Abstract
The self-association of misfolded or damaged proteins into ordered amyloid-like aggregates characterizes numerous neurodegenerative disorders. Insoluble amyloid plaques are diagnostic of many disease states. Yet soluble, oligomeric intermediates in the aggregation pathway appear to represent the toxic culprit. Molecular chaperones regulate the fate of misfolded proteins and thereby influence their aggregation state. Chaperones conventionally antagonize aggregation of misfolded, disease proteins and assist in refolding or degradation pathways. Recent work suggests that chaperones may also suppress neurotoxicity by converting toxic, soluble oligomers into benign aggregates. Chaperones can therefore suppress or promote aggregation of disease proteins to ameliorate the proteotoxic accumulation of soluble, assembly intermediates.
Key words: chaperone, heat shock protein, protein aggregation, amyloid, Hsp70, Hsp40, prion
Introduction
Many neurodegenerative disorders are characterized by the misfolding and subsequent aggregation of toxic proteins. Alzheimer, Parkinson, and the glutamine-encoding expansion diseases are examples of conformational disorders which show late onset symptoms of progressive neuronal dysfunction and eventual neuronal loss within certain brain regions.1 The disease-causing protein can adopt aberrant conformations due to age or stress which enables its aggregation and ultimate accumulation in amyloid fibrils (Fig. 1).2,3 Amyloids share a β-sheet-rich, fibrillar protein conformation in which the β-sheets stack parallel or anti-parallel to one another and run perpendicular to the fiber axis.4,5 Numerous pathogenic substrates as well as yeast prion proteins can exist in alternate, amyloid-like states that are characterized by a defined set of biochemical parameters including the ability to bind indicator dyes such as Thioflavin or Congo Red as well as resistance to both protease digestion and detergent solubilization.6 Though the appearance of amyloid is diagnostic of the disease state, the extent of amyloid accumulation often does not correlate with the disease symptoms.5,7 In fact, a wide variety of organisms utilize functional forms of amyloid-like structures to perform important cellular processes such as phenotypic adaptation in yeast8–10 and melanin synthesis in humans.11 These and other lines of evidence suggest that amyloid formation per se is not toxic but may represent a benign byproduct or even a protective mechanism.12–14
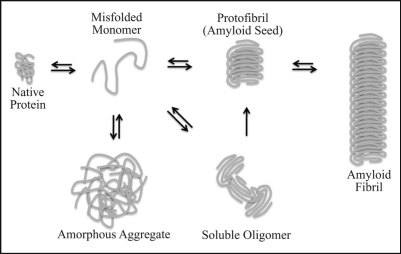
Figure 1.
Schematic representation of the amyloid assembly pathway. A disease causing protein can adopt a non-native conformation due to intrinsic and/or environmental stress. The misfolded, disease protein can then self-associate into different intermediate structures such as detergent-soluble oligomers and/or protofibrils whose intracellular accumulation correlate with cell death. In this model, we speculate about the structural properties of the ill-defined intermediates. Alternatively, the misfolded protein can accumulate as non-specific, amorphous aggregates. The formation of amyloid seeds drives the autocatalytic conversion of native and misfolded protein monomers into β-sheet-rich, amyloid aggregates.
Soluble intermediates in the amyloid assembly pathway appear to represent the toxic species.7,15 Relatively little is known about the exact nature of the assembly intermediates due to their dynamic structure (Fig. 1). Antibodies generated against the Aβ42-peptide (proteolytically cleaved peptide from the Amyloid Precursor Protein, APP, and a causative agent for Alzheimers disease) could recognize assembly intermediates formed by numerous amyloidogenic substrates and buffer their cytotoxicity.15 Subsequent work demonstrated that the same conformational-specific antibodies could selectively inhibit spontaneous amyloid formation of yeast prions in vitro.16,17 Thus, amyloid assembly intermediates, which include soluble oligomers, do exhibit common conformation-dependent structures that are unique to the intermediates regardless of the amino acid sequence.
