Preface
Attachment of ubiquitin-like proteins (UBLs) to their targets via multienzyme cascades constitutes a central mechanism through which protein functions are modulated. This process is initiated by a family of mechanistically and structurally-related E1 (or activating) enzymes. The E1s serve to activate UBLs through C-terminal adenylation and thiol transfer and to coordinate the utilization of UBLs in specific downstream pathways by charging cognate E2 enzymes, which then interact with the downstream ubiquitylation machinery to coordinate modification of the target. A broad understanding of how E1s activate UBLs and how they selectively coordinate UBLs with downstream function has come from enzymatic, structural and genetic studies.
A major mechanism for regulating protein function in eukaryotes involves covalent attachment of ubiquitin or ubiquitin-like proteins (referred to collectively as UBLs) to the primary amino group of a target, often from a Lys side-chain, through an isopeptide bond. Post-translational modification by UBLs regulates numerous processes, which include cell division, immune responses and embryonic development. Accordingly, defects in UBL pathways are associated with various diseases, particularly cancer, neurodegenerative disorders, and muscle atrophy or ‘cachexia’1,2.
The UBL carboxyl termini are attached to other proteins, or in some cases lipids, generally through E1-E2-E3 multienzyme cascades (FIG. 1a). At the apex of each UBL cascade is an E1 enzyme, which activates the UBL and then directs the UBL to downstream pathways (FIG. 1). The ubiquitin system itself is the best understood UBL pathway. In the first step of ubiquitin activation, the E1 enzyme binds MgATP and ubiquitin, and catalyzes ubiquitin C-terminal acyl-adenylation. In the second step, the catalytic Cys in the E1 attacks the ubiquitin~adenylate to form the activated ubiquitin~E1 complex (the tilde “~” represents a high-energy thioester bond between the C-terminal carboxylate of the ubiquitin-like protein and the conjugation machinery or AMP, while a dash “-” represents a non-covalent complex). Ultimately, an E1 engages one of up to tens of cognate E2 conjugating enzymes to initiate downstream signalling, typically through the coordinated function of E3 ubiquitin ligases (FIG. 1). E3s contain binding sites for both charged E2s and ubiquitylation substrates3. For the largest class of E3s (the Really Interesting New Gene, RING, and the RING-related U-box family), an ε-amino group of a Lys residue in the associated substrate attacks the thioester of the transiently associated charged E2 to make an isopeptide bond with ubiquitin. The discharged E2 then dissociates from the E3, allowing a second charged E2 to interact with the E3, facilitating a second round of ubiquitin transfer, either by attack of a Lys residue in ubiquitin itself or by attack of a different Lys in the substrate. Multiple E2 cycles of E1-mediated ubiquitin loading and subsequent unloading – through a variety of mechanisms (reviewed in 4) - lead to polyubiquitylation of the substrate (FIG. 1a, b).
Figure 1. Diverse functions of Ubiquitin-like proteins (UBLs).
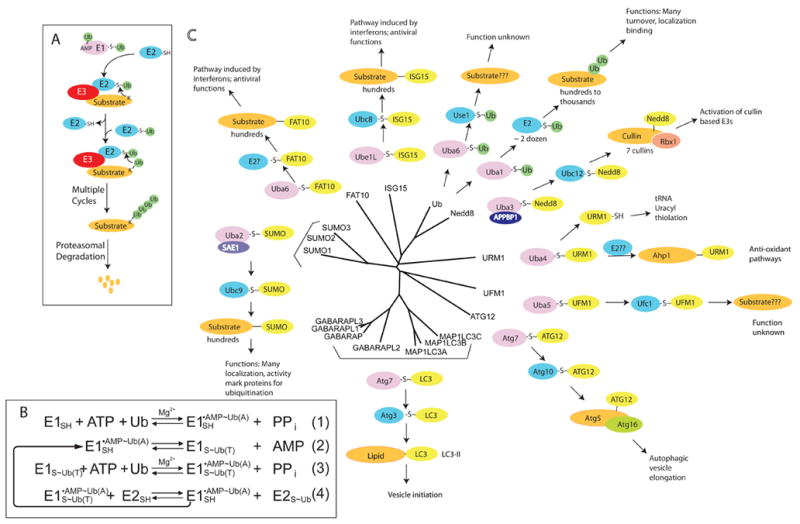
a | The canonical conjugation pathway for ubiquitin (Ub). The E1 enzyme ubiquitin activating enzyme 1 (UBA1) reacts with ubiquitin to form a ubiquitin-adenylate intermediate. Ubiquitin is transferred to a Cys in the catalytic domain of UBA1 to form the activated ubiquitin~UBA1 complex. A second molecule of ubiquitin binds to the adenylation domain and is converted to ubiquitin-adenylate. The doubly loaded E1 complex is then recognized by a cognate E2 ubiquitin conjugating enzyme, which receives ubiquitin to form a ubiquitin-charged E2. E2s recognize E3s that are associated with substrates and transfer ubiquitin to the substrate. Multiple cycles of binding to charged E2s leads to the formation of ubiquitin chains, which are recognized by the 26S Proteasome, facilitating substrate degradation. b | Enzymatic mechanism of the ubiquitin activation and conjugation cycle. E1 – UBA1; Ub(A) – ubiquitin associated noncovalently at the adenylation active site; Ub(T) – ubiquitin covalently linked to the catalytic Cys of UBA1 through a thioester bond. c |The pathways that employ ubiquitin-like proteins (UBLs) (yellow) are arranged around the phylogenetic tree for the 17 UBLs that are known to be conjugated to other molecules via their carboxyl-terminal Gly residue. E1 proteins (purple) for specific UBLs can be monomeric (UBA1, UBA6, UBE1L), heterodimeric (UBA2/SAE1, UBA3/APPBP1), or homodimeric (UBA4, ATG7, and likely UBA5). E2 proteins (green) associate with E1 proteins and receive the activated UBL via a trans-thioesterification reaction. E2s then transfer their UBLs to substrates (orange), typically via association is an E3 ubiquitin ligase.
Substrates with four or more linked ubiquitins are often targeted to the 26S proteasome for degradation. However, ubiquitin, itself, can serve multiple functions, depending at least in part on how it is attached to targets. In addition to polyubiquitin chains directing targets to proteasome-dependent degradation, mono-ubiquitylation or polyubiquitin chains linked via a variety of Lys residues in the ubiquitin molecule can alter the localization or activity of the target protein, generally through recruitment of ubiquitin binding proteins5–9
In total, 17 human UBLs, within 9 phylogenetic classes, have been reported to be conjugated to other molecules (FIG. 1c)1,10–17. All UBLs display a common overall fold, but, in general, the different UBLs have their own discrete E1-E2-E3 cascades, and they impart distinct functions to their targets12,13,18. Besides ubiquitin, other well-studied UBLs are neuronal-precursor-cell-expressed developmentally downregulated protein-8 (NEDD8) and small ubiquitin-related modifier (SUMO)-family members, both of which are essential for viability in organisms ranging from fission yeast to mammals. NEDD8 conjugation is initiated by its dedicated E1, the heterodimeric NEDD8 activating enzyme 1 (NAE1)-ubiquitin activating enzyme 3 (UBA3) complex. Attachment of NEDD8 to its predominant targets, cullins, enhances enzymatic activity of cullin-RING E3 ubiquitin ligases19–22 (FIG. 1c). In contrast, three human SUMOs (SUMO-1, -2, -3) are conjugated to many diverse proteins. All three are activated by a common E1, the heterodimeric SUMO activating enzyme 1 (SAE1)-UBA2 complex (FIG. 1c). SUMO attachment often alters a the interaction of a target with other proteins, via interactions between SUMO and SUMO-binding motifs23.
A number of other UBLs function in diverse biological pathways (FIG. 1c). The conjugation of two UBLs – interferon stimulated gene 15 (ISG15) and FAT10 - are under the control of the interferon system, which responds to viral signals. ISG15 – the product of an interferon inducible gene - is activated by the E1 UBA7 (FIG. 2) and is transferred to dozens of targets in a wide range of pathways through a specific E2, ubiquitin conjugating enzyme 8 (UBCH8), whose expression is also under interferon control (FIG. 1c). Additional UBLs, including the autophagy related protein 8 (ATG8) and ATG12 families, ubiquitin fold modifier 1 (UFM1) and ubiquitin-related modifier 1 (URM1) are activated by their own E1 enzymes (ATG7, UBA5 and UBA4, respectively) (FIG. 1c). ATG8 and ATG12 are involved in multiple steps in autophagy, a process by which cells degrade their cytoplasmic organelles through the lysosome (see below). In contrast, URM1 is functionally distinct in that it is used in biosynthetic reactions that involve sulfur transfer.
Figure 2. Activation of a prokaryotic ubiquitin-like protein, MoaD, by MoeB.
