Abstract
Propagation of yeast prions requires normal abundance and activity of many protein chaperones. Central among them is Hsp70, a ubiquitous and essential chaperone involved in many diverse cellular processes that helps promote proper protein folding and acts as a critical component of several chaperone machines. Hsp70 is regulated by a large cohort of co-chaperones, whose effects on prions are likely mediated through Hsp70. Hsp104 is another chaperone, absent from mammalian cells, that resolubilizes proteins from aggregates. This activity, which minimally requires Hsp70 and its co-chaperone Hsp40, is essential for yeast prion replication. Although much is known about how yeast prions can be affected by altering protein chaperones, mechanistic explanations for these effects are uncertain. We discuss the variety of effects Hsp70 and its regulators have on different prions and how the effects might be due to the many ways chaperones interact with each other and with amyloid.
Key words: Hsp70, Hsp40, chaperone, prion, yeast
Yeast Prions
Yeast prions are believed to propagate as amyloid forms of cellular proteins. Amyloid is a fibrous aggregate of a single type of protein in which each monomer in the fiber has a similar non-native fold that allows uniform linear stacking. Amyloid propagates by recruiting the soluble form of the protein and converting it into the same misshapen conformation as it adds to the end of the fiber. Although inheritable prion entities are widely believed to be amyloid fibers (an assumption we make to simplify our discussion), they have been termed propagons1 because it is not yet clear whether some other oligomeric intermediate or byproduct of amyloid propagation represents the actual infectious elements. The most widely studied yeast prions [URE3], [PSI+] and [PIN+]/[RNQ1+] are altered forms of the proteins Ure2p, Sup35p and Rnq1p, respectively.2–7 Each has a region rich in polar residues referred to as a prion domain as it confers prion behavior. Purified Ure2p and Sup35p have high propensity to assemble into amyloid spontaneously and their prion domains form an amyloid core leaving the remaining portion of the protein properly folded.8–10 The modular property of prion domains suggests this independence of domain structures is a general characteristic.7,11–13 The depletion of most of the soluble native protein into insoluble prion aggregates can confer a phenotype useful for monitoring the prion.
Despite having a highly organized structure, amyloid is an aggregate of misfolded protein so it is not surprising that protein chaperones strongly influence prion propagation in yeast. In order to be maintained in a growing yeast population, prions must grow, replicate and be transmitted to daughter cells. In vitro, purified Sup35p, Ure2p and Rnq1p spontaneously assemble into infectious forms of amyloid, and once formed the amyloid catalyzes its own assembly from soluble protein.14–17 Likewise, once a prion is present in a yeast cell, continued growth of prion polymers probably occurs without assistance from chaperones. Transmission of prions occurs very efficiently from mother to daughter and between mating partners, and this step is also thought to be spontaneous through diffusion of cytoplasmic propagons, of which there can be hundreds per cell.18 Sonicating in vitro reactions accelerates amyloid assembly by fragmenting fibers, which generates more “seeds.” While growth or transmission of prions can be influenced by altering chaperone abundance or activity, replication of yeast prions, which is believed to be associated with fragmentation, depends critically on normal chaperone function.
Essential Role of Hsp104/70/40 Machinery for Prion Replication
Hsp104 is a hexameric ring-shaped AAA+ ATPase that resolubilizes proteins from aggregates by extruding proteins through its central channel. It was identified in a search for proteins that inhibited [PSI+] propagation when expressed at elevated levels.19 Paradoxically, deleting the non-essential Hsp104 also eliminated [PSI+]. Subsequent studies showed that Hsp104 acts in prion propagation by extruding monomers from prion fibers,20,21 which destabilizes them allowing them to break into pieces that each can act as a template for continued prion propagation.23,100 This activity is required for propagation of the known amyloidogenic prions (see Fig. 1).
Figure 1.
Chaperone assisted prion replication. Prion polymers (shown as stacks of rectangles) grow by addition of monomers (shown as ovals) to the ends. Monomers are converted into the prion conformation when they join the polymer. Fragmentation of an individual polymer by chaperone action results in formation of two polymers, each of which can grow and continue the cycle. Hsp70/40 probably acts directly on polymers, facilitating Hsp104 interaction. Hsp104 extracts monomers from the polymers by extruding them through its central channel, which results in destabilization and fragmentation of the polymer. Hsp70/40 might also act where peptide exits Hsp104 to help extrude monomers and assist refolding. Many co-chaperones that are known to regulate Hsp70 influence prion propagation, possibly by affecting this reaction. Chaperones might also influence prion propagation by affecting rate of incorporation of monomers into polymers.
