J Biol Chem. 2005;280:9083–9087. doi: 10.1074/jbc.M407832200. [DOI] [PubMed] [Google Scholar]
- 5.Wang JQ, Kon J, Mogi C, Tobo M, Damirin A, Sato K, Komachi M, Malchinkhu E, Murata N, Kimura T, Kuwabara A, Wakamatsu K, Koizumi H, Uede T, Tsujimoto G, Kurose H, Sato T, Harada A, Misawa N, Tomura H. TDAG8 is a proton-sensing and psychosine-sensitive G-protein-coupled receptor. J Biol Chem. 2004;279:45626–45633. doi: 10.1074/jbc.M406966200. [DOI] [PubMed] [Google Scholar]
- 6.Im DS, Heise CE, Nguyen T, O'Dowd BF, Lynch KR. Identification of a molecular target of psychosine and its role in globoid cell-formation. J Cell Biol. 2001;153:429–434. doi: 10.1083/jcb.153.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radu CG, Cheng D, Nijagal A, Riedinger M, McLaughlin J, Yang LV, Johnson J, Witte ON. Normal immune development and glucocorticoid-induced thymocyte apoptosis in mice deficient for the T-cell death-associated gene 8 receptor. Mol Cell Biol. 2006;26:668–677. doi: 10.1128/MCB.26.2.668-677.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CW, Tzeng JN, Chen YJ, Tsai WF, Chen CC, Sun W-H. Nociceptors of dorsal root ganglion express proton-sensing G- protein coupled receptors. Mol Cell Neurosci. 2007;36:195–210. doi: 10.1016/j.mcn.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Sheriff S, Hauger RL, Mulchahey JJ, Pisarska M, Chance WT, Balasubramaniam A, Kasckow JW. Interaction of neuropeptide Y and corticotropin-releasing factor signaling pathways in AR-5 amygdalar cells. Peptides. 2001;22:2083–1089. doi: 10.1016/s0196-9781(01)00549-6. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Casey G. Identification of human OGR1, a novel G protein-coupled receptor that maps to chromosome 14. Genomics. 1996;35:397–402. doi: 10.1006/geno.1996.0377. [DOI] [PubMed] [Google Scholar]
- 11.Seuwen K, Ludwig MG, Wolf RM. Receptors for protons or lipid messengers or both? J Rec Signal Trans Res. 2006;26:599–610. doi: 10.1080/10799890600932220. [DOI] [PubMed] [Google Scholar]
- 12.Lanuza E, Novejarque A, Martínez-Ricós J, Martínez-Hernández J, Agustín-Pavón C, Martínez-García F. Sexual pheromones and the evolution of the reward system of the brain: the chemosensory function of the amygdala. Brain Res Bull. 2008;75:460–466. doi: 10.1016/j.brainresbull.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 13.Shimokawa N, Dikic I, Sugama S, Koibuchi N. Molecular responses to acidosis of central chemosensitive neurons in brain. Cell Signal. 2005;17:799–808. doi: 10.1016/j.cellsig.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Wemmie JA, Askwith CC, Lamani E, Cassell MD, Freeman JH, Welsh MJ. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang C, Rojas A, Wang R, Wang X. CO2 central chemosensitivity: why are there so many sensing molecules? Respir Physiol Neurobiol. 2005;145:115–126. doi: 10.1016/j.resp.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Krishtal OA. The ASICs: signaling molecules? Modulators? Trends Neurosci. 2003;26:477–483. doi: 10.1016/S0166-2236(03)00210-8. [DOI] [PubMed] [Google Scholar]
- 17.Semtner M, Schaefer M, Pinkenburg O, Plant TD. Potentiation of TRPC5 by protons. J Biol Chem. 2007;282:33868–33878. doi: 10.1074/jbc.M702577200. [DOI] [PubMed] [Google Scholar]
