Abstract
The ability to design drugs (so-called ‘rational drug design’) has been one of the long-term objectives of chemistry for 50 years. It is an exceptionally difficult problem, and many of its parts lie outside the expertise of chemistry. The much more limited problem – how to design tight-binding ligands (rational ligand design) – would seem to be one that chemistry could solve, but has also proved remarkably recalcitrant. The question is ‘Why is it so difficult?’ and the answer is ‘We still don't entirely know’. This perspective discusses some of the technical issues – potential functions, protein plasticity, enthalpy/entropy compensation, and others – that contribute, and suggests areas where fundamental understanding of protein–ligand interactions falls short of what is needed. It surveys recent technological developments (in particular, isothermal titration calorimetry) that will, hopefully, make now the time for serious progress in this area. It concludes with the calorimetric examination of the association of a series of systematically varied ligands with a model protein. The counterintuitive thermodynamic results observed serve to illustrate that, even in relatively simple systems, understanding protein–ligand association is challenging.
Rational ligand design
Molecular recognition is, in a sense, the most important process in molecular biology. Most of the reactions and interactions that dominate the operation of living systems – recognition of sequences in nucleic acids by complementary nucleic acids and by proteins; folding of proteins; recognition of ligands by proteins; interaction of drugs and target proteins; differential stabilization of the transition state and ground state of biochemical reactions by enzymes; many others – involve molecular recognition. Understanding molecular recognition is centrally important to understanding – and controlling – the set of molecular processes that make up the cell, and life.
The problem of understanding molecular recognition translates, in utilitarian terms, into being able to design ligands that bind to specific sites on biomacromolecules (most often, proteins). This problem is intellectually enormously interesting, since solving it would provide ways of modulating the activities of proteins in vivo. More practically, it is central to the idea of so-called ‘rational drug design’. Generating drugs has been, and remains, a difficult, expensive, failure-prone, and highly empirical activity. The difficulty and cost of developing new drugs has become prohibitive for the pharmaceutical industry, and without improved efficiency and productivity from the R&D groups of this industry, there is a widespread concern that the rate at which new drugs will be developed in the future will be substantially less than it has been in the past. The process of generating a drug is, of course, much more complicated than simply designing a ligand that targets a protein: ‘hits’ and ‘leads’ are an important part of the process, but ADME/Tox/PK/PD (adsorption, distribution, metabolism, excretion, toxicology, pharmacokinetics, pharmacodynamics), and performance in humans, are more important than just high affinity. Nonetheless, most of the processes involved in the complex chain of gates through which a molecule must pass before it becomes a successful drug involve elements of molecular recognition, and generating leads (and safety assessment candidates) is an essential part of the process.
For 50 years, controlling molecular recognition has been an objective of organic chemistry. Biochemistry and medicinal chemistry, in particular, have shared a grand vision: to be able to go directly from the DNA sequence coding for a protein, to the design and synthesis of ligands that bind tightly and specifically to that protein. Although there has been enormous progress toward accomplishing parts of the vision, the core problem – the design of ligands to bind proteins – has remained intractable.
Four areas have succeeded beyond any of the early expectations of this field: (i) reading sequences of nucleic acids, (ii) producing proteins using recombinant DNA methods, (iii) determining the structures of proteins by X-ray and NMR methods, and (iv) synthesizing hypothesized ligands once these ligands are specified. What has largely failed is designing ligands based on known protein structure. (Predicting the tertiary structure of proteins based on their amino-acid sequence is also still far from producing useful results, and crystallizing proteins for X-ray analysis remains slow and difficult, but these problems are not as central to ‘rational drug design’ as is the problem of ligand design.)
Why is it so difficult to design molecules that associate with high affinity (low dissociation constants) to the active site of a protein of known structure? We are, we believe, beginning to understand what the issues are that make this problem so difficult. Understanding the problem is not, of course, understanding its solution, but it is a start. The following list includes some of the important issues with which the field concerned with molecular recognition in biology is now struggling.