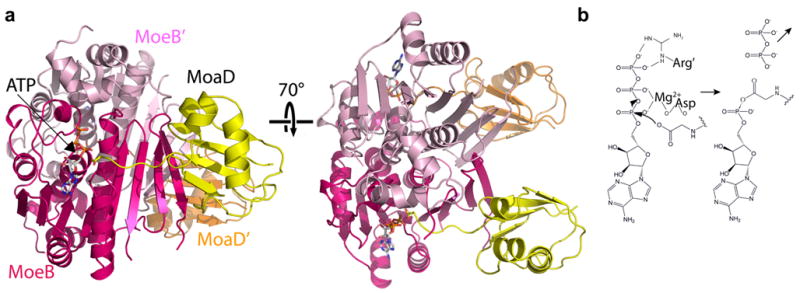
a | Overall structure of the Escherichia coli molybdopterin biosynthetic enzyme B (MoeB) (magenta, pink)-MoaD (yellow, orange)-ATP homodimer (1JWA.PDB)25, shown in two orientations rotated by 70° around the x-axis. The left view highlights the MoaD carboxyl-terminus approaching ATP for adenylation, and the right view highlights the dimeric structure of an adenylation domain. b | Schematic view of MoeB-catalyzed adenylation of MoaD, highlighting the roles of the Mg2+-coordinating Asp of MoeB and the MoeB’ Arg finger.
This Review summarizes our current understanding of the mechanisms by which E1s initiate UBL conjugation.
Prokaryotic antecedents of E1s and UBLs
UBLs and E1s have their origins in prokaryotic biosynthetic pathways. The bacterial proteins molybdopterin converting factor, subunit 1 (MoaD) and thiamine biosynthesis protein S (ThiS), which share the UBL fold24,25 (FIG. 2a), carry sulfur for incorporation into molybdopterin and thiazole, respectively. Molybdopterin is a small molecule cofactor found in proteins that bind molybdenum while thiazole is a precursor in the production of thiamine. Furthermore, in a reaction resembling that catalyzed by eukaryotic E1s, MoaD and ThiS are activated by C-terminal acyl-adenylation by the bacterial enzymes MoeB and ThiF, respectively26,27. MoeB and ThiF share sequence homology with the domain of eukaryotic E1s that is responsible for UBL binding and adenylation, which is the common building block for all E1s (FIG. 3). Thus, MoeB and ThiF embody minimal modules harbouring UBL-E1 recognition and adenylation activities12,13.
Figure 3. E1 domain structures and enzymatic mechanism.
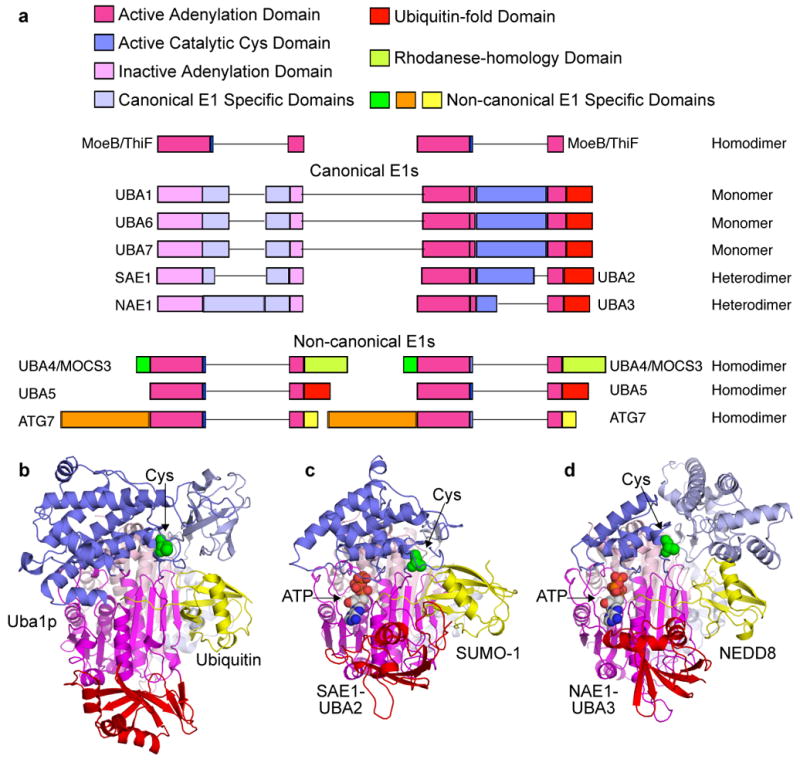
a | Primary structures of human canonical and noncanoncal E1 enzymes, with domains indicated and coloured according to the legend (bottom), and adenylation domains aligned according to molybdopterin biosynthetic enzyme B (MoeB) and thiamine biosynthesis enzyme F (ThiF) primary structures shown above. Lines reflect insertions in sequences between conserved domains. b | Cartoon view of canonical E1 crystal structures with ubiquitin (yellow) associated noncovalently at the adenylation active site of Saccharomyces cerevisiae Uba1 (3CMM.PDB)51, c | with small ubiquitin-related modifier (SUMO)- 1 (yellow) and ATP associated noncovalently at the adenylation active site of SAE1-UBA2 (1Y8R.PDB)50, and d | with NEDD8 (yellow) and ATP noncovalently at the adenylation active site of NAE1-UBA3 (1R4M.PDB)67. The domains are coloured according to the schematic view in panel a, and oriented as the left view of MoeB in FIG. 2a.
The crystal structures of the MoeB-MoaD complex, and the ATP-bound and MoaD-acyl-adenylated forms, and of ThiF-ATP and ThiF-ThiS complexes, have provided key insights into the mechanism of UBL recognition and activation25,28,29. The MoeB and ThiF structures are homodimeric (FIG. 2a, 3A), with two symmetric catalytic centres (Fig 2). Each contains a four-stranded β-sheet, with a hydrophobic surface that binds the region of MoaD or ThiS corresponding to the essential Leu8/Ile44/His68/Val70 hydrophobic patch of ubiquitin30. This UBL binding site of MoaD or ThiS is structurally stabilized by homodimerization of MoeB, or ThiF, (FIG. 2). The C-terminal tail of MoaD or ThiS extends toward the ATP that is located in a nucleotide-binding site. A conserved ATP-binding Arg finger comes from the opposite monomer in the complex and a conserved Asp coordinates Mg2+, which in turn alleviates electrostatic repulsion during the attack of the UBL’s C-terminal carboxylate oxygen on the α-phosphate of ATP·Mg2+. The basic side-chains of MoaD or ThiS would stabilize the developing negative charge from the ensuing pentacovalent phosphate intermediate, allowing the generation of MoaD or ThiS~adenylate and PPi. It remains an open question as to whether there is any cross-talk between the two catalytic centres in a MoeB or ThiF homodimer, or also in homodimeric eukaryotic E1s.
Additional bacterial MoeB and ThiF homologues suggest that E1 -like proteins might initiate a range of prokaryotic biosynthetic or post-translational modification pathways. Perhaps the most distinctive variant is Escherichia coli Microcin C7 Biosynthesis gene B (MccB), which initiates biosynthesis of the antimicrobial peptide MicrocinC7. Notably, MccB is unique among MoeB/ThiF/E1 relatives in that its substrate is not a UBL, but instead is a heptapeptide with an atypical C-terminal sequence31. At the opposite end of the spectrum, the small prokaryotic ubiquitin-like protein (Pup) protein from Mycobacterium tuberculosis becomes conjugated to 26S proteasome targets32. However, to date, no MoeB or ThiF homologue has been identified in this pathway. Thus, it seems likely that a range of unexpected variations on UBL activation mechanisms will as yet be discovered in prokaryotes.
Diversity and specificity of E1s
In humans, eight E1s are known to initiate UBL conjugation (FIGs 1, 3)33–47. Each is nucleated by a homodimeric or heterodimeric ‘adenylation domain’, which is responsible for initial UBL recognition and acyl-adenylation, and which resembles MoeB and ThiF48 (FIG. 3).
In addition to their common adenylation domain, E1s also display a range of other domains. For clarity, we refer to E1s for ubiquitin (UBA1, UBA6), SUMO (SAE1-SAE2), NEDD8 (NAE1-UBA3), and ISG15 (UBA7) as ‘canonical’, due to their related domain structures and enzymatic mechanisms. Canonical E1s have two MoeB and ThiF-homologous repeats either in a single polypeptide or two subunits corresponding to the amino- and C-terminal halves of homomeric E1s (FIG. 3A)12,13,48. The adenylation domain of canonical E1s is pseudosymmetric, with one MoeB or ThiF repeat binding MgATP and UBL, and the other primarily providing structural stability (FIG. 3)49–51. Canonical E1s also share two additional domains, for UBL transfer to E2s (see below). We refer to E1s for URM1 (UBA4), UFM1 (UBA5), and ATG12 and ATG8 isoforms (ATG7) as ‘non-canonical’. Available evidence indicates that non-canonical E1s contain homodimeric adenylation domains (FIG. 3c), which presumably adopt symmetric structures like MoeB and ThiF43,52–54. Each of these E1s also has unique sequences (FIG. 3a).