Curiously, [PSI+] is the only native yeast prion eliminated efficiently by elevating expression of Hsp104.5,19,63,101,102 Nevertheless, this curing effect has been widely viewed as resulting from complete dissolution of prion aggregates by increased Hsp104 disaggregating activity. However, several Hsp104 mutants that function normally in [PSI+] propagation and protein disaggregation cannot cure cells of [PSI+] when overexpressed, showing that Hsp104 disaggregation activity is not enough to eliminate the prion.20 Other possibilities are that Hsp104 possesses an Hsp70/40-independent amyloid severing activity that inactivates infectivity of Sup35p amyloid specifically,22 although increasing Hsp104 in vivo increases size of prion polymers,23 or that elevating Hsp104 when it is not normally induced somehow specifically inhibits transmission of [PSI+] prions during cell division.1,24 Evidence showing a role of Hsp104 in restricting inheritance of damaged proteins, and an involvement of the actin cytoskeleton in this process and in [PSI+] propagation are consistent with this latter idea.25–29
While the protein resolubilizing activity of Hsp104 is required for its function in prion propagation,20 Hsp70 and its obligatory Hsp40 partner are necessary for Hsp104 and similar AAA+ chaperones to resolubilize proteins from aggregates.30–32 Thus, although Hsp104 functions as a primary factor involved in prion replication, Hsp70 and Hsp40 likely play critical roles in this process as components of the Hsp104 machinery. The binary Hsp70/40 system can cooperate to refold proteins from small aggregates so it is possible that these chaperones can influence prion replication independently of Hsp104. The degree to which the Hsp70 effects on prions are mediated by an effect on the Hsp104 machinery is not yet clear. Regardless of the answer, it is evident that Hsp70 is a focal point influenced by many factors that affect prion propagation (see Figs. 1 and 2).
Figure 2.
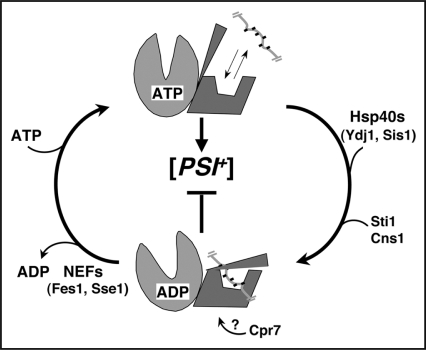
Hsp70 reaction cycle and its regulation by co-chaperones. When ATP is bound to the nucleotide-binding domain (light gray) the substrate-binding domain (dark gray) is in an “open” conformation and rapidly exchanges substrate, which is indicated as a short hydrophobic segment of a larger protein. Hydrolysis of ATP to ADP (downward arching arrow) induces a conformational change in the substrate-binding and C-terminal domains that traps substrate. Exchange of nucleotides (upward arrow, ADP release allows rebinding of ATP) restores the open conformation and release of substrate. ATP hydrolysis is rate limiting and is stimulated by Hsp40s, Sti1 and Cns1. ADP release is enhanced by nucleotide exchange factors (NEFs). How Cpr7 influences Hsp70 is unknown but its effects on [PSI+], which are additive with those of Sti1, are consistent with Cpr7 stabilizing the ADP-bound state. Altering the cycle to favor the ADP-bound state (e.g., stimulating ATP hydrolysis or inhibiting nucleotide exchange) impairs [PSI+].
Hsp70 and its Regulators
Hsp70s are universally conserved and essential chaperones. They facilitate protein folding directly and act as key components of cellular protein folding and disaggregation machines.30,33–35 Hsp70 has a role in all aspects of protein “quality control” including synthesis, translocation across membranes, protection from aggregation during conditions of cellular stress, recovery from aggregates, and facilitating degradation or sequestration of irrecoverably misfolded or aggregated proteins.36–38 Hsp70 acts by binding and releasing exposed hydrophobic stretches of incompletely folded proteins (see Fig. 2). This cycle relies on conformational changes regulated by ATP hydrolysis and ADP/ATP exchange. ATP-bound Hsp70 has low substrate affinity and rapid exchange, while ADP-bound Hsp70 binds substrate with high affinity and slow exchange. Thus, ATP hydrolysis helps Hsp70 trap substrates while ADP release and rebinding of ATP allows substrate release. Binding of partially folded proteins masks hydrophobic surfaces to prevent aggregation or keep proteins extended during translocation. Cycling between low and high affinity states provides opportunities for incompletely folded proteins to fold properly when released, or to be re-bound. Eukaryotes typically encode several Hsp70 paralogs. S. cerevisiae encodes six canonical cytosolic Hsp70s, four of the essential Ssa subfamily (Ssa1p-Ssa4p) and two of the ribosome-associated Ssb subfamily.39–41
Hsp70 has very slow intrinsic ATP hydrolysis and ADP release, which provide key points for regulation of its reaction cycle. Accordingly, Hsp70 function is intimately associated with co-chaperones that regulate its function at these steps and recruit it to various sub-cellular locations or chaperone machineries.37 The most abundant and obligatory regulators are Hsp40s, which bind substrate with specificity overlapping that of Hsp70 and stimulate Hsp70 ATP hydrolysis. This combination of activities allows Hsp40 to present substrates to Hsp70 and coordinate Hsp70 ATP hydrolysis with substrate binding.42,43 S. cerevisiae encodes over a dozen Hsp40 homologs, the major ones being Ydj1p and Sis1p. Among the others, Apj1p was identified as antagonizing [PSI+] when overexpressed.44 Efficient cycling of Hsp70 also requires nucleotide exchange factors (NEFs), which accelerate ADP release and subsequent rebinding of ATP so Hsp70 can release substrate. Fes1p and Sse1p (Hsp110) are the major cytosolic S. cerevisiae NEFs.45–47 Additionally, the tetratricopeptide repeat (TPR)-containing Hsp90 co-chaperones Sti1p (Hop1) and Cns1p stimulate Hsp70 ATPase, and Cpr7p affects Hsp70 function in an uncertain way.48–50 This large number of Hsp70 regulators allows many combinations of interactions that can affect Hsp70 activity in distinct ways to fine-tune its function (see Fig. 2).