The accumulation of oligomeric amyloid assembly intermediates closely correlates with cell death7 and distinct clearance pathways maintain the proteotoxic oligomers at low levels.18 This can be accomplished by either solubilizing the aggregation-prone substrate or driving its assembly into higher-ordered amyloid-like aggregates. The ability to promote aggregation does not necessarily represent the cell's primary means of clearance but may provide backup pathways when solublization machinery has become compromised by age or environmental insult.18 The nature and identity of the unfavorable interactions between the soluble oligomers and other cellular components is just being defined and may result in transcriptional deregulation,19–21 proteasomal inactivation,22 aberrant signaling23 or membrane damage.24–26 It should be noted that disease proteins associated with conformational disorders perform a wide variety of cellular functions and scenarios may exist where protein species other than amyloid assembly intermediates represent the predominant toxic species.
All cells contain quality control machinery termed molecular chaperones to maintain the appropriate folding state of proteins. Many of these molecular chaperones have been termed Heat-Shock Proteins (HSP) due to their increased expression upon heat treatment of cell and have been divided into six major families including HSP100, HSP90, HSP70, HSP60, HSP40 and the small HSPs.27 Chaperones recognize misfolded protein conformers and ultimately coordinate the appropriate folding or degradation pathways. As the first line of defense against conformational disorders, chaperones maintain the solubility of misfolded, disease-related substrates by directly binding to non-native conformers or disassembling aggregates formed by the disease protein.27 Alternatively, recent evidence suggests that chaperones can also promote aggregation and thereby prevent the toxic accumulation of amyloid assembly intermediates.13,28 Herein we discuss how molecular chaperones manage proteotoxic misfolding events of numerous amyloidogenic substrates.
Molecular Chaperones Antagonize Aggregation of Disease Proteins
Chaperones act by multiple means to maintain the solubility of disease-causing proteins. Chaperone-mediated solubilization can occur in the initial stages of the amyloid assembly pathway by preventing the self-association of misfolded, disease proteins into toxic, oligomeric intermediates. Alternatively, chaperones also act later in the assembly pathway to dismantle amyloid-like aggregates and resolubilize the disease protein. Thus chaperone intervention can either maintain or generate disease protein monomers which can be properly refolded or marked for degradation by the ubiquitin-proteasome system or autophagy pathways. This reduces the flux of misfolded, disease proteins through different stages of the amyloid assembly pathway and decreases the cellular concentration of proteotoxic assembly intermediates.
Chaperones prevent disease proteins from assembling into amyloid-like aggregations.
Chaperone machinery can act early in the amyloid assembly pathway by recognizing misfolded, disease proteins and subsequently preventing their aggregation. The Hsp70 chaperone system has been well-defined in its role of preventing the assembly of different, disease-causing proteins into amyloid-like aggregates.27,29 The Hsp70 family of chaperone proteins is abundantly expressed throughout the cell and contains 11 known isoforms in humans.30 Though not as abundant as its Hsp70 partner, the Hsp40 co-chaperone (also referred to as J-proteins due to their highly conserved J-domain) represent a more diverse family of proteins which contain 41 isoforms in humans and are present in most every intracellular compartment.31 The structural and functional diversity of Hsp40 co-chaperones provide specific substrate recognition properties for the Hsp70 work horse.32
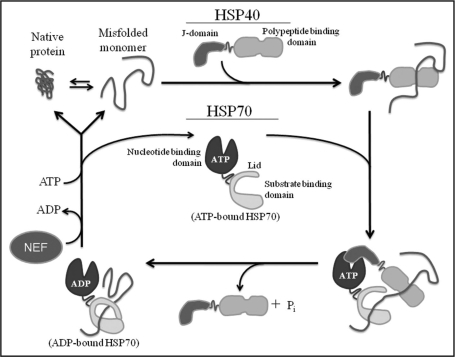
The misfolding of disease proteins can arise from numerous intrinsic or environmental stresses3 and Hsp70 cooperates with different Hsp40 co-chaperones to ultimately recognize and refold the non-native protein conformers (Fig. 2). Hsp40 can bind hydrophobic peptides which are normally buried in the core of natively folded proteins but become solvent-exposed in misfolded states.33 The polypeptide binding domain of Hsp40 binds non-native proteins and delivers them to Hsp70.34,35 The ability of Hsp70 to bind the misfolded substrate is tightly regulated through an ATP hydrolytic cycle. Once in a complex with Hsp70, the J-domain of Hsp40 stimulates ATP hydrolysis on Hsp70 which promotes a conformational change in the Hsp70 substrate-binding domain and increases its binding affinity for the misfolded substrate.36–38 Hsp40 is liberated from the Hsp70:substrate complex following ATP hydrolysis yet it is unclear how co-chaperone release occurs. The substrate in complex with Hsp70 can be properly refolded, marked for degradation via Hsp70 coordination with degradation machinery, or released to re-enter this binding cycle. The substrate remains tightly bound to Hsp70 until ADP is exchanged for ATP via a diverse class of nucleotide exchange factors (NEF).39 It is in this manner that cooperation between Hsp40 and Hsp70 prevents the proteotoxic accumulation of misfolded, aggregation-prone proteins.