Despite overall related architectures, several UBL pathways display their own idiosyncrasies (FIG. 1) and we describe specialized features of the ubiquitin, SUMO and autophagy pathways as examples. In vertebrates and sea urchins, two different E1s initiate conjugation of ubiquitin44–46, despite prior notions that each E1 activates unique UBL(s). Indeed, for more than two decades, the central dogma in the field was that a single ubiquitin activating enzyme, UBA1, functions to charge E2s dedicated to the ubiquitin system, as this is the case in lower eukaryotes. However, recent studies identifying UBA6 (also known as Ube1L2) as a second E1 enzyme for ubiquitin challenged this dogma44–46. UBA1 and UBA6 are distantly related (~40% identical); in fact, UBA1 is phylogenetically more closely related to UBA7 (the E1 for ISG15)44. Nevertheless, UBA6 is charged by ubiquitin in vitro and in cells. Furthermore, UBA6 is uniquely responsible for transferring ubiquitin to UBA6’s selective E2, USE1 (Uba6-specific E2)44. UBA6 and USE1 are found from humans to zebrafish, as well as sea urchin, and are ubiquitously expressed, but are absent from worms, flies and yeast, indicating a selective role in certain multicellular organisms. Indeed, deletion of the mouse UBA6 gene results in embryonic lethality46. Intriguingly, at high protein concentrations, UBA6 can form a complex with the UBL FAT10 in vitro46, and depletion of UBA6 can block conjugation of FAT10 to unknown proteins. However, mice lacking FAT10 are viable, suggesting that the essential functions of UBA6 are unlinked to FAT10 activation.
The SUMO pathway has an opposite, perplexing feature, with the one E1 in higher eukaryotes (the SAE1-UBA2 heterodimer)38,39 activating at least three SUMOs. Similarly, the single E1 involved in autophagy, Atg7, activates multiple structurally-distinct UBL proteins - Atg8 and Atg12 in budding yeast, and even more Atg8 relatives in higher eukaryotes (FIG. 1)47,55. Thus, how E1s recognize their distinct UBLs, and when and why different E1s activate ubiquitin are important questions.
Catalytic mechanisms of canonical E1s
The catalytic mechanism is best characterized for the ubiquitin E1, UBA1 (FIG. 1b). After UBL C-terminal adenylation56–58, the ubiquitin~AMP is attacked by the catalytic Cys of UBA1, producing a covalent thioester linkage between the Cys sulfhydryl of UBA1 and the C terminus of ubiquitin56,57,59,60. Subsequently, UBA1 catalyzes the adenylation of a second ubiquitin molecule56,57. Thus, UBA1 becomes asymmetrically loaded with two ubiquitin molecules at distinct active sites: ubiquitin(T) is covalently linked to the E1’s Cys via a thioester bond; ubiquitin(A) is associated non-covalently at the adenylation active site. Ultimately, UBA1 physically associates with a cognate E2 conjugating enzyme, and a thioester transfer reaction ensues whereby the C terminus of ubiquitin(T) is transferred to the E2’s catalytic Cys3,57,61–63. Individual steps of the E1 reaction are reversible. Progression of the cascade is driven by release of the small molecule products PPi and AMP in the first and second steps, respectively56–58,62,63.
A curious feature of the E1-E2 cycle is the asymmetric double-loading of E1 with two ubiquitin molecules56,57. Why double load? Although a previous study showed that a partially purified UBA1~ubiquitin complex containing only the single ubiquitin(T) is capable of transferring the ubiquitin to an E2, this transfer is accelerated by ubiquitin(A)~adenylate or MgATP64. Also, UBA1 adenylation active site mutants generate an E2~Ub complex more efficiently than would be predicted based on their crippled adenylation of ubiquitin, further indicating cross-talk between the adenylation active site and E1-E2 thioester transfer65. Coupling the second adenylation reaction (FIG. 1b, line 3) with ubiquitin transfer to E2 (FIG. 1b, line 4) might make the cascade energetically or conformationally favourable, or might prevent the E1 from becoming trapped in an unfavourable conformation.
A mechanism similar to the one of UBA1 has been confirmed for the heterodimeric E1 of NEDD8, NAE1-UBA3, through biochemical and structural studies66–68. Furthermore, most features of this scheme (adenylation, E1~UBL thioester intermediate, E2 charging) have also been observed for SAE1-UBA2, UBA6 and UBA7, and the similarities of their primary sequences and, where determined, tertiary structures, are also in keeping with the notion of parallel reaction mechanisms40, 41, 44, 50, 51, 69. Thus, it is likely that mechanistic and structural studies of a subset of canonical E1s provide broad insights into this E1 class as a whole.
Structural insights into canonical E1s
Structural studies of human NAE1-UBA3, SAE1-UBA2, and Saccharomyces cerevisiae Uba1, alone and/or in complexes with ATP, UBL(s) and/or cognate E2 have provided many insights into general features of canonical E1s, and also features unique to the different pathways49–51,67,68,70–72. Briefly, the structures all display an ‘adenylation domain’ that resembles MoeB and ThiF and two canonical-E1-specific domains: a ‘catalytic Cys domain’ harbouring the Cys residue that is involved in the E1~UBL thioester linkage and a C-terminal ubiquitin-fold domain (UFD) that resembles ubiquitin that binds E2 (FIG. 3a).
Canonical E1 adenylation domain
In the E1s for ubiquitin, SUMO and NEDD8, a single adenylation active site is focused around the C-terminal MoeB and ThiF-like repeat in Uba1, UBA2 and UBA3, respectively. Structural superposition of E1 adenylation domains shows extensive similarity to MoeB and ThiF, and their interactions with ATP and UBLs (FIG. 3). Mutational analyses agree with the notion of E1-catalyzed UBL-acyl-adenylation through mechanisms similar to that proposed for MoeB25,49,50,65.
A catalytic Cys domain
After the adenylation reaction, the catalytic Cys sulfhydryl of the E1 enzyme attacks the UBL~acyl-adenylate, resulting in a E1~UBL(T) complex that is covalently linked by a thioester bond between the C terminus of UBL and the catalytic Cys of E1. In the structures with NEDD8, SUMO-1 or ubiquitin at the UBL(A) site, a ~35 Å gap between the C terminus of UBL(A) and the catalytic Cys of E1 raises questions as to UBL and E1 conformations during the formation of the thioester intermediate (FIG. 3c)50,51,67. Part of the gap might be closed if the UBL’s C-terminal tail were freed from interaction with E1 after the adenylation reaction. However, several pieces of data suggest that the E1 might adopt as yet unseen conformations for the catalytic Cys attack of UBL~AMP. First, ubiquitin~AMP binds UBA1 with higher affinity than free ubiquitin, UBA1 seems to harbour a single nucleotide binding site, and a non-hydrolyzable analogue of ubiquitin~adenylate is a potent UBA1 inhibitor in part by competing with ATP binding56–58,3. Second, following the adenylation reaction, the Cys of UBA1 can form a thioester complex with a fluorescently-labelled peptide that corresponds to ubiquitin’s C terminus74. Third, the UBA1 catalytic Cys domain is structurally similar in isolation and in the context of full-length UBA151,75, and domain rotation is consistent with subtle conformational differences between apo and UBL(A)-bound structures50,67. Insights into the thioester intermediate come from the crystal structure of a trapped doubly UBL-loaded/E2-bound NEDD8 E1 complex (FIG. 1b, line 4), where NEDD8(T) is covalently bound at the Cys of UBA3 through a thioester bond, and NEDD8(A) is noncovalently associated at the adenylation active site68. Here the catalytic Cys is in the same relative location as in apo and UBL(A)-bound E1 structures (FIG. 4). Thus, if the catalytic Cys domain undergoes conformational changes while attacking the UBL(A) C terminus, it apparently can subsequently return to a central orientation after forming the thioester bond. Although no structure has identified a general base poised to deprotonate the catalytic Cys of E1, an important role for a network of polar and charged side-chains surrounding the thioester bond was revealed by mutational analysis68.