Biochemical Analysis of Amyloid Formation and Chaperone Interactions
Studies using a candidate approach or co-purification of Sup35p or Rnq1p showed that Ssa and Ssb Hsp70s and their Ydj1p, Sis1p and Sse1p co-chaperones individually or cooperatively bind preferentially to the prion form of Sup35p.51–53 Although many of the purified chaperones bind Sup35p independently, the most abundant protein(s) co-purifying from [PSI+] cells is Ssa1/2p (indicated this way since Ssa1p and Ssa2p are nearly identical and distinguishing if either or both were present was not determined), suggesting that Hsp70 interacts directly with Sup35p in vivo and that its co-factors might associate with prions through interaction with Hsp70. Consistent with this view, binding of Ssa1p and its co-factors Sse1p and Sis1p to Sup35p in vitro is enhanced when they are combined. Hsp104 was also identified among the less abundant proteins co-purifying with Sup35p preferentially from [PSI+] cells, suggesting that some Hsp70 and co-chaperone effects on prion phenotype, particularly with respect to prion replication, could be due to alterations of the disaggregation machinery. These studies provide evidence that the influence of altered expression of these chaperones on prions is due to alteration of growth or replication of prions by direct interactions of the chaperones.
Several groups have addressed this possibility by monitoring action of chaperones on amyloid and on the kinetics of assembly of Sup35p and Ure2p, or their prion-determining regions, into amyloid.54–57 Overall the biochemical data showing fragmentation of amyloid or inhibition of amyloid assembly are consistent with binding interactions of the different chaperones with ex vivo soluble and prion forms of proteins. However, results from different groups conflict and some conclusions based on data from in vitro experiments are not consistent with phenotypic effects shown from genetic studies. These discrepancies illustrate the difficulty in recapitulating the complex interplay of chaperones in vivo using purified components, and the actual mechanisms of how chaperones alter prion propagation remain uncertain.
Insights Provided by Hsp70 Mutations
We isolated a mutant of the Hsp70 Ssa1p with the L483W substitution, designated Ssa1-21p, in an unbiased screen for cellular factors that inhibit [PSI+].58 Residue L483 is in the substrate-binding domain (SBD) away from the binding pocket but near the ATPase domain interface. Although it is a universally conserved residue, the mutation has no detectable effect on stress tolerance or cell growth under all conditions tested, suggesting it affects a specific and non-essential Hsp70 activity. To gain insight into how the mutation alters Hsp70 function we mutagenized wild type Ssa1p to identify additional mutations that caused a similar anti-prion effect, and Ssa1-21p to identify second-site mutations that restore normal prion propagation despite the presence of L483W.50,59 All of the new prion-inhibiting mutations were in the ATPase domain, suggesting regulation of chaperone activity was more important than direct substrate interactions for the inhibitory effect, an interpretation supported by a subsequent screen.60 Locations of the second-site mutations indicated they suppress anti-prion effects of L483W by reducing ATP hydrolysis through inhibiting interaction with Hsp40 or TPR co-chaperones, or by weakening substrate trapping. These mutations also suppress the other [PSI+]-inhibiting mutations, implying that all the Hsp70 mutations that impair [PSI+], regardless of the domain in which they are located, do so by a similar mechanism that involves alteration of the Hsp70 reaction cycle to favor the ADP-bound state (see Fig. 2).