Figure 2.
Hsp70-Hsp40 hydrolytic cycle for the binding and refolding of non-native proteins. A native protein can adopt a non-native conformation. Hsp40 co-chaperones can recognize the misfolded protein and bind it via its polypeptide binding domain. Hsp40 delivers the non-native substrate to Hsp70 and its J-domain stimulated ATP hydrolysis on Hsp70. This promotes a conformational change in Hsp70 via closure of the lid domain and increases the binding affinity of Hsp70 for the substrate. Hsp70 along with other cellular components not shown in this model can assist in the proper refolding of the substrate. Nucleotide exchange factors (NEF) release ADP and load ATP back onto the nucleotide binding domain of Hsp70 which promotes substrate release.
The involvement of nucleotide exchange factors (NEF) adds yet another level of regulation to the Hsp70 system. This diverse family of exchange factors is critical for the functional cycle of Hsp70 because they replace ADP for ATP on Hsp70 and trigger substrate release.39 Well characterized NEFs include the bacterial GrpE, which facilitates nucleotide dissociation from the Hsp70 (DnaK),37,40 and the BAG family of eukaryotic proteins which are NEFs for different cytosolic Hsp70s.41 A third family of proteins which possess nucleotide exchange activity including the cytosolic Fes1 and ER luminal Sls1 in Saccharomyces cerevisiae,42,43 and the human ortholog HspBP1 (Hsp70 binding protein 1).44 Interestingly, other molecular chaperones are capable of nucleotide exchange on Hsp70. The yeast Hsp110, Sse1, regulates nucleotide exchange on different cytosolic Hsp70s,45 while the Hsp170, Lhs1, stimulates nucleotide exchange of the ER Hsp70, Kar2.46 Sse1 is a particularly interesting exchange factor because its activity in yeast is required for the propagation of the [URE3] prion,47 as well as the formation and propagation of the [PSI+] prion.48 The recent findings which implicate Sse1 in the formation and propagation of amyloid-like prions may establish a better understanding of how nucleotide exchange on Hsp70 affects amyloid dynamics. Thus, the cell can utilize this diverse family of NEFs to regulate the different activities of Hsp70 in distinct subcellular compartments.
The ability of Hsp70 and Hsp40 to suppress the assembly of disease proteins into amyloid-like aggregates has been well documented in numerous model systems,27 yet only a few examples will be discussed. Glutamine-encoding repeats within a set of unrelated proteins are the cause of at least nine different late onset neurodegenerative disorders.49,50 Elevating intracellular pools of Hsp70 as well as Hsp40 reduce expanded-polyQ aggregate formation and suppressed toxicity in cultured cells.51–53 Due to the-high conservation of chaperone machinery between eukaryotic cells, Saccharomyces cerevisiae has emerged as a model system to study protein aggregation and toxicity. Overexpression of the yeast Hsp70, Ssa1, or the Hsp40, Ydj1, inhibit the formation of large, detergent-insoluble aggregates by an expanded-polyQ protein containing the N-terminal sequence of the huntingtin protein, the causative gene for Huntington disease.54 Ydj1 also reduced the aggregation of the Q/N-rich prion domain of the yeast prion, Rnq1, which correlated with its ability to suppress prion toxicity.55 Elevated expression of another yeast Hsp40, Sis1, reduced the aggregation of expanded-polyQ Htt protein and suppressed toxicity in [psi−] yeast, which are void of the amyloid-like [PSI+] prion.56 Hsp70 and Hsp40 have also been shown to suppress the aggregation and toxicity of numerous other disease-causing proteins including mutant superoxide dismutase 1 (SOD),57 the causative gene for familial amyotrophic lateral sclerosis, and α-synuclein,58 the causative gene for Parkinson disease. Thus, the ability of Hsp70 and Hsp40 to prevent assembly of amyloid-like aggregates and suppress cytotoxicity extends to numerous disease-causing proteins in a variety of model organisms.