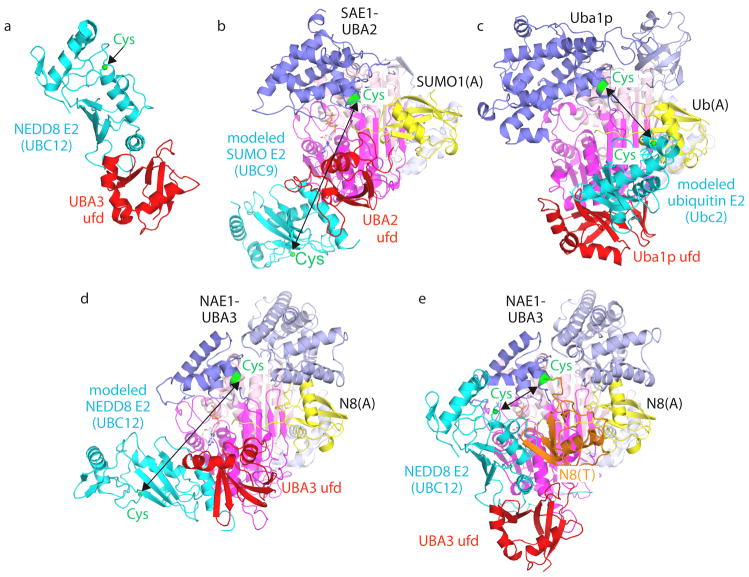
Figure 4. Canonical E1 domain rotation in ubiquitin-like protein transfer to E2s.
a | E1–E2 interactions shown by the crystal structure of the isolated ubiquitin-fold domain (UFD) from the UBA3 subunit of the E1 of NEDD8 (red) complexed with the core domain from UBC12 (cyan), the E2 of NEDD8 (1Y8X.PDB)71. The sulfhydryl of the catalytic Cys in Ubc12 is shown as a green sphere. b–d, Models of E2s (cyan) bound to structures of E1-UBL(A) complexes, with arrows highlighting E1-to-E2 Cys-to-Cys distances, for the E1 of SUMO (1Y8R.PDB)50 and Ubc9 (1U9B.PDB)119 (b), the yeast ubiquitin E1 (3CMM.PDB)51 and Ubc2 (2AYZ.PDB)120 (c), and the E1 of NEDD8 (1R4M.PDB)67 and Ubc12 (1Y8X.PDB)71 (d). e | UFD rotation revealed from the structure of the doubly UBL-loaded/E2-bound NEDD8 E1 [NAE1-UBA3~NEDD8(T)-NEDD8(A)-MgATPUbc12(catalytically inactive mutant] (2NVU.PDB)68. NEDD8(T) is in orange, NEDD8(A) is in yellow, and the location that would correspond to a Ubc12 catalytic Cys is shown as a green sphere. E1-UBL structures are oriented and coloured as in FIG. 3, and E1 and E2 catalytic Cys residues are shown in green.
The E2-binding ubiquitin-fold domain
The UFD binds to the E2 enzyme49–51, 71. Structural studies for the NEDD8 pathway revealed the binding of the UFD’s concave β-sheet of UBA3 to the N-terminal helix and β1β2-loop from the core domain of UBC12, the E2 of NEDD8 (FIG. 4a)71. Mutational analysis indicates that other canonical E1s bind their E2s similarly44, 50, 51, 76–78.
Docking the isolated UFD-E2 structure onto the UFD of full-length NAE1-UBA3, SAE1-UBA2 or Uba1 does not provide a model for UBL transfer from E1 to E2. In the models, E1-to-E2 Cys-to-Cys distances are ~30–65 Å (FIG. 4b–d)49, 50, 67, 71, 79, whereas these residues must be juxtaposed during UBL transfer. A striking, ~110° rotation of the UFD of UBA3 was observed by comparing different NEDD8 E1 structures (FIG. 4d, e)68. This difference revealed the propensity for the UFD to undergo dramatic rotation, and brings the UBA3 donor and Ubc12 acceptor Cys residues within ~20 Å of each other. Additional rotation of the UFD and/or catalytic Cys domain would allow the E1 and E2 Cys residues to be in close proximity for UBL transfer. Consistent with an important role for UFD rotation, mutations hindering rotation of the loop that connects the adenylation and ubiquitin-fold domains diminish E2 charging by NAE1-UBA3 and Uba151,71. Interestingly, different E1s might display different UFD rotations: whereas apo and UBL(A)-bound structures of NAE1-UBA3 and SAE1-UBA2 showed the E2-binding surface of UFD facing away from the E1 Cys49,50,67, the corresponding UBA1 surface faces toward the Cys of UBA151. Although future studies will be required to understand when and how the UFD adopts different orientations, it is possible that conformational differences among the E1s might reflect structural differences in their cognate E2 enzymes that might limit orientations for approaching UBL(T).
The UFD rotation also unmasks an additional E2-binding surface in the adenylation domain of E1, adjacent to the adenine moiety of ATP68. It is tempting to speculate that E2 binding adjacent to the nucleotide binding site could have a role in coupling between the adenylation and thioester transfer reactions64,65.
A thioester switch directs E1 cycling
Several studies indicated that covalent thioester UBL linkage alters E1-E2 noncovalent interaction properties (FIGs 4, 5). First, UBA1 and ubiquitin E2s display different relative affinities for each other in their free and ubiquitin-thioester-linked states. Free UBA1 separates from E2s during gel filtration, suggesting low affinity3. By contrast, doubly-ubiquitin-loaded UBA1 binds to uncharged E2 substrates with nanomolar affinities63. After charging, a thioester-linked E2~UBL product is apparently released from E1, as E1 undergoes multiple reaction cycles in the presence of excess E2 Second, different affinities for free and UBL-bound enzymes might promote progression through E1-E2-E3 cascades. E1-E2 and E2-E3 interactions are mutually-exclusive, due to structural overlap between E1 and E3 binding sites on E2 molecules71,77,80,81. Thus, the E2~UBL product would need to be released from E1 prior to binding to E3. Third, striking differences in E1 conformation, and concomitant unmasking of cryptic E2-binding sites, are observed upon comparing crystal structures of the NEDD8 E1 with and without NEDD8(T) (FIG. 1B)67,68.
Figure 5. Model for a thioester switch modulating E1–E2 interactions68.
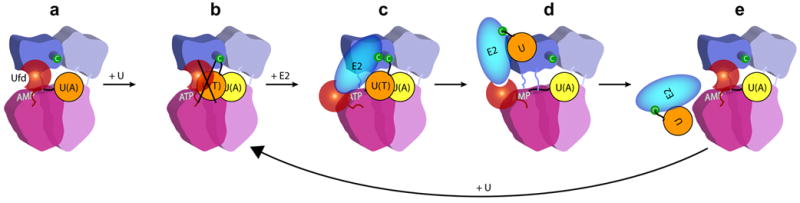
a | E1 conformation as for the NAE1-UBA3-NEDD8(A) complex67, modelled with the carboxyl terminus of the adenylation active site (U(A)) covalently linked to AMP. b | After double-UBL-loading of the E1, the thioester-bound UBL would clash with the ubiquitin-fold domain (UFD) of E1 in the position observed in the NAE1 -UBA3-NEDD8(A) structure67. Thus, double-UBL-loading might restrict positions accessible by the thioester active site of UBL (UBL(T)) and the UFD of E1. c | With the rotation of the E1’s UFD observed in the NAE1-UBA3~NEDD8(T)-NEDD8(A)-MgATP-Ubc12(catalytically inactive mutant) structure68, two cryptic E2-binding sites are unmasked, facilitating the doubly-UBLloaded E1 binding to or positioning E2 for the UBL transfer reaction. d | Following UBL transfer to the catalytic Cys of E2, the covalent tether of UBL(T) to E1 is eliminated. e | Steric clashing between E1 and E2~UBL might further facilitate the release of this product, and reset the E1 for another activation cycle. The first UBL to enter the cascade is coloured in orange, and the second in yellow.
Taken together, the available data suggest that canonical E1 reaction cycles might be driven by ‘thioester switch’ mechanisms toggling E1–E2 affinities (FIG. 5)63,68. When E1 is doubly-loaded and thioester-linked to a UBL(T), affinity for E2 is increased due to additional binding sites, including the UBL(T) itself. Transfer of the covalent linkage of UBL to the catalytic Cys of E2 generates the E2~UBL thioester product. At this point, the UBL is no longer covalently tethered to the E1, eliminating one E1-associated E2 binding site, which might facilitate the release of the product.
Different E1 characteristics raise the possibility that distinct E1–E2 pathways utilize diverse thioester-switching mechanisms50,51,67. First, the single UBA1 charges numerous ubiquitin E2 enzymes, whereas the NEDD8 and SUMO pathways are restricted to selected E2 enzymes36,37,61,82. Second, whereas free UBA1 displays low affinity for E2s, uncharged NAE1-UBA3 and SAE1-UBA2 (and their isolated UFDs) can bind their E2s49,50,77,83. Thus, NEDD8 and SUMO E1s might require additional mechanisms for ensuring release of the E2~UBL product, and E1 recycling. Accordingly, different UFD orientations in the E1-UBL(A) structures might reflect distinct thioester switches. In the UBA1-ubiquitin crystal structure, the UFD is oriented such that an associated E2 would face the catalytic Cys of Uba1. Formation of the UBA1~Ub thioester complex might primarily drive E2 binding by increasing affinity through noncovalent interactions that are mediated by the thioester-bound ubiquitin. By contrast, the higher basal E1-E2 affinities in the NEDD8 and SUMO pathways might suggest that a primary role of the thioester switch would be to promote release of the E2~UBL thioester product. In these cases, after the formation of the E2~UBL product, the rotation of UFD into the orientation observed in NAE1-UBA3-NEDD8(A) and SAE1-UBA2SUMO(A) might facilitate the release of the product due to clashing between the E1 and the E2~UBL complex.