Subsequent genetic analysis confirmed this conclusion showing that prion propagation could be inhibited by altering NEF and TPR regulators of Ssa1p in a way that accelerates Hsp70 ATP hydrolysis or slows ADP release.50 For example, either increasing ATPase by increasing abundance of Sti1p or decreasing ADP release by deleting Fes1p weakens [PSI+] phenotype, while deleting Sti1p or increasing Fes1p improves it (see Fig. 2). Thus, perturbing the Hsp70 reaction cycle in a similar way, either directly by mutation or indirectly by altering its regulators, has the same anti-prion effect. The TPR co-chaperones Cns1p and Cpr7p also affect prions in this system in predictable ways, showing that all of these co-chaperones affect prions through their ability to regulate Hsp70. The data suggest that enhanced binding of the soluble or insoluble form of Sup35p by Ssa1p interferes with prion replication. Further studies showed Ssa1-21p does not affect fiber fragmentation but promotes accretion of individual fibers into large aggregates or interferes with recovery of propagons from such aggregates.61
In line with the genetic results, biochemical analysis showed intrinsic ATP hydrolysis of Ssa1-21p is elevated ten-fold.62 It is unexpected that such a large change of this essential protein does not confer a growth or thermo-tolerance phenotype. It is probable that Hsp70 activity is regulated more stringently in vivo and that prions are particularly sensitive to the specific alteration of Hsp70 function. Additional surprises were that second site suppressing mutations that restore [PSI+] phenotype do not restore normal ATP hydrolysis, and that other prion-impairing Ssa1p mutants have nearly normal hydrolysis rates. Moreover, L483W is in the SBD, and the other mutants with normal ATPase activity are altered in the ATPase domain, although some of these latter mutations are in locations expected to affect functional interactions with co-factors that influence ATPase. Together with data showing that the mutations cause conformational changes in Ssa1p, the results suggest L483W inappropriately stimulates ATPase by altering conformation at the inter-domain interface, and the ATPase domain mutations affect regulation of ATP hydrolysis by co-factors or ATPase signaling to the SBD in ways that promote substrate binding. These studies are consistent with the observation that the prion-impairing effects are not caused by altering direct interaction of Hsp70 with substrate as much as by altering the regulation of this interaction, and by extension, the complexes of which Hsp70 is a component.
Ydj1
Among the yeast prions [URE3] is most sensitive to Ydj1p expression level. Although [URE3] does not require Ydj1p, overexpression of Ydj1p weakens [URE3] phenotype and causes gradual loss of [URE3] from dividing cells.63–65 Ydj1p binds to soluble Ure2p with or without its prion domain, and when added with purified Ure2p it inhibits formation of Ure2p amyloid in a concentration dependent manner.57,66 It was therefore believed that Ydj1p cured cells of [URE3] by binding soluble Ure2p and preventing fibril formation. However, when Ydj1p is added after Ure2p polymerization begins it does not affect amyloid formation. Moreover, cells overproducing Ydj1p have longer prion polymers.67 If this increased polymer size was due simply to faster growth, then elevating Ydj1p would not weaken [URE3] phenotype. Thus, in vivo Ydj1p seems to interfere with fragmentation of Ure2p polymers rather than with polymer assembly.
Interaction of Ydj1p with Ssa Hsp70 is critical for protein translocation and other processes important for cell growth.68,69 We recently showed the Ydj1p J-domain, which mediates interactions with Hsp70 and is needed for Ydj1p to stimulate Hsp70 ATPase, is sufficient for eliminating [URE3]. This result indicates that Ure2p binding by Ydj1p is dispensable for curing and suggests that regulation of Hsp70 by Ydj1p is crucial for the curing process. In agreement with these conclusions the J-domain mutant Ydj1-H34Q, which can dimerize and bind substrate but cannot functionally interact with Hsp70, does not cure [URE3]. Additionally, although elevating Sis1p or other Hsp40s has no effect on [URE3], the J-domains alone of Sis1p and another Hsp40, Jjj1p, do cure [URE3], which is consistent with overlapping functions of many yeast Hsp40 J-domains.64,65,70 Together, the data suggest that Ydj1p has a unique, non-J-domain Hsp40 activity and that it cures [URE3] by affecting prion replication through an interaction with Hsp70.
Overexpressing Ydj1p also cures some but not all variants of [PIN+]71 and it co-immuno-precipitates with the prion form of Rnq1p.72,73 As with [URE3], deleting Ydj1p does not affect propagation of typical [PIN+] prions.73 Ydj1p binding to Rnq1p might protect cells from prion toxicity, as overexpression of the Rnq1p prion domain is lethal to [PIN+] cells lacking Ydj1p. Binding of Ydj1p to Rnq1p requires Ydj1p farnesylation, but how the farnesyl chain influences Hsp40 function is unclear. It could be involved in regulation of Ydj1p substrate binding by altering Ydj1p conformation, in direct interaction with Rnq1p prion folding intermediates, or in regulation of other cellular factors that might influence [PIN+].73,74
Overexpressing Ydj1p also antagonizes artificial versions of [PSI+] prions. For example, variants of [PSI+PS], a hybrid prion with the S. cerevisiae Sup35p prion domain replaced by the analogous domain of Pichia methanolica Sup35p, are cured by increased Ydj1p with different efficiencies.11 Ssa1p also has modest curing activity on both [PSI+PS] and [URE3],11,75 and anti-prion effects are considerably enhanced when Ssa1p and Ydj1p are simultaneously overexpressed, suggesting Ydj1p also cures these prions indirectly through its action on Hsp70. As with Ure2p, combining Ydj1p and Ssa1p blocks fibrilization of purified Sup35p and Sup35-NM.55,56 When overexpressed, however, neither Ydj1p nor Ssa1p, alone or in combination, cures cells of typical [PSI+] prions so the significance of this interaction with Sup35p in vivo is uncertain.