Molecular chaperones solubilize protein aggregates.
Chaperones can act later in the aggregation pathway to disassemble amyloid-like particles. Although it is unclear whether mammalian cells possess this disaggregation activity, fungi, plants and bacteria express the AAA ATPase protein remodeling factor, Hsp104 (ClpB in E. coli).59 The hexameric Hsp104 works synergistically with the Hsp70 chaperone system to not only resolubilize misfolded substrates, but to also restore their proper function.60 Interestingly, normal Hsp104 function is required in a yeast model to maintain different prion proteins in their amyloid-like prion conformation61–64 by generating small prion seeds via fragmentation of larger prion aggregates which enables new rounds of prion propagation.65 Further in vitro studies demonstrated that Hsp104 couples ATP hydrolysis with the rapid disassembly of amyloid-like fibers and oligomeric intermediates formed by the yeast prions [PSI+] and [URE3].17,66 Thus, it is not surprising that Hsp104 overexpression was able to efficiently solubilize amyloid-like aggregates formed by the expanded-polyQ huntingtin (Htt) fragment and reduce cell death in a yeast model.67 Yet removal of Hsp104 via gene deletion also subdued the toxicity caused by overexpression of the expanded-polyQ Htt fragment.68 Thus, Hsp104 may indirectly affect polyQ aggregation and toxicity in yeast by acting through the yeast prions, [PSI+] and/or [RNQ+] (also termed [PIN+]63), which can serve as Q/N-rich templates to alter the conformation of the expanded-polyQ protein into an aggregation-prone, toxic form.56,68,69
To clarify the role of Hsp104 in polyQ aggregation and toxicity, the yeast Hsp104 chaperone was introduced into a number of higher eukaryotes and retained its ability to remodel amyloid-like assemblies. In a C. elegans model, expression of Hsp104 reduced the aggregation of expanded-polyQ proteins and abrogated developmental delays.70 Similarly in a rat model, the presence of Hsp104 suppressed expanded-polyQ toxicity in a manner which correlated with altered distribution and number of the polyQ assemblies.71 The small heat shock proteins, Hsp26 in yeast and Hsp27 in rats, were identified as potentiators for Hsp104-mediated suppression of polyQ aggregation and toxicity.67,71 Utilizing a Parkinson disease model in rats, Hsp104 was also shown to antagonize the formation oligomeric intermediates and amyloid fibers by α-synuclein which reduced dopaminergic degeneration.72 Thus, molecular chaperones can act at multiple stages in the aggregation pathway to both coordinate the disassembly of amyloid-like aggregates and prevent the self-association of non-native protein monomers.
Molecular chaperones participate in the degradation of proteotoxic substrates by the ubiquitin-proteasome system or chaperone-mediated autophagy.
Under certain conditions, chaperone machinery cannot repair the misfolded protein and must coordinate its degradation by either the ubiquitin-proteasome pathway or lysosomal-mediated autophagy. CHIP (carboxy terminus of Hsc70-interacting protein) is a versatile protein that acts as a co-chaperone to Hsc70 (Heat shock cognate 70) or Hsp70, possesses intrinsic chaperone activity in its own right and functions as an E3 ligase to mediate the transfer of polyubiquitin chains to misfolded substrates.73–75 The Hsp70 interacting protein, Bag2 (Bcl2-associated athanogene 2), can inhibit the ubiquitin ligase activity of CHIP and promote substrate refolding by Hsp70.76,77 Overexpression of CHIP increased the ubiquitination and degradation of expanded-polyQ huntingtin and ataxin-3, the causative gene for Spinocerebellar ataxia type 3 (SCA3).78 This subsequently reduced polyQ aggregation levels and suppressed cell death. The suppressive activity by CHIP overexpression became more prominent when Hsc70 levels were also elevated. Similar results were obtained which demonstrated CHIP's ability to reduce aggregation of expanded-polyQ substrates and suppress cell death in transfected cell lines, primary neurons and zebrafish models.79 Therefore, CHIP along with other cofactors can partition Hsp70 substrates between refolding and degradation pathways.