Canonical E1 UBL and E2 specificity
In addition to their chemical roles in initiating UBL conjugation cascades, E1 enzymes also establish specificity, by matching a particular UBL with only cognate E2s. Rules for how this specificity is achieved are only beginning to emerge, but it is clear that specificity is achieved at multiple levels.
E1s are highly specific for their cognate UBLs66,84. The best understood example involves UBA1 and UBA3 distinguishing between the C-terminal tail residue 72 in ubiquitin and NEDD8 (Arg in ubiquitin, Ala in NEDD8)66,67,74,84. In both cases, residue 72 makes positive contacts with the E149,51,67. However, the majority of the UBA3’s selectivity comes from a unique Arg that repels the Arg72 of ubiquitin72.
The E1–E2 selectivity for the ubiquitin, NEDD8, SUMO and ISG15 cascades seems to involve at least two noncovalent interaction surfaces. First, selectivity is dictated by interactions between an E1’s UFD and the N-terminal sequence of a E2 catalytic domain. Swapping the UFD of UBA6 or UBA7 with that of UBA1 alters E2 specificity, and transplanting N-terminal sequences of UbcH8 (the E2 enzyme of ISG15) into UbcH7 (the E2 of ubiquitin) resulted in roughly 20-fold improved binding to UBA7, the E1 of ISG1544,78.
Pathway-specific E1–E2 interactions also impart selectivity. A unique N-terminal extension on NEDD8’s E2 (Ubc12) docks in a groove exclusive to UBA368,70. By contrast, relative to the smaller catalytic Cys domain from UBA3, UBA1 and UBA2 have unique insertions near the catalytic Cys that have been implicated in binding to their cognate E2s51,85,86. These distinctive interaction surfaces also select against off-pathway mis-charging: these are key features that mask the vestigial ability of the NEDD8 E2, Ubc12, to bind the ubiquitin E1 UBA187.
In vivo regulation of canonical E1s
While the mechanisms underlying recognition of ubiquitin-like proteins and the specificity of transfer to E2s are beginning to be understood, far less is known about how E1s are regulated in vivo.
The E1 enzymes for ubiquitin
In the early 1980’s, landmark studies identified temperature sensitive (ts) mutations in mammalian UBA1, and ts UBA1 cells arrest the cell cycle at the G2/M phase transition and display dramatically decreased ubiquitin conjugation88,89. RNA interference (RNAi)-mediated knockdown of UBA1 also diminished human cell proliferation90. Cells harbouring ts UBA1 variants have been used to test roles for ubiquitin in vivo, such as in the turnover of an unstable protein or in a specific pathway, for example endocytosis or phagocytosis91,92.
Hypomorphic93 and ts94 alleles of Saccharomyces cerevisiae and Caenorhabditis elegans95 Uba1 will allow further functional analysis of the ubiquitin E1 in these genetically tractable eukaryotes. Several Uba1 mutant alleles have also been identified in Drosophila melanogaster79,96. Reduced Uba1 activity in flies led to tissue overgrowth, raising the possibility that Uba1 is regulated during development, and/or tumour suppressive functions for Uba1. UBA1 knockdown in C. elegans led to enhanced aggregation of a polyglutamine-containing protein, consistent with the involvement of the ubiquitin system in removing misfolded and polyglutamine-containing proteins97. In addition, missense mutations in UBA1 are associated with the motor neuron disorder Spinal Muscular Atrophy, a disease involving spinal motor neuron protein degradation via the ubiquitin system98.
The use of RNAi helped to distinguish the functions for the multiple ubiquitin E1s that are found in higher eukaryotes44. UBA1 knockdown virtually eliminated ubiquitin charging of two UBA1-specific E2s, cell division cycle regulatory protein 34 A (CDC34A) and CDC34B. By contrast, depletion of UBA6 essentially eliminated ubiquitin charging of USE1 but not Cdc34 E2s44. UBA1 and UBA6 seem to have different catalytic efficiencies in vivo. Essentially all the UBA1 molecules present in proliferating mammalian cells, as well as the tested UBA1 substrate E2s, Cdc34A and Cdc34B, are in their activated/charged forms at steady-state44. This suggests that there is sufficient E1 activity to maintain fully charged pools of E2s, and that the interaction of charged E2s with E3s is rate-limiting for target ubiquitylation. Under similar conditions, UBA6 and its specific E2 USE1 are only ~50% activated/charged44.
Are the E1 enzymes for ubiquitin regulated? Although to date, UBA6 regulation remains uncharacterized, UBA1 has been found to be phosphorylated. Proposed roles of UBA1 phosphorylation include increasing nuclear import and/or retention99, and modulation of nucleotide excision repair during macrophage differentiation100. Furthermore, distinct isoforms of UBA1 display different subcellular localizations101, although the functions of different UBA1 modifications and isoforms remain poorly understood.
The E1 enzyme for ISG15
The expression of the ISG15 E1, UBA7, is induced by interferon-α and β, consistent with the notion that the ISG15 pathway has a role in the interferon-α/β-induced antiviral response41. Further evidence for a role in the antiviral response comes from the finding that UBA7-dependent activation is prevented by the influenza protein NS1B binding to the ISG15 E141.
The SUMO E1 enzyme
SUMO E1 activity seems to be regulated at many levels. The UBA2 subunit of the heterodimeric SUMO E1 has a C-terminal nuclear localization sequence (NLS). NLS deletion from S. cerevisiae Uba2 results in nuclear-to-cytoplasmic relocalization and altered SUMO conjugation102
SAE1-SAE2 activity is also altered in the presence of reactive oxygen species: in the presence of H2O2 the catalytic Cys residues of UBA2 and the SUMO E2, Ubc9, become covalently linked by a disulfide bond, thereby inhibiting SUMOylation103.
SUMO activation is also inhibited by the avian adenovirus chicken embryo lethal orphan (CELO) Gam1 protein104. Gam1 possesses a suppressor of cytokine signalling (SOCS)-box motif, allowing assembly into an E3 that targets SAE1 for ubiquitin-mediated proteolysis105.
The NEDD8 E1 enzyme
An important role for NEDD8 activation was uncovered with the identification of a mammalian cell line that contains a ts mutation in the NAE1 subunit (amyloid β precursor protein binding protein 1, APPBP1).. At the non-permissive temperature where NAE1 is inactive, there are defects in coupling of DNA synthesis and mitosis106, 107. Further, the ultimate function of NAE1-UBA3 to transfer NEDD8 to its E2 Ubc12 is also inhibited by reactive oxygen species that are induced by bacteria, which apparently inactivate the catalytic Cys of Ubc12 without promoting cross-linking to UBA3108.
Additional tools, which include specific small-molecule inhibitors of various E1 (BOX 1), are under development and have the potential to greatly facilitate functional analysis of the E1 enzymes in various processes.
BOX 1: E1 inhibitors
The development of specific inhibitors of E1 enzymes, including cell-permeable small-molecule inhibitors, represents an important area of current research. Several E1 inhibitors have been reported, the majority targeting ubiquitin activating enzyme 1 (UBA1). The first, adenosyl-phospho-ubiquitinol (APU), is a non-hydrolyzable analogue of ubiquitin~acyl-adenylate73. Replacement of a ubiquitin carboxyl-terminal Gly76 oxygen with a methylene group prevents reactivity, rendering APU a high-affinity intermediate analogue, which apparently occupies both the ubiquitin and ATP-binding sites. The 35 nM Ki value of APU is substantially lower than Km values for ubiquitin and ATP. Small molecule UBA1 inhibitors include a natural product, panepophenanthrin, which inhibits UBA1~ubiquitin thioester formation in vitro by an uncharacterized mechanism117, and a commercially-available cell-permeable inhibitor, PYR-41, identified in high-throughput screening of an E1–E2–E3 cascade, which might block the catalytic Cys of UBA1118
NEDD8 activating enzyme 1 (NAE1)-ubiquitin activating enzyme 3 (UBA3) can also be inhibited by multiple mechanisms. ATP-competitive inhibitors of NAE1-UBA3 are in development as anti-cancer therapeutics. A second mechanism for NAE1-UBA3 inhibition takes advantage of the unique interaction with the E2, Ubc12: a synthetic peptide corresponding to the 26-residue amino-extension of Ubc12 (Ubc12N26) competitively inhibits the binding of Ubc12 to NAE1-UBA370. Despite the poor Ki (20 μM), its high specificity makes Ubc12N26 a useful in vitro NAE1-UBA3 inhibitor, allowing the quenching of Ubc12~NEDD8 charging for characterizing downstream steps in the NEDD8 pathway using pulse-chase assays.