Free energy, enthalpy, and entropy
The association of a protein and a ligand can be described by its free energy through the well-known relation, ΔG = ΔH − TΔS. Chemistry has developed an excellent sense for enthalpies (ΔH) in covalent chemistry: the combination of quantum mechanics and thermochemistry has been very successful in building a body of theory and experiment that makes it possible to predict the enthalpies of many covalent reactions. Predicting enthalpies of non-covalent reactions is at a much more primitive level, and the ability to predict the entropy (ΔS) of almost any reaction – and especially any reaction in water – is poor. Since life proceeds largely in water, the poor intuition of chemists about the thermodynamics of molecular recognition in water – and the almost complete inability to predict the entropies of these processes other than by analogy – pose a fundamental barrier to the problem of ligand design. (Developing the understanding necessary to solve this very difficult problem has not been helped by the erosion of the place of thermodynamics and statistical mechanics in the educational system, and by the replacement of these quantitative ways of thinking about molecular interactions by relatively non-mathematical subjects such as organic synthesis, genomics, combinatorial/high-throughput methodologies, and cell biology.)
Interactions: water and the hydrophobic effect
Water remains the most astonishing of liquids. It has everything required to cause complexity: an ability to hydrogen bond that gives it a strong tendency to form ordered structures, a uniquely small molar volume, a high heat capacity, a high dielectric constant, and a high temperature-dependence of its dielectric constant. Although it is the most important liquid on our planet, it is also the most complicated chemically. There still is no comprehensive theory of pure water, although that of Dill (the ‘Mercedes Benz model’) (Southall et al. 2002; Dill & Bromberg, 2003) provides a very important start, and alternatives by others (Lazaridis, 2001; Chandler, 2002, 2005) are also important contributions. These theories, however, are still at levels that are too abstract to give useful predictions at the level of molecular detail required for the design of ligands. Moreover, pure water, of course, is well removed from the very complex media that are the cytosol, the intracellular fluids of tissues, and serum – the fluids in which biological reactions occur.
Water is also the home of the hydrophobic effect (Lazaridis, 2001; Chandler, 2002, 2005; Southall et al. 2002; Dill & Bromberg, 2003). The nature of the insolubility of hydrocarbons in water – the hydrophobic effect – is central to biology: as a guess, 80% of the free energy of an average molecular recognition event – whether protein folding or protein–ligand association – is due to the hydrophobic effect. Although there is a general belief that the hydrophobic effect is an entropic contribution to the free energy reflecting the structuring of water around hydrophobic surfaces, there is not much understanding of what this statement means in molecular/atomic detail. Further, there remain large, troublesome facts that are difficult to explain. The solubility of hydrophobic hydrocarbons in water as a function of temperature is one example. This solubility – for example, of cyclohexane, a representative hydrocarbon – is remarkably indifferent to temperature, although between 0 °C and 100 °C the free energy of solution changes from being almost entirely entropic to almost entirely enthalpic (Southall et al. 2002). This type of compensation – an example of a so-called enthalpy/entropy compensation (EEC) – remains poorly understood.
Interactions: Coulomb's law in water
Electrostatic interactions in water often defy simple intuition: like charges can attract, and unlike charges repel (Honig & Nicholls, 1995; Gitlin et al. 2006). Underlying electrostatic interactions in water is Coulomb's law, but this most familiar of expressions must be modified by solvation. Even when Coulomb's law holds, and unlike charges attract, it may not be for the expected reason. Several types of analyses now indicate that even ‘simple’ electrostatic interactions in water are dominated by entropy, not determined by enthalpy (Gitlin et al. 2006). It is not, of course, unexpected that ions, in water, are highly solvated. What is not understood at a level of detail useful in the prediction of dissociation constants is how the enthalpy of ion–dipole interactions, and the entropy of ordering or disordering of water as ions interact, combine to give a free energy.
There are also further levels of complexity. Essentially all proteins include charged amino acids. Understanding how these charges interact among themselves, and with charges present in the ground state of ligands, or generated in the transition state of catalytic reactions, is beginning to be understood semi-quantitatively, and is more complicated than expected. In particular, the simplest picture of charge–charge interactions in proteins – that the charges were largely isolated from one another by the high dielectric constant and high ionic strength of most biological fluids – is clearly incomplete or incorrect. The charges in a system of protein and ligand must be considered as an electrostatic network, with the possibility of interactions over long distances and with the ability to regulate charge through mechanisms that involve changing the values of pKa of amino acids such as histidine (or, equivalently, the local pH) in response to changes in the local electrostatic environment (Honig & Nicholls, 1995; Gitlin et al. 2006).