The autophagy pathway represents a mechanism independent of the ubiquitin-proteosome system which participates in the intracellular bulk degradation of organelles as well as misfolded and aggregated proteins. Autophagy entails the packaging/engulfment of cytoplasmic contents into autophagosome vesicles which fuse with lysosomes, wherein the contents are degraded.80 Loss of autophagy in mice via disruption of ATG5 manifested features characteristic of neurodegeneration even in the absence of any disease-associated mutant proteins.81 Molecular chaperones have been shown to participate in the recognition and subsequent packaging of misfolded proteins into autophagasomes which has become termed chaperone-mediated autophagy (CMA).82 CMA involves the selective targeting of proteins containing a KFERQ peptide motif to lysosomes.83 The rate-limiting step in CMA involves chaperone/cargo binding to the lysosomal receptor, Lamp2a.84 In this manner, chaperones can recognize misfolded or aggregated proteins and target them via the KFERQ motif to lysosomes for degradation. The causative agent in Parkinson disease, α-synuclein, possesses a pentapeptide sequence which is consistent with the CMA recognition motif and targets the disease protein to lysosomes.85 The pathogenic A53T and A30P mutations in α-synuclein were capable of binding the lysosomal receptors but inhibited both their own degradation as well as other substrates. Additionally, CMA has also been linked to polyQ-expanded huntingtin toxicity through the small heat shock protein, HspB8 and Bag3. In cultured cells, elevated expression of HspB8 prevented the intracellular accumulation of polyQ-expanded huntingtin protein.86 Subsequent work demonstrated that HspB8 works in complex with Bag3 to stimulate the degradation of polyQ-expanded huntingtin protein by macroautophagy.87 Thus, chaperones can couple their activity with multiple degradation pathways to remove toxic, disease proteins.
Chaperones Promote Aggregation of Toxic Proteins
Chaperones are also capable of packaging disease proteins into tight, amyloid-like aggregates and thereby reducing the accumulation of soluble, oligomeric intermediates. Elevated expression of the human Hsp40, Hdj2, increased the aggregation of expanded-polyQ huntingtin protein in COS-7 cells.88 Subsequent in vitro studies demonstrated that purified forms of Hsp70 and Hsp40, Hdj1, were able to attenuate polyQ oligomer assembly and drive the formation of amyloid-like fibrillar structures.89 These data suggest that chaperones can partition disease proteins between oligomeric and fibrillar aggregation states. Chaperones other than Hsp70 and Hsp40 can take an active role in promoting protein aggregation. The proteasomal chaperones, Rpt4 and Rpt6 (AAA ATPase subunits of the 19S proteasomal particle), facilitated the aggregation of different expanded-polyQ disease proteins, huntingtin and ataxin-3, without affecting proteasomal degradation.90 Furthermore, in vitro reconstitution experiments showed that purified 19S proteasomal particles enhanced aggregation of expanded-polyQ huntingtin protein.90 Thus, a number of molecular chaperones can promote the assembly of disease proteins into amyloid-like aggregates.
Chaperone-mediated aggregation reduces proteotoxicity.
Recent work has begun to link chaperone-mediated aggregation of toxic proteins with suppression of toxicity. The chaperonin TriC complex was identified as potent suppressor of polyQ-mediated toxicity in C. elegans.91 This family of chaperonins form multi-subunit, cage-like structures that sequester non-native proteins92 and work alongside Hsp70 chaperone systems to promote substrate refolding in the cytosol upon rounds of ATP hydrolysis.93,94 Substrate refolding occurs within the central cavity of the chaperonin complex and substrate release occurs after folding is complete.95 Interestingly, reducing the cellular concentration of TriC resulted in the accumulation of low-molecular weight, soluble oligomers by polyQ-expansions which correlated with toxicity in cell culture lines and yeast.28,96,97 One group demonstrated by size exclusion chromotagraphy that toxic oligomers were approximately 200 kDa in size,98 yet the chaperonin complex remodeled the polyQ-expansions into higher molecular weight aggregates around the 500 kDa size range.28 The ability of the chaperonin complex to remodel polyQ-expanded disease proteins worked synergistically with Hsp70, Ssa1 and Hsp40, Ydj1. In contrast, studies in [psi−] yeast demonstrate that excess Ydj1 increased aggregation of expanded-polyQ Htt, yet enhanced toxicity.56 Though these data seems contrary to the work done with TriC, the nature and size of the polyQ aggregates may differ between experimental conditions and account for whether the polyQ protein elicits a toxic or benign outcome. Additionally, cellular factors such as endogenous prions add to complexity of how chaperone networks buffer the accumulation of expanded-polyQ aggregates in a yeast model. Nevertheless, this provides correlative evidence that chaperonin complexes can not only solubilize their substrates but also promote the efficient packaging of proteotoxic species into benign aggregates.