Non-canonical E1 enzymes
Non-canonical E1s seem to initiate their UBL cascades through related but distinct mechanisms. The E1s UBA4 (molybdopterin coenzyme synthase 3, MOCS3, in humans) and ATG7 are conserved among all eukaryotes, but have been best characterized in S. cerevisiae, for activating their respective UBL proteins URM1, and the two UBL proteins that are involved in autophagy (ATG8 and ATG12)43,52–54. Like MoeB and ThiF, UBA4 and ATG7 both homodimerize (FIG. 3c). In a related vein, UBA5 has not been found to partner with another protein harbouring homology to MoeB and ThiF, also suggesting homodimerization (FIG. 3c). Thus, as with MoeB and ThiF, these E1s might simultaneously bind two UBLs, one at each adenylation active site. The sequences of each of these E1 enzymes reflects unique features, likely for distinct functions downstream of UBL adenylation that are described below.
UBA4, a non-canonical E1 functions in sulfur transfer reactions
Mutations in Uba4 and Urm1 in yeast lead to defects in invasive and pseudohyphal growth, in the response to oxidative stress, in the response to nutrient starvation, and in signalling via the target of rapamycin (TOR) pathway. Yeast Uba4 was initially shown to promote Urm1 conjugation to at least one protein target, the alkyl hydroperoxide reductase Ahp1, in a manner that requires the catalytic Cys of Uba4, which suggests parallels to traditional UBL conjugation cascades (FIG. 1, 6)42,109. Whereas Urm1 modification of Ahp1 has been linked to oxidative stress, cells lacking Uba4 or Urm1 display additional phenotypes that are not observed with Ahp1 deletion, suggesting additional roles for Urm1. Moreover, no E2 has been identified as either forming a thioester bond with Urm1 or as binding to Uba4, and it is not clear presently whether MOCS3 also promotes conjugation of URM1 to proteins.
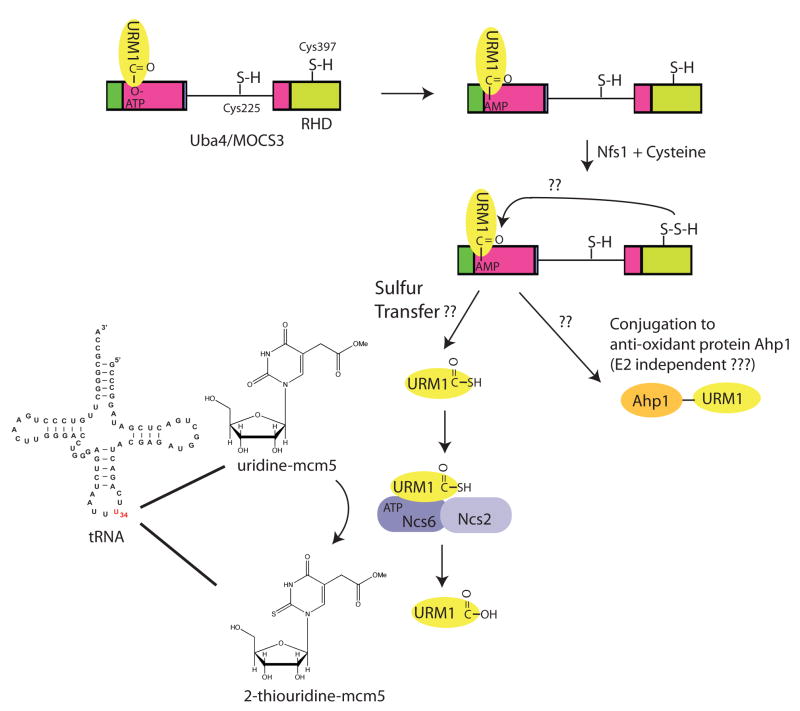
Figure 6. The UBA4/MOCS3 pathway.
Uba4 and its mammalian homologue MOCS3 function to promote both sulfur transfer reactions through URM1 as well as the conjugation of URM1 to other proteins via an isopeptide bond. URM1 forms an adenylate intermediate with the adenylation domain of MOCS3 or Uba4. A reactive Cys (residue 397) in the Rhodanese homology domain (RHD) of UBA4 becomes persulfurated likely via the action of nitrogen fixing bacteria S-like protein (Nfs1) in a reaction that requires Cysteine (Cys). Through a process that is poorly understood from a mechanistic perspective, the adenylate on URM1 is replaced by a sulfur from the persulfide on Cys397, forming URM1 thiocarboxylate. URM1-thiocarboxylate associates with the ATP binding subunit of a heterodimeric enzyme complex required for conversion of uridine-mcm5 in the anti-codon of U-rich tRNAs to the 2-thiouridine-mcm5 derivative. In yeast, this complex is composed of Needs CLA4 for Survival 2 (Ncs2), Ncs6, and a previously unstudied protein Yor251c. The ATP associated with Ncs6 has been proposed to make an adenylate with uridine, as an intermediate for the trans-thiolation reaction. 2-Thiouridine-mcm5 is crucial for the fidelity of translation. In a separate process, URM1 has been proposed to be conjugated to the alkyl hydroperoxide reductase Ahp1 in yeast but the molecular intermediates in this process are not defined.
Several recent findings reveal that Uba4 and MOCS3 function in a manner more related to E. coli MoeB and ThiF, than to canonical E1s. As with MoeB and ThiF activation of MoaD and ThiS, respectively, Uba4 and MOCS3-mediated thiocarboxylated URM1 might serve as a sulfur donor within the thiolation pathways110,111 (FIG. 6). First, Uba4 and MOCS3 have a ~120-residue rhodanese-homology domain (RHD) C-terminal to their adenylation domain. Rhodanese-domain enzymes, which include some MoeB orthologues and the thiamine biosynthesis enzyme ThiI protein that is downstream of ThiF in the thiamin biosynthesis pathway, are thought to catalyze sulfur transfer. Second, Uba4 catalyzes thiocarboxylation of human URM1 in vitro54, 111, and URM1 is thiocarboxylated in vivo110. Third, a downstream role for URM1 as a sulfur carrier is implicated by the findings that both URM1 and Uba4 or MOCS3 are required for uridine thiolation in certain tRNA molecules (tRNALys with anticodon sequence UUU, tRNAGln(UUG) and tRNAGlu(UUC)) in yeast and mammalian cells110,111. These tRNA modifications also involve the interaction of URM1 with multimeric ATP-binding protein complexes composed of Ncs6 (needs CLA4 to survive 6), Ncs2, and an uncharacterized open reading frame Yor251c in yeast and the ATP binding domain containing protein ATPBD3-LOC348180 in humans, presumably in a step of the cascade downstream of the activation by the MOCS3 E1 (FIG. 6)110–113 In mammals, the analogous complex has recently been identified and is composed of the ATP-binding domain containing protein ATPBD3 with similarity to Ncs2, and a previously uncharacterized open reading frame (LOC348180) with similarity to Ncs6 (FIG. 6) 110 (Peter Lee and J.W.H., unpublished data).
Ncs6 and ATPBD3 contain a PP-loop ATPase domain, which is known to function in adenylation of tRNAs, and Ncs6, and its C. elegans orthologue Tut-1, were shown to bind directly to tRNAs111. Genetic data suggest that Ncs6 specifically binds to tRNAs that have been modified on their wobble uridine34 moiety by a methoxy-carbonyl-methyl (mcm5) group in a reaction catalyzed by the elongator (ELP) complex. Indeed, mutations in the ELP complex are synthetically lethal with URM1Δ and UBA4Δ mutants111. Consistent with the involvement in this pathway, deleting Uba4 or Urm1 in yeast, or depleting them in mammalian cells, blocks uridine thiolation in vivo110,111. Moreover, a reactive analogue of URM1 (URM1-vinyl-methylester) can be directly crosslinked to ATPBD3, suggesting that this subunit serves to both bind the tRNA and the URM1 sulfur carrier110.
Many questions remain to be answered, including: How does URM1 lead to tRNA thiouridinylation? Is URM1-thiocarboxylate the direct sulfur donor for tRNA, or is there a sulfur-transfer pathway involving additional enzymes? Does URM1 carry sulfur for other pathways? And how might dual functions of URM1 as both a protein modifier (for proteins such as Ahp1) and a sulfur carrier be established? Can Uba4 or MOCS3 transfer URM1 to target proteins in a manner that is independent of an E2 intermediate? Finally, depletion of URM1 in mammalian cells leads to defects in cytokinesis110. Thus, what role does URM1 have in cell cycle control?
ATG7 regulates autophagy
ATG7 functions at the apex of a signalling system required for autophagy, the process whereby the cell directs the digestion of cytoplasmic components in response to reduced nutrients in the environment. Despite its essential role in autophagy, many aspects of the function of ATG7 remain poorly understood. First, how does ATG7 recognize its multiple UBL substrates (ATG8 and ATG12), as they are rather distantly related to each other (the yeast Atg8 and Atg12 share 18% amino acid sequence identity) (FIG. 1C)? Second, how does ATG7 mediate specific ATG8 charging of its dedicated E2 enzyme (ATG3), and ATG12 charging of its dedicated E2 enzyme (ATG10)? Third, how does ATG7 specifically recognize these two E2s? Fourth, does ATG7 also participate in ATG3-mediated transfer of ATG8 to its target, the lipid phosphatidylethanolamine, or ATG10-mediated transfer of ATG12 to its protein target, ATG5? Finally, mammals have a greatly expanded repertoire of ATG8 orthologues, with 7 genes in mammals (FIG. 1c). Although it is presumed that each of these is charged through the same pathway, whether these proteins have distinct functions remains unknown.