Sis1p
Sis1p is an essential cytosolic Hsp40 whose association with Rnq1p in [PIN+] cells led to the identification of its role in prion propagation.51 Sis1p also possesses a unique Hsp40 activity in that it is the only cytosolic Hsp40 required for propagation of [PIN+], [PSI+] and [URE3].64,76 The non-essential G/F region confers Sis1p functional specificity, and this region is necessary for Sis1p function in [PIN+] propagation.51,77 Purified Sis1p binds specifically to a region of Rnq1p outside the prion-determining domain, and this binding is important for [PIN+] propagation.72,78 Whether the G/F region influences this binding is unknown. However, [RPS+], a hybrid prion formed from Sup35p with its prion domain replaced by that from Rnq1p, also requires the Sis1p G/F region while [PSI+] does not. These results show that although the Rnq1p prion domain lacks the Sis1p binding site, it specifies function of the Sis1p G/F region for propagation of [PIN+] and [RPS+]. Ssa1p also interacts with Rnq1p, and the Sis1p G/F region is believed to aid cooperation between Sis1p and Ssa1p in substrate interactions. The interaction of Sis1p and Ssa1p with Rnq1p occurs only in [PIN+] cells,51 and elimination of the ability of Sis1p to stimulate Hsp70 ATPase also inhibits [PIN+]. Together these data suggest that Sis1p operates as an Hsp70 co-chaperone with respect to [PIN+] propagation.64,72,76
When Sis1p or Hsp104 expression is repressed, the size of Rnq1p polymers increases and the number of [PIN+] propagons decreases.76 Both effects appear to be due to altered cooperation of Sis1p with Hsp70 and Hsp104, and imply that Sis1p is involved in fragmentation of Rnq1p polymers required for [PIN+] prion replication.76 Sup35p polymers in [PSI+] cells also increase in size when Sis1p is depleted or when Hsp104 is inhibited.23,64 Additionally, although a recent study to decipher the functional relationship between Sis1p and Hsp104 in [PSI+] propagation does not unequivocally distinguish between cause and effect, the data strongly suggest a role for Sis1p, in conjunction with its Hsp70 partner, to deliver prion substrates to the Hsp104 complex.79
A cytotoxic effect of overexpressing intact Rnq1p in [PIN+] cells is alleviated by increasing Sis1p abundance just 3-fold.78 Unexpectedly, increased expression of Sis1p promotes [PIN+] assembly rather than inhibiting it. These results led to the proposal that Sis1p plays a cytoprotective role by promoting formation of Rnq1p amyloid, which reduces accumulation of toxic oligomeric intermediates.78 The data therefore point to a dual function for Sis1p in [PIN+] propagation whereby Sis1p promotes growth as well as fragmentation of prion polymers.
In addition to the specific effects Ydj1p and Sis1p have on yeast prions, it is worth noting that these two Hsp40s have opposite effects on polyglutamine aggregation and toxicity.80 While Sis1p decreases both aggregation and toxicity, Ydj1 increases both. These Hsp40s regulate Hsp70 differently81 but whether their effects are direct or mediated by Hsp70 was not investigated. Hsp40 structural domains are interchangeable and expression of hybrid proteins has been used to determine domains important for prion-specific effects.72,82 Such analysis would be useful for identifying Hsp40 activities and possible chaperone interaction specificities that mediate the effects on polyglutamine.
Prions Respond Differently to Different Hsp70s
In addition to the various ways different Hsp40s and other co-chaperones influence prions, highly homologous Hsp70 proteins also affect different prions differently. Constitutively expressed cytosolic Ssa1p and Ssa2p are 98% identical, and stress-inducible Ssa3p and Ssa4p are 88% identical to each other and 80% identical to Ssa1/2p. Remarkably, elevating expression of Ssa1p cures [URE3] but similarly elevating Ssa2p does not, and neither treatment affects [PSI+] stability.75 Additionally, [URE3] is weaker in cells lacking Ssa2p, but not in cells lacking Ssa1p,83,84 while [PSI+] is weaker in cells lacking Ssa1p but not in cells lacking Ssa2p.20,84 These data show that despite their nearly identical structures Ssa1p and Ssa2p have distinct activities, and suggest that propagation of the different prions relies on different Hsp70 functions.