Related studies in yeast have recently demonstrated the protective effects of Hsp40-mediated aggregate formation. The Hsp40, Sis1, is an essential chaperone99 that participates in protein refolding,100 protein translocation101 and translation initiation.102 Sis1 activity is also required to maintain the Rnq1 yeast prion in its [RNQ+] amyloid-like, prion conformation.103 Endogenous Rnq1 is not toxic to yeast yet modest elevation of Rnq1 levels induced cell death.13 Rnq1 toxicity is dependent on the presence of pre-existing [RNQ+] prions as elevated Rnq1 levels are not toxic to [rnq−] cells in which the [RNQ+] prion conformer is absent. Thus, [RNQ+] prion seeds provide a template which can alter the conformation of native Rnq1 into a toxic form.13 Elevating Sis1 levels was able to suppress Rnq1 toxicity in a manner that correlated with increased formation of [RNQ+] prion assemblies and decreased amounts of soluble Rnq1. Point mutations (Rnq1 L94A) within the Sis1 binding site of Rnq1 enabled it to assume an aberrant conformation which formed SDS-soluble aggregates and became toxic in [rnq−] cells. Interestingly, Sis1 overexpression was able to detoxify excess Rnq1 L94A in [RNQ+] cells yet was unable to suppress Rnq1 L94A toxicity in [rnq−] cells. Therefore, Sis1 detoxification requires [RNQ+] prion seeds so that nascent Rnq1 can be efficiently packaged into benign, amyloid-like aggregates. Inefficiencies in the [RNQ+] prion assembly pathway due to compromised Sis1 activity exacerbated cell death.
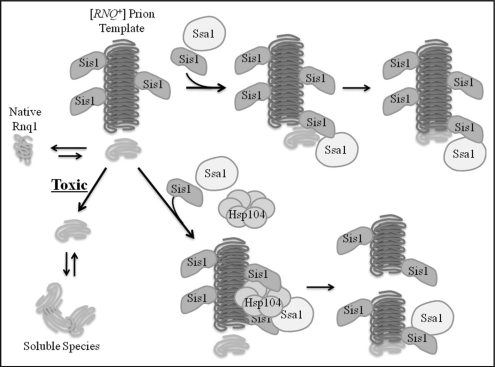
The molecular mechanism by which chaperones facilitate Rnq1 assembly into amyloid-like prions is not clear. Sis1 was previously shown to work in concert with Hsp104 to mediate the fragmentation of larger [RNQ++] prion aggregates.104 This generates more [RNQ+] prion seeds and more nucleation points to further drive the autocatalytic conversion of nascent Rnq1 into the prion conformer. The Ssa subclass of Hsp70 chaperones positively affects the formation and propagation of amyloid-like [PSI+] prions in yeast105–107 (discussed in a previous Prion review article).108 Yet the role of Hsp70 in [RNQ+] prion assembly remains elusive. It does appear that [RNQ+] prion shearing alone is not sufficient to suppress Rnq1 toxicity as overexpression of Hsp104 or different Hsp70 members was unable to suppress Rnq1 toxicity like excess Sis1.13 Therefore, we suggest that Sis1 may perform an additional function as an elongation factor in the [RNQ+] prion assembly process (Fig. 3). Sis1 was shown to bind a region of Rnq1 outside the Q/N-rich prion domain13 in equimolar ratios.109 Thus Sis1 may coat [RNQ+] prions and facilitate the stable association of nascent Rnq1 with [RNQ+] prions which would subsequently promote its efficient conversion into the growing amyloid-like [RNQ+] aggregate. Yet it is difficult to judge the relative importance of Sis1 action in fragmentation versus elongation of [RNQ+] prions.
Figure 3.
Roles for Sis1 in [RNQ+] prion assembly. The [RNQ+] prion acts as a template to alter the conformation of native Rnq1 into an assembly competent monomer. Sis1 promotes the appropriate packaging of templated Rnq1 monomers into higher-ordered [RNQ+] prions via a fragmentation and elongation model. Inefficiencies in this assembly process by Rnq1 overexpression or Sis1 depletion enable templated monomers to accumulate as soluble, proteotoxic species.