It is likely that some of these unique ATG7-mediated functions are carried out by the ~300 N-terminal and ~50 C-terminal residues flanking the ThiF-like adenylation domain. These ATG7-specific sequences lack detectable homology to suggest possible functions (FIG. 3a)47,52,114. Nonetheless, it is likely that as with other E1 enzymes, these sequences beyond the adenylation domain bind the autophagy-specific E2s, and promote UBL transfer. It seems that ATG3, the E2 for ATG8, binds to ATG7 in a unique way, through an ~80-residue flexible region not shared with other E2s, including the other autophagy-specific E2, ATG10115.
UBA5, a poorly understood non-canonical E1
UBA5, the E1 of UFM1, is found in multicellular organisms but not yeast and is perhaps the least characterized E143. The sequence of UBA5 suggests that the catalytic Cys is located in a loop, rather than in a discrete domain. The C-terminal domain of UBA5 is predicted to adopt a ubiquitin-fold and bind to UFC1, the E2 of UFM1 (FIG. 1c), analogous to that seen in the crystal structure of UFD-E2 in the NEDD8 pathway116. Future studies will be required to understand whether UBA5 functions as a homodimer, whether UBA5 represents a relatively minimal version of a canonical E1, whether it can simultaneously transfer two UBL molecules to associated E2(s) or whether like canonical E1s it has evolved an asymmetric mechanism for initiating UFM1 activation. Moreover, the targets of UFM1 will need to be identified in order to begin to reveal biological roles for the UFM1-UBA5-UFC1 pathway.
Future directions
Our understanding of the important roles of the E1 enzymes has increased dramatically since the discovery of UBA1 in the early 1980s. Nevertheless, there is still much to be discovered. The detailed mechanism of UBL transfer to the Cys residue of an E1 enzyme, and the subsequent transfer to E2 are still poorly understood, as is our knowledge of conformational changes associated with these reactions. Additionally, it is unclear why the number of E2s working with UBA1 has been so dramatically expanded relative to the other cascades, which use a limited repertoire of E2s. In principle, this could reflect the wider array of ubiquitin targets, but it also indicates the special nature of UBA1 as a master activator of most E2s.
Further questions concern why particular UBL classes have expanded into multiple members. As examples, the distinct functions of the 3 SUMO and 7 Atg8 family members in humans remain incompletely understood (FIG. 1), as do any differences in their mechanisms of activation. Finally, E1 enzymes are unique in the UBL conjugating pathway in that they are the only components that employ ATP, and this property is being exploited to develop selective inhibitors. The potential for the use of such inhibitors in therapy is high, given the links seen between components of UBL cascades and human disease.
On-line Summary
- At the apex of each ubiquitin-like protein (UBL) cascade is an E1 enzyme, which activates the UBL and then directs the UBL to downstream pathways. UBL proteins and E1 enzymes have their origins in prokaryotic biosynthetic pathways.
- In humans, eight E1 enzymes are known to initiate UBL conjugation. We refer to E1s for ubiquitin (UBA1, UBA6), SUMO (SAE1-SAE2), NEDD8 (NAE1-UBA3) and ISG15 (UBA7) as ‘canonical’, due to their related domain structures and enzymatic mechanisms, and to the more divergent E1s for URM1 (UBA4), UFM1 (UBA5) and ATG12 and ATG8 isoforms (ATG7) as ‘non-canonical’.
- Canonical E1 structures all display an ‘adenylation domain’ that resembles prokaryotic ancestors and two canonical-E1-specific domains: a ‘catalytic Cys domain’ harbouring the Cys that is involved in the E1~UBL thioester linkage and a carboxyl-terminal ubiquitin-fold domain (UFD) that resembles ubiquitin that binds E2.
- In addition to their chemical roles in initiating UBL conjugation cascades, E1s also establish specificity, by matching a particular UBL with only cognate E2s. Rules for how this specificity is achieved are only beginning to emerge, but it is clear that specificity is achieved at multiple levels.
- A particularly intriguing non-canonical E1 is UBA4. Recent data indicate that UBA4 initiates a sulfur-transfer pathway, which is related to bacterial E1 -like enzymes.
- E1 enzymes are unique in the UBL conjugating pathway in that they are the only components that employ ATP, and this property is being exploited to develop selective inhibitors. The potential for the use of such inhibitors in therapy is high, given the links seen between components of UBL cascades and human disease.
Acknowledgments
We thank the NIH (J.W.H. and B.A.S.) and Millennium Pharmaceuticals, Inc (J.W.H.) for funding our work on E1 enzymes and Catherine Regni for assistance with Figure 2. B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Rhodanese-homology domain
A domain sharing sequence and structural homology to rhodanese, a mitochondrial enzyme that catalyzes Cys-mediated sulfur transfer reactions
- Adenylation
Synthesis of a phosphodiester bond between a hydroxyl group and the phosphate group of AMP. In the case of ubiquitin-like (UBL) protein conjugation cascades, the hydroxyl is from the C terminus of the UBL protein
- Thioester bond
Covalent linkage of a sulfur with an acyl group. In the case of ubiquitin-like (UBL) cascades, the Cys sulfur of an enzyme is linked to the terminal carbon of a UBL protein
- 26S Proteasome
Large multisubunit protease complex that selectively degrades multiubiquitylated proteins. It contains a 20S particle that carries the catalytic activity and two regulatory 19S particles
- UBL fold
A structural motif also known as a β-GRASP fold that has a domain has a β(2)-α-β(2) structure
- PP- loop ATPase domain
A structural motif found in a family of conserved ATP binding proteins that serve as tRNA modifying enzymes and activate the tRNA through adenylation
- elongator (ELP) complex
An enzyme complex that replaces the hydrogen atom on the 5 position of uridine at position 34 of tRNAs by a methoxy-carbonyl-methyl (mcm5) group
- Thiouridinylation
An enzymatic process in which the 2′-oxygen of uridine within the anti-codon of a tRNA is replaced by sulfur
Biographies
Brenda Schulman is an Investigator of the Howard Hughes Medical Institute and a Member in the Departments of Structural Biology and Genetics/Tumor Cell Biology at St. Jude Children’s Research Hospital, and she also holds an affiliate faculty position at the University of Tennessee College of Medicine. She received her bachelor’s degree in Biology from the Johns Hopkins University in 1989. Following obtaining her Ph.D. in Biology from M.I.T. in 1996, she did postdoctoral studies in cell cycle research at Massachussetts General Hospital, and in X-ray crystallography at Memorial Sloan-Kettering Cancer Center. Her current research is focused on understanding the conjugation pathways for ubiquitin and ubiquitin-like proteins, and on the roles of these pathways in controlling cell division.
J. Wade Harper is the B and N Vallee Professor of Molecular Pathology in the Department of Pathology at Harvard Medical School. He obtained a Ph.D. in Chemistry from the Georgia Institute of Technology in 1984 and performed post-doctoral work at Harvard Medical School, prior to starting an independent faculty position in the Department of Biochemistry and Molecular Biology at Baylor College of Medicine in 1988. In 1996, he was promoted to full professor and in 2003, moved to his current position. His work focuses on cell cycle control by protein phosphorylation and the ubiquitin system from biochemical and genetic perspectives.