Unlike the opposing effects Ssa1p and Ssa2p have on [URE3], increasing expression of any of the four Ssa proteins inhibits curing of [PSI+] by overexpressed Hsp104 and increases frequency of appearance of [PSI+] when Sup35p prion domain is overexpressed.52,85 These data point to a general [PSI+]-promoting activity of Ssa protein by antagonizing Hsp104 or enhancing assembly of Sup35p amyloid, although biochemical data suggest Ssa1p antagonizes amyloid formation. As an exception, certain variants of [PSI+] that form atypically larger aggregates respond oppositely to increases in abundance of Ssa1p and Hsp104. Efficient propagation of these prion variants under non-selective conditions requires increased Hsp104, which was interpreted to mean they are less susceptible to disaggregation by the Hsp104 machinery and thus require increased Hsp104 activity.86,87 Increasing Ssa1p was proposed to adversely affect propagation of these variants by further enhancing aggregation, which leads to increased destabilization. A very recent report shows that prions of a Sup35-GFP fusion protein can be cured by overexpressing Ssa1p.88 What makes the GFP fusion protein prion sensitive to inhibition by Ssa1p, unlike the native Sup35p prion used to induce its formation, was not investigated, but it is possible it also forms larger than normal prion aggregates. In contrast to the Ssa proteins, increasing or depleting Ssb Hsp70 subfamily protein shows Ssb protein consistently as a [PSI+] prion antagonist.89,90
One observation from these studies is that under different conditions the same Hsp70 can inhibit or promote propagation of the same prion. Since individual chaperones are components of complex machineries, and some even regulate expression of others, care should be used in interpreting data from overexpression studies as being due to simple mass action effects of increasing a particular enzymatic activity. In fact, with regard to Hsp104 and Hsp40 the effects of overexpression are inconsistent with a direct effect of the elevated chaperone activities on prion aggregates. Opposing effects could reflect differences in how a single elevated protein alters different machineries that influence processes of nucleation, assembly and fragmentation of prion polymers. As chaperones are involved in many cellular processes, the effects on prions could be through alteration of cellular processes, possibly connected to the normal functions or interacting partners of the prion protein, that specifically affect propagation of certain prions.
We developed a more simplified system to assess function of any individual Hsp70 by constructing yeast strains lacking all the Ssa proteins. We found that certain Hsp70s from primates and plants support both cell growth and prion propagation, and that they varied widely in these abilities.84,91 In line with phenotypes of cells lacking individual Ssa proteins, [PSI+] is normal in cells expressing only Ssa1p and somewhat weaker in cells expressing only Ssa2p. Although Ssa3p does not support growth as well as Ssa1/2p, [PSI+] is stronger in cells expressing only Ssa3p. [PSI+] is considerably weak but stable in cells expressing Ssa4p, which is weakest at supporting cell growth. In contrast, [URE3] was strong but mitotically unstable in Ssa1p-expressing cells, normal in cells expressing Ssa2p, weak and unstable in cells expressing Ssa3p, and weak but more stable in Ssa4p-expressing cells. Thus, for these Ssa Hsp70s there is no correlation between ability to support growth and prion propagation, and all but Ssa4p have opposite effects on the two prions. The former observation could mean that the different Ssa proteins have different abilities to perform in essential processes that don't affect prion propagation. The latter reveals subtle functional differences among the Hsp70s that might be due to differences in specificity of interactions with substrates or various co-chaperones. These data also show that despite the similar [PSI+] promoting effect of elevating abundance of any of these four Hsp70s, when expressed at physiological levels each has unique activities that influence prions in both positive and negative ways.
Prion “strength” and mitotic stability are related to the ratio of soluble to insoluble prion protein and the number of prions per cell, respectively.92,93 The solubility is determined by rate of polymer growth and depends on the speed that soluble protein adds to prion polymers and the number of ends that recruit it. The number of prions per cell, which determines the number of ends, reflects the rate of replication due to fragmentation of prion polymers. Therefore, noticeable changes in strength and stability phenotypes provide clues to whether growth or replication processes are affected.93 Although factors that affect prions can influence both growth and replication, these processes can be affected separately. For example, the unstable but strong [URE3] phenotype in cells expressing only Ssa1p probably reflects reduced prion replication, which leads to fewer prions per cell and an increased chance that daughter cells fail to inherit prions, yet the strong phenotype likely indicates normal or even accelerated polymer growth despite the reduced number of polymer ends. Conversely, the weak but stable phenotype of [URE] cells expressing only Ssa4p could reflect reduced growth of prion polymers but normal replication that maintains a high number of prions per cell. Here the implication is that Ssa1p inhibits or functions less efficiently at processes involving Ure2p polymer fragmentation, while Ssa4p inhibits addition of Ure2p monomers to polymers or is less efficient at promoting polymer growth.
Chaperone alterations that lead to inhibition of fragmentation, and therefore prion replication, likely occur through an effect on the Hsp104 machinery, which could be direct or by limiting interaction of Hsp104 with prions as substrates. In prion aggregates purified from [PSI+] cells, a molar ratio of Hsp70 (Ssa1/2p) to Sup35p of 1:2 indicates Hsp70 binds along entire Sup35p polymers and might help stabilize them.53 Hsp70s also could act to promote or inhibit growth of prions by binding at the ends of polymers.
The specificity of Hsp70 action on the different prions might be direct and determined by differences in affinity of Hsp70s for the different prion proteins as substrates, although the homology of Ssa1/2p substrate binding domains argues against it. Alternatively, as mentioned, many observations suggest Hsp70 effects on prions are due more to differences in the way they interact with other components of the chaperone machinery than to differences in intrinsic Hsp70 activity. Hsp40s also vary in substrate specificity and they interact differently with different Hsp70s and in their capacity to influence Hsp70 activity. For example, the differences in ways Sis1p and Ydj1p influence [PIN+] could be because they bind different regions of Rnq1p or because they differ in ability to regulate Ssa protein activity.73,78,81 Thus, while both Hsp70 and Hsp40 recognize prions as substrates, it is likely that specific Hsp70/40 combinations determine specificity for effects on different prions. These combinations are further amplified as different NEFs also affect the system. The curing effect of the NEFs Fes1p and Sse1p on [PSI+] and [URE3], respectively, appear to be mediated through Hsp70,50,67 as does a requirement of Sse1p for induction of [PSI+] when Sup35p is overexpressed.94 TPR co-chaperones also could affect prions through interactions with Hsp90, which has been shown to reduce induction of [PSI+] by Sup35p overexpression.94 In considering the distinctions in chaperone-prion interactions, it has been suggested that prions can be “typed” on the basis of the ways the different chaperones affect their propagation.67 However, the large number of combinations makes categorizing all the effects tricky, and since each prion has certain unique responses to chaperones, correlations are difficult to define (see Table 1).