Distinct signaling pathways facilitate protein detoxification via opposing mechanisms.
Cells can manage the proteotoxic accumulation of soluble, oligomeric species by both disaggregation and aggregation methods. C. elegans have become a valuable tool to dissect the dynamics of amyloid assembly pathways in aging organisms. Distinct signaling pathways exist which both prevent the formation of amyloid aggregates as well as promote efficient amyloid assembly. The heat shock factor-1 (HSF-1) signaling pathway promoted the solubilization of toxic, Aβ1–42 assemblies.18 HSF-1 activity is induced by a variety of stress signals including heat-shock which subsequently promotes the expression of numerous chaperones.110–112 In contrast, the DAF-16 signaling pathway appears to promote efficient assembly of Aβ1–42 peptides into amyloid particles. The FOXO transcription factor, DAF-16,113,114 is a downstream target of insulin/insulin growth factor (IGF)-1-like signaling pathway which regulated life span and youthfulness in worms, flies and mammals.115 Although HSF-1 and DAF-16 signaling pathways promote longevity and cell survival, they appear to act by opposing mechanisms to accomplish this feat.
Conclusions
Molecular chaperones can intervene at multiple points in the amyloid assembly pathway to buffer the proteotoxic accumulation of misfolded protein intermediates. As a first line of defense, chaperones networks can coordinate the solubilization and subsequent refolding or degradation of different toxic proteins. Alternatively, the cell possesses a second line of defense which can package misfolded proteins into benign aggregates and thereby minimize the cell's exposure to the toxic assembly intermediates. Although the formation of amyloid-like aggregates appears to protect cells against protein misfolding events, the gradual stockpiling of amyloid fibrils into intra- and extra-cellular plaques can clearly have a negative impact on normal cellular processes over extended periods of time.116–118 Additionally, the general decline in protein homeostasis or proteostasis3 by age or chronic stress may compromise chaperone networks2 resulting in the destruction of benign aggregates and the generation of soluble, toxic oligomers. Two distinct signaling cascades appear to regulate the flux of misfolded, amyloidogenic proteins between solubilization and aggregation pathways.18 HSF-1 regulates the expression of numerous molecular chaperones111 and can antagonize aggregation through control of the integrated chaperone network. However, it is unclear in the DAF-16 signaling pathway which downstream components are involved in the remodeling of misfolded, disease proteins into ordered aggregates. Sis1 and the chaperonin complex were shown to drive protective aggregate formation, yet both proteins historically act to suppress non-specific aggregation.100,119 Therefore, different signaling pathways such as DAF-16 and HSF-1 may enable an individual chaperone such as Sis1 or the TriC chaperonin complex to perform opposing functions on misfolded proteins. Alternatively, the disease substrate may possess intrinsic, structural properties which determine the action of the interacting chaperone. Further studies will elucidate the means by which opposing chaperone activities are regulated at a molecular and cellular level.
Acknowledgements
We thank Dr. Michael Douglas for the critical reading of this manuscript. This work was supported in whole or in part, by a predoctoral fellowship from the American Heart Association (to P.M.D), National Institutes of Health Pre-doctoral Training Grant 5T32GM008581-09 (to D.W.S.), and National Institutes of Health Grant 5R01GM067785-06 (to D.M.C.).