- 18.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 19.Radu CG, Nijagal A, McLaughlin J, Wang L, Witte ON. Differential proton sensitivity of related G protein-coupled receptors T cell death-associated gene 8 and G2A expressed in immune cells. Proc Nat Acad of Sci. 2005;102:1632–1637. doi: 10.1073/pnas.0409415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg R, Reshef-Bankai E, Coleman R, Green J, Maor G. Chronic acidosis-induced growth retardation is mediated by proton-induced expression of Gs protein. J Bone Miner Res. 2006;21:703–713. doi: 10.1359/jbmr.060210. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg JL, Barres BA. The relationship between neuronal survival and regeneration. Ann Rev Neurosci. 2000;23:579–612. doi: 10.1146/annurev.neuro.23.1.579. [DOI] [PubMed] [Google Scholar]
- 22.Walton MR, Dragunow M. Is CREB a key to neuronal survival? Trends Neurosci. 2000;23:48–53. doi: 10.1016/s0166-2236(99)01500-3. [DOI] [PubMed] [Google Scholar]
- 23.Bito H, Takemoto-Kimura S. Ca(2+)/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium. 2003;34:425–430. doi: 10.1016/s0143-4160(03)00140-4. [DOI] [PubMed] [Google Scholar]
- 24.Malone MH, Wang Z, Distelhorst CW. The glucocorticoid-induced gene TDAG8 encodes a pro-apoptotic G Protein-coupled receptor whose activation promotes glucocorticoid-induced apoptosis. J Biol Chem. 2004;279:52850–52859. doi: 10.1074/jbc.M408040200. [DOI] [PubMed] [Google Scholar]
- 25.Kintner DB, Anderson MK, Fitzpatrick JH, Jr, Sailor KA, Gilboe DD. 31P-MRS-based determination of brain intracellular and interstitial pH: its application to in vivo H+ compartmentation and cellular regulation during hypoxic/ischemic conditions. Neurochem Res. 2000;25:1385–1396. doi: 10.1023/a:1007664700661. [DOI] [PubMed] [Google Scholar]
- 26.Chesler M, Kaila K. Modulation of pH by neuronal activity. Trends in Neurosci. 1992;15:396–402. doi: 10.1016/0166-2236(92)90191-a. [DOI] [PubMed] [Google Scholar]
- 27.Krishtal OA, Osipchuk YV, Shelest TN, Smirnoff SV. Rapid extracellular pH transients related to synaptic transmission in rat hippocampal slices. Brain Res. 1987;436:352–356. doi: 10.1016/0006-8993(87)91678-7. [DOI] [PubMed] [Google Scholar]
- 28.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 29.Friedman SD, Mathis CM, Hayes C, Renshaw P, Dager SR. Brain pH response to hyperventilation in panic disorder: preliminary evidence for altered acid-base regulation. Am J Psy. 2006;163:710–715. doi: 10.1176/ajp.2006.163.4.710. [DOI] [PubMed] [Google Scholar]
- 30.Shekhar A, Sajdyk TJ, Gehlert DR, Rainnie DG. The amygdala, panic disorder, and cardiovascular responses. Ann N Y Acad Sci. 2003;985:308–325. doi: 10.1111/j.1749-6632.2003.tb07090.x. [DOI] [PubMed] [Google Scholar]
- 31.Wemmie JA, Coryell MW, Askwith CC, Lamani E, Leonard AS, Sigmund CD, Welsh MJ. Overexpression of the acid-sensing ion channel 1a in transgenic mice increases acquired fear-related behavior. Proc Nat Acad Sci. 2004;101:3621–3626. doi: 10.1073/pnas.0308753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wemmie JA, Price MP, Welsh MJ. Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 2006;29:578–586. doi: 10.1016/j.tins.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 33.Ugawa S, Ueda T, Yamamura H, Shimada S. In situ hybridization evidence for the coexistence of ASIC and TRPV1 within rat single sensory neurons. Brain Res Mol Brain Res. 2005;136:125–133. doi: 10.1016/j.molbrainres.2005.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.