Table 1.
Effects of altered chaperones on prion phenotypes
| Chaperone/co-chaperonea | Elevation | Alterationb Depletion | Mutation | [PSI+] (Sup35p) | [URE3] (Ure2p) | [PIN+] (Rnq1p) | Reference |
| Hsp70s | |||||||
| Ssa1 | x | ⇑ | ⇓ | ? | 75, 85 | ||
| Ssa1 | x | ⇓ | - | ? | 20, 84 | ||
| Ssa1 | dominant neg. | ⇓ | ⇓ | ? | 58, 84 | ||
| Ssa1 (only) | - | ⇓ | ? | 84 | |||
| Ssa2 | x | ⇑ | - | ? | 52 | ||
| Ssa2 | x | - | ⇓ | ? | 84 | ||
| Ssa2 | dominant neg. | ⇓ | ⇓ | ? | 83S, 84 | ||
| Ssa2 (only) | ⇑ | - | ? | 84 | |||
| Ssa3 | x | ⇑ | ? | ? | 52 | ||
| Ssa3 (only) | ⇑ | ⇓ | ? | 84 | |||
| Ssa4 | x | ⇑ | ? | ? | 52 | ||
| Ssa4 (only) | ⇓ | ⇓ | ? | 84 | |||
| Hsp40s | |||||||
| Ydj1 | x | - | ⇓ | ⇓d | 63–65 | ||
| Ydj1 | x | - | ⇓e | - | 64, 65 | ||
| Ydj1 | J-domain | ? | ⇓ | ? | 64, 65 | ||
| Sis1 | x | - | - | ⇑ | 51, 64, 78 | ||
| Sis1 | x | ⇓ | ⇓ | ⇓ | 64 | ||
| Sis1 | G/F alteration | - | ? | ⇓ | 51, 72 | ||
| Sis1 | J-domain | ? | ⇓ | ? | 65 | ||
| Apj1 | x | ⇓f | - | - | 44, 64 | ||
| Jjj1 | J-domain | ? | ⇓ | ? | 64 | ||
| NEFs | |||||||
| Fes1 | x | ⇑ | - | ? | 50 | ||
| Fes1 | x | ⇓ | ⇓ | ? | 50, 67 | ||
| Sse1 | x | ⇑ | ⇓ | ? | 67, 94 | ||
| Sse1 | x | ⇓ | ⇓ | ? | 67, 94 | ||
| TPRs | |||||||
| Sti1 | x | ⇓g | ? | ? | 50, 94 | ||
| Sti1 | x | ⇑ | ? | ? | 50 | ||
| Cns1 | x | ⇓ | ? | ? | 50 | ||
| Cpr7 | x | ⇑ | ? | ? | 50 | ||
| Hsp82 (Hsp90) | x | ⇓ | ? | ? | 94 |
(only) means indicated Ssa protein is the only Ssa expressed in the cell.
x indicates alteration producing the effects. c⇑, enhances prion strength and/or stability; ⇓, weakens phenotype or eliminates prion; -, alteration has no effect; ?, unknown.
Cures cells of some [PIN+] variants, not others.
Cures cells of some [PSI+PS] variants, not others (see text).
Finally, although all amyloid-forming yeast prions tested assemble into in-register parallel beta-sheet conformations,95–99 it should be pointed out that differences in structural organization of polymers formed by different prion proteins, as well as different strains of the same prion, could influence their susceptibility to being recognized or acted on by chaperones. For example Ure2p might form more rigid structures than Sup35p, and thus rely more heavily on particular combinations of chaperone components that provide strongest fragmentation activity.
Defining the precise molecular mechanisms underlying prion propagation is challenging because altering chaperones affects prions in very many ways and the complex and often unpredictable ways chaperones interact with each other and with substrates in the cell make it difficult to simplify. Moreover, chaperones have roles in many diverse cellular processes so they can be expected to affect prions in indirect ways. Although the complexity is daunting, the strict requirement of Hsp104 and it's well-defined role in prion replication provides a basis for inferring that most chaperone alterations that influence the prion replication process are likely to be mediated through an effect on the Hsp104 machinery. At least for [PSI+], solubilization activity of Hsp104, which depends on Hsp70 and Hsp40, is critical for prion propagation, implying that Hsp104 does not act alone in [PSI+] replication. There is only one Hsp104 but many chaperones that influence the Hsp104 machinery. As a central player and regulatory target, Hsp70 links these chaperones to this machine.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes, Digestive and Kidney diseases.