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/8587
References
- 1.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caughey B, Lansbury PT. Protofibrils, pores, fibrils and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 7.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 8.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 9.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 10.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 11.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 13.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 15.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 16.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 17.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 18.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 19.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 21.Burke JR, Enghild JJ, Martin ME, Jou YS, Myers RM, Roses AD, et al. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 22.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 23.Small DH, Mok SS, Bornstein JC. Alzheimer's disease and Abeta toxicity: from top to bottom. Nat Rev Neurosci. 2001;2:595–598. doi: 10.1038/35086072. [DOI] [PubMed] [Google Scholar]
- 24.Arispe N, Rojas E, Pollard HB. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc Natl Acad Sci USA. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volles MJ, Lee SJ, Rochet JC, Shtilerman MD, Ding TT, Kessler JC, et al. Vesicle permeabilization by protofibrillar alpha-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry. 2001;40:7812–7819. doi: 10.1021/bi0102398. [DOI] [PubMed] [Google Scholar]
- 26.Kremer JJ, Sklansky DJ, Murphy RM. Profile of changes in lipid bilayer structure caused by beta-amyloid peptide. Biochemistry. 2001;40:8563–8571. doi: 10.1021/bi010417x. [DOI] [PubMed] [Google Scholar]
- 27.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 28.Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Bonini NM. Chaperoning brain degeneration. Proc Natl Acad Sci USA. 2002;99:16407–16411. doi: 10.1073/pnas.152330499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- 35.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- 37.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 39.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Zylicz M, Ang D, Georgopoulos C. The grpE protein of Escherichia coli. Purification and properties. J Biol Chem. 1987;262:17437–17442. [PubMed] [Google Scholar]
- 41.Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabani M, Beckerich JM, Gaillardin C. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol. 2000;20:6923–6934. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raynes DA, Guerriero V., Jr Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J Biol Chem. 1998;273:32883–32888. doi: 10.1074/jbc.273.49.32883. [DOI] [PubMed] [Google Scholar]
- 45.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 47.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andreasson C, Lindquist S, Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 2008;3:1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 50.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 51.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 53.Kobayashi Y, Sobue G. Protective effect of chaperones on polyglutamine diseases. Brain Res Bull. 2001;56:165–168. doi: 10.1016/s0361-9230(01)00593-7. [DOI] [PubMed] [Google Scholar]
- 54.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Summers DW, Douglas PM, Ren HY, Cyr DM. The type I HSP40 YDJ1 utilizes a farnesyl moiety and zinc finger like-region to suppress prion toxicity. J Biol Chem. 2008 doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi H, Kobayashi Y, Yoshihara T, Niwa J, Doyu M, Ohtsuka K, Sobue G. Hsp70 and Hsp40 improve neurite outgrowth and suppress intracytoplasmic aggregate formation in cultured neuronal cells expressing mutant SOD1. Brain Res. 2002;949:11–22. doi: 10.1016/s0006-8993(02)02568-4. [DOI] [PubMed] [Google Scholar]
- 58.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 Reduces alpha-Synuclein Aggregation and Toxicity. J Biol Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 59.Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 60.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 61.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 62.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 63.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, et al. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington's disease. Mol Ther. 2007;15:903–911. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 72.Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, et al. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 74.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 75.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 76.Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16:5891–5900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- 78.Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 79.Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, et al. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 81.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 82.Terlecky SR, Olson TS, Dice JF. A pathway of lysosomal proteolysis mediated by the 73-kilodalton heat shock cognate protein. Acta Biol Hung. 1991;42:39–47. [PubMed] [Google Scholar]
- 83.Olson TS, Terlecky SR, Dice JF. Targeting specific proteins for lysosomal proteolysis. Biomed Biochim Acta. 1991;50:393–397. [PubMed] [Google Scholar]
- 84.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 85.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 86.Carra S, Sivilotti M, Chavez Zobel AT, Lambert H, Landry J. HspB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum Mol Genet. 2005;14:1659–1669. doi: 10.1093/hmg/ddi174. [DOI] [PubMed] [Google Scholar]
- 87.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 88.Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, et al. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 90.Rousseau E, Kojima R, Hoffner G, Djian P, Bertolotti A. Misfolding of proteins with a polyglutamine expansion is facilitated by proteasomal chaperones. J Biol Chem. 2009;284:1917–1929. doi: 10.1074/jbc.M806256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fenton WA, Horwich AL. Chaperonin-mediated protein folding: fate of substrate polypeptide. Q Rev Biophys. 2003;36:229–256. doi: 10.1017/s0033583503003883. [DOI] [PubMed] [Google Scholar]
- 93.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 94.Farr GW, Scharl EC, Schumacher RJ, Sondek S, Horwich AL. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and non-native forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 95.Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- 96.Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 97.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 99.Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 101.Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 102.Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 103.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- 111.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 112.Wu C, Wilson S, Walker B, Dawid I, Paisley T, Zimarino V, Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987;238:1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- 113.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 114.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 115.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 116.Fiala JC. Mechanisms of amyloid plaque pathogenesis. Acta Neuropathol. 2007;114:551–571. doi: 10.1007/s00401-007-0284-8. [DOI] [PubMed] [Google Scholar]
- 117.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 118.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 119.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]