References
- 1.Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5944–5967. doi: 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. Describes the chromatographic separation of E1, E2 and E3 activities and demonstrates reconstitution of the first ubiquitin ligase reaction. [PubMed] [Google Scholar]
- 4.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. CurrOpin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser M. Ubiquitin signalling: what’s in a chain? Nature Cell Biol. 2004;6:571–572. doi: 10.1038/ncb0704-571. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Biochemistry. All in the ubiquitin family. Science. 2000;289:563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 13.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature Cell Biol. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 14.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 15.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nature Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 18.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 19.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 20.Yamoah K, et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Xi J, Begley TP, Nicholson LK. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat Struct Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 25.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. Describes the structure of the MoeB-MoaD complex and reveals the mechanism of MoaD adenylation. This is the only structure to date of an E1 -like enzyme with an adenylate. [DOI] [PubMed] [Google Scholar]
- 26.Taylor SV, et al. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 27.Leimkuhler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276:34695–34701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 28.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. J Mol Biol. 2005;349:774–786. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 31.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc. 2008;130:3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 32.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-Like Protein Involved in the Proteasome Pathway of Mycobacterium tuberculosis. Science. 2008 doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath JP, Jentsch S, Varshavsky A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. Embo J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handley PM, Mueckler M, Siegel NR, Ciechanover A, Schwartz AL. Molecular cloning, sequence, and tissue distribution of the human ubiquitin-activating enzyme E1. Proc Natl Acad Sci U S A. 1991;88:258–262. doi: 10.1073/pnas.88.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatfield PM, Callis J, Vierstra RD. Cloning of ubiquitin activating enzyme from wheat and expression of a functional protein in Escherichia coli. J Biol Chem. 1990;265:15813–15817. [PubMed] [Google Scholar]
- 36.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. Embo J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 38.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 39.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 40.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. Embo J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu M, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. Embo J. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. Together with references 45 and 46, this paper identified UBA6/UBE1L2 as a second activating enzyme for ubiquitin, and identified USE1 as a specific E2 conjugating enzyme for UBA6. [DOI] [PubMed] [Google Scholar]
- 45.Pelzer C, et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. J Biol Chem. 2007;282:23010–23014. doi: 10.1074/jbc.C700111200. [DOI] [PubMed] [Google Scholar]
- 46.Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. Identifies UBA6 as a candidate E1 for FAT10. [DOI] [PubMed] [Google Scholar]
- 47.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 48.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23:1958–1971. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 49.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. Describes the first crystal structure of the canonical E1 protein complex responsible for activating NEDD8, and defines the domain structure of canonical E1s. [DOI] [PubMed] [Google Scholar]
- 50.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. Describes the first structure of the heterodimeric SUMO activating enzyme bound to SUMO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. Describes the first structure of an activating enzyme for ubiquitin in complex with ubiquitin. [DOI] [PubMed] [Google Scholar]
- 52.Komatsu M. etal. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- 53.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 54.Schmitz J, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 55.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 56.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- 57.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–2548. Demonstrates that UBA1 becomes doubly loaded with ubiquitin, with one molecule in a thioester with UBA1’s catalytic cysteine and the second ubiquitin as an adenylate. [PubMed] [Google Scholar]
- 58.Haas AL, Warms JV, Rose IA. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry. 1983;22:4388–4394. doi: 10.1021/bi00288a007. [DOI] [PubMed] [Google Scholar]
- 59.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci U S A. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. Suggested a two step mechanism for ubiquitin activation, the first involving adenylation and the second involving thioester formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciechanover A, Elias S, Heller H, Hershko A. Covalent affinity” purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–2542. Describes the purification of E1 ubiquitin activating enzyme, UBA1, which used the thioester as a tool for recovery of active enzyme. The ability to purify active UBA1 revolutionized the field. [PubMed] [Google Scholar]
- 61.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–1581. Demonstrated the existence of a family of E2 conjugating enzymes that are charged by UBA1, foreshadowing the discovery that distinct E2s are involved in distinct biological pathways. [PubMed] [Google Scholar]
- 62.Haas AL, Bright PM. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J Biol Chem. 1988;263:13258–13267. [PubMed] [Google Scholar]
- 63.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988;263:13268–13275. [PubMed] [Google Scholar]
- 64.Pickart CM, Kasperek EM, Beal R, Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1) J Biol Chem. 1994;269:7115–7123. [PubMed] [Google Scholar]
- 65.Tokgoz Z, Bohnsack RN, Haas AL. Pleiotropic effects of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J Biol Chem. 2006;281:14729–14737. doi: 10.1074/jbc.M513562200. [DOI] [PubMed] [Google Scholar]
- 66.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 67.Walden H, et al. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 68.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. Reports the first structure of a doubly loaded canonical E1 enzyme, and defines a thioester switch responsible for promoting transfer of the ubiquitin like protein NEDD8 to its E2, UBC12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao C, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang DT, et al. A unique E1–E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 72.Souphron J, et al. Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8’s E1. Biochemistry. 2008;47:8961–8969. doi: 10.1021/bi800604c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkinson KD, et al. A specific inhibitor of the ubiquitin activating enzyme: synthesis and characterization of adenosyl-phospho-ubiquitinol, a nonhydrolyzable ubiquitin adenylate analogue. Biochemistry. 1990;29:7373–7380. doi: 10.1021/bi00484a004. [DOI] [PubMed] [Google Scholar]
- 74.Madden MM, et al. Substrate properties of ubiquitin carboxyl-terminally derived peptide probes for protein ubiquitination. Biochemistry. 2008;47:3636–3644. doi: 10.1021/bi702078m. [DOI] [PubMed] [Google Scholar]
- 75.Szczepanowski RH, Filipek R, Bochtler M. Crystal structure of a fragment of mouse ubiquitin-activating enzyme. J Biol Chem. 2005;280:22006–22011. doi: 10.1074/jbc.M502583200. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan ML, Vierstra RD. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991;266:23878–23885. [PubMed] [Google Scholar]
- 77.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 78.Durfee LA, Kelley ML, Huibregtse JM. The basis for selective E1–E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee TV, et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 81.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tatham MH, et al. Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry. 2003;42:9959–9969. doi: 10.1021/bi0345283. [DOI] [PubMed] [Google Scholar]
- 84.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 85.Wang J, et al. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol Cell. 2007;27:228–237. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haas AL. Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol Cell. 2007;27:174–175. doi: 10.1016/j.molcel.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 87.Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15:280–287. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- 88.Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. Identifies UBA1 as the protein mutated in the ts85 cell line and demonstrates that UBA1 is responsible for the vast majority of ubiquitin conjugation in mammalian cells. [DOI] [PubMed] [Google Scholar]
- 89.Ciechanover A, Finley D, Varshavsky A. Mammalian cell cycle mutant defective in intracellular protein degradation and ubiquitin-protein conjugation. Prog Clin Biol Res. 1985;180:17–31. [PubMed] [Google Scholar]
- 90.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. Embo J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- 92.Rocca A, Lamaze C, Subtil A, Dautry-Varsat A. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor beta chain to late endocytic compartments. Mol Biol Cell. 2001;12:1293–1301. doi: 10.1091/mbc.12.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swanson R, Hochstrasser M. A viable ubiquitin-activating enzyme mutant for evaluating ubiquitin system function in Saccharomyces cerevisiae. FEBS Lett. 2000;477:193–198. doi: 10.1016/s0014-5793(00)01802-0. [DOI] [PubMed] [Google Scholar]
- 94.Ghaboosi N, Deshaies RJ. A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol Biol Cell. 2007;18:1953–1963. doi: 10.1091/mbc.E06-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kulkarni M, Smith HE. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 2008;4:e1000131. doi: 10.1371/journal.pgen.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme Uba1 causes tissue overgrowth in Drosophila. Fly (Austin) 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- 97.Nollen EA, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramser J, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet. 2008;82:188–193. doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stephen AG, Trausch-Azar JS, Ciechanover A, Schwartz AL. The ubiquitin-activating enzyme E1 is phosphorylated and localized to the nucleus in a cell cycle-dependent manner. J Biol Chem. 1996;271:15608–15614. doi: 10.1074/jbc.271.26.15608. [DOI] [PubMed] [Google Scholar]
- 100.Nouspikel T, Hanawalt PC. Impaired nucleotide excision repair upon macrophage differentiation is corrected by E1 ubiquitin-activating enzyme. Proc Natl Acad Sci U S A. 2006;103:16188–16193. doi: 10.1073/pnas.0607769103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grenfell SJ, Trausch-Azar JS, Handley-Gearhart PM, Ciechanover A, Schwartz AL. Nuclear localization of the ubiquitin-activating enzyme, E1, is cell-cycle-dependent. Biochem J. 1994;300 (Pt 3):701–708. doi: 10.1042/bj3000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dohmen RJ, et al. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 103.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 104.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 105.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 106.Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- 107.Chen Y, McPhie DL, Hirschberg J, Neve RL. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J Biol Chem. 2000;275:8929–8935. doi: 10.1074/jbc.275.12.8929. [DOI] [PubMed] [Google Scholar]
- 108.Kumar A, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. Embo J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2:930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schlieker C, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the Ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0808756105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009 doi: 10.1038/nature07643. in press. [DOI] [PubMed] [Google Scholar]
- 112.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 113.Huang B, Lu J, Bystrom AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. Rna. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanida I, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yamada Y, et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- 116.Mizushima T, et al. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem Biophys Res Commun. 2007;362:1079–1084. doi: 10.1016/j.bbrc.2007.08.129. [DOI] [PubMed] [Google Scholar]
- 117.Sekizawa R, et al. Panepophenanthrin, from a mushroom strain, a novel inhibitor of the ubiquitin-activating enzyme. J Nat Prod. 2002;65:1491–1493. doi: 10.1021/np020098q. [DOI] [PubMed] [Google Scholar]
- 118.Yang Y, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 119.Tong H, Hateboer G, Perrakis A, Bernards R, Sixma TK. Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J Biol Chem. 1997;272:21381–21387. doi: 10.1074/jbc.272.34.21381. [DOI] [PubMed] [Google Scholar]
- 120.Worthylake DK, Prakash S, Prakash L, Hill CP. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J Biol Chem. 1998;273:6271–6276. doi: 10.1074/jbc.273.11.6271. [DOI] [PubMed] [Google Scholar]