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/9134
References
- 1.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox BS. “Y” a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 3.Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 4.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 5.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 7.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 8.King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–50030. doi: 10.1074/jbc.M406612200. [DOI] [PubMed] [Google Scholar]
- 11.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 12.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 13.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 16.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel BK, Liebman SW. “Prion-proof ” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne LJ, Cole DJ, Cox BS, Ridout MS, Morgan BJ, Tuite MF. The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS ONE. 2009;4:4670. doi: 10.1371/journal.pone.0004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 20.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 22.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 24.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 25.Bailleul PA, Newnam GP, Steenbergen JN, Chernoff YO. Genetic study of interactions between the cytoskeletal assembly protein sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81–94. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailleul-Winslett PA, Newnam GP, Wegrzyn RD, Chernoff YO. An antiprion effect of the anticytoskeletal drug latrunculin A in yeast. Gene Expr. 2000;9:145–156. doi: 10.3727/000000001783992650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, et al. Modulation of prion formation, aggregation and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessarz P, Schwarz M, Mogk A, Bukau B. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol Cell Biol. 2009 doi: 10.1128/MCB.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 31.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Nishikawa S, Brodsky JL, Nakatsukasa K. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD) J Biochem. 2005;137:551–555. doi: 10.1093/jb/mvi068. [DOI] [PubMed] [Google Scholar]
- 34.Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 37.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 38.Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 39.Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 41.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 42.Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42:4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- 44.Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- 45.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- 49.Hainzl O, Wegele H, Richter K, Buchner J. Cns1 is an activator of the Ssa1 ATPase activity. J Biol Chem. 2004;279:23267–23273. doi: 10.1074/jbc.M402189200. [DOI] [PubMed] [Google Scholar]
- 50.Jones GW, Song Y S C, Masison DC. Propagation of yeast [PSI+] is prion impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem. 2004;279:52319–52323. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- 55.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savistchenko J, Krzewska J, Fay N, Melki R. Molecular chaperones and the assembly of the prion Ure2p in vitro. J Biol Chem. 2008;283:15732–15739. doi: 10.1074/jbc.M800728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loovers HM, Guinan E, Jones GW. Importance of the Hsp70 ATPase domain in yeast prion propagation. Genetics. 2007;175:621–630. doi: 10.1534/genetics.106.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Y, Wu YX, Jung G, Tutar Y, Eisenberg E, Greene LE, et al. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell. 2005;4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Needham PG, Masison DC. Prion-impairing mutations in Hsp70 chaperone Ssa1: Effects on ATPase and chaperone activities. Arch Biochem Biophys. 2008;478:167–174. doi: 10.1016/j.abb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moriyama H, Edskes HK, Wickner RB. [URE3] Prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma D, Stanley RF, Masison DC. Curing of Yeast [URE3] Prion by the Hsp40 Cochaperone Ydj1p Is Mediated by Hsp70. Genetics. 2009;181:129–137. doi: 10.1534/genetics.108.098699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lian HY, Zhang H, Zhang ZR, Loovers HM, Jones GW, Rowling PJ, et al. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem. 2007;282:11931–11940. doi: 10.1074/jbc.M606856200. [DOI] [PubMed] [Google Scholar]
- 67.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hon T, Lee HC, Hach A, Johnson JL, Craig EA, Erdjument-Bromage H, et al. The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Mol Cell Biol. 2001;21:7923–7932. doi: 10.1128/MCB.21.23.7923-7932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Summers DW, Douglas PM, Ren HY, Cyr DM. The type I Hsp40 Ydj1 utilizes a farnesyl moiety and zinc finger-like region to suppress prion toxicity. J Biol Chem. 2009;284:3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol Biol Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 81.Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 82.Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts BT, Moriyama H, Wickner RB. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107–117. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- 84.Sharma D, Masison DC. Functionally redundant isoforms of a yeast hsp70 chaperone subfamily have different antiprion effects. Genetics. 2008;179:1301–1311. doi: 10.1534/genetics.108.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Borchsenius AS, Muller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–29. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 88.Mathur V, Hong JY, Liebman SW. Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chacinska A, Szczesniak B, Kochneva-Pervukhova NV, Kushnirov VV, Ter-Avanesyan MD, Boguta M. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr Genet. 2001;39:62–67. doi: 10.1007/s002940000180. [DOI] [PubMed] [Google Scholar]
- 91.Tutar Y, Song Y, Masison DC. Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics. 2006;172:851–861. doi: 10.1534/genetics.105.048926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hall D, Edskes H. Silent prions lying in wait: a two-hit model of prion/amyloid formation and infection. J Mol Biol. 2004;336:775–786. doi: 10.1016/j.jmb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 93.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 94.Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, et al. Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 97.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register beta-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 98.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wickner RB, Shewmaker F, Kryndushkin D, Edskes HK. Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures. Bioessays. 2008;30:955–964. doi: 10.1002/bies.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]