Protein plasticity
Early models of protein structure rested on the implicit assumption that proteins were relatively structurally static: that is, that the structure revealed by X-ray crystallography was directly relevant to the problem of molecular recognition. We now understand that proteins can be remarkably plastic. This plasticity appears at a number of scales from subtle amino-acid rearrangements to gross conformational changes (Carlson, 2002). Many proteins undergo large-scale conformational movement, and some seem (based on single-molecule studies) to flicker between active and inactive conformations. Extensive structural rearrangements at the active site of a protein on binding of a ligand are common. Detailed analyses of interactions among the amino acids of a protein – revealed, for example, in studies such as the monumental study by Benkovic and co-workers on dihydrofolate reductase (Benkovic & Hammes-Schiffer, 2003) – show entirely unexpected interactions occurring at large distances. The old model of a protein as a rigid structure is slowly being replaced by one in which the protein (and particularly the active site of a protein) is considered to be, perhaps, closer to a structured liquid crystal than to a crystalline solid.
The ‘lock-and-key’ hypothesis
The model for protein–ligand interactions that permeates the intuition of many biochemists and medicinal chemists is the ‘lock and key’, which has been modified, in more recent thinking, to the ‘induced fit’ model. The ‘lock-and-key’ model has, as its implied core, the idea of a rigid receptor – the ‘lock’ – being fit by a rigid ligand of complementary shape – the ‘key’. We all know, of course, that this model is oversimplified; still, the idea of complementarity in shape, and perhaps in hydrogen bonding or charge–charge interactions, continues to dominate intuition. The extension to the ‘induced fit’ model, where the protein undergoes a conformational change induced by ligand binding, takes into account protein plasticity (see previous section) but the idea still remains that complementarity in shape between protein (in its active formation) and ligand is still important for binding.
Simple ideas are good: they can guide experiment and thinking. Ideas that are too simple can be bad: they can mislead. ‘Lock and key’ and ‘induced fit’ may be in the latter category. By emphasizing ‘shape’ and ‘charge complementarity’ and related concepts, they implicitly emphasize enthalpy. An important part of molecular recognition – in fact, often the dominant part – can be entropy, and ‘lock-and-key’ and ‘induced-fit’ models with their emphasis on a ‘tight’ fit can lead in the wrong direction. It now increasingly seems that the fit that gives the tightest association is, in fact, a ‘sloppy’ fit: one that is sufficiently complementary in shape to allow some favorable enthalpy, but loose enough not to be entropically too unfavorable (Williams et al. 2004). Unfortunately, even this description is conceptually inadequate, since it focuses on the protein and the ligand. In fact, the water solvating the surfaces of the protein and the ligand may dominate the association through combinations of enthalpy of desolvation and entropy of solvent release and disordering, and we have no way of building solvation/desolvation into the idea of a fit, whether tight or sloppy.
Enthalpy/entropy compensation (EEC)
EEC has emerged as a focus of concern in molecular recognition. In examining the binding of a series of ligands to a protein, it has been observed (not infrequently) that changing the structure of the ligand produces unexpectedly small changes in the value of the dissociation constant (Gilli et al. 1994; Dunitz, 1995; Searle et al. 1995; Williams et al. 2004). On examination, small changes in ΔG often mask much larger compensating changes in ΔH and −TΔS. EEC was (perhaps correctly) dismissed for many years as an artifact, reflecting errors in analysis (Cornish-Bowden, 2002; Houk et al. 2003). Early studies of the thermodynamics of processes involving proteins were, in general, usually carried out by examining the temperature-dependence of the dissociation constant (van't Hoff analysis). These studies are notoriously prone to errors of types that would give the appearance of EEC (Liu & Sturtevant, 1995, 1997; Naghibi et al. 1995). The development of isothermal titration calorimetry (ITC) has, however, largely removed these errors, but EEC has not disappeared. It seems that it is, in fact, a real and important part of interactions in biology.
Dunitz, Williams, and others have postulated a qualitative explanation for EEC (Dunitz, 1995; Searle et al. 1995; Williams et al. 2004). In this hypothesis, enthalpically more favorable (tighter) binding (in the ‘lock-and-key’ or ‘induced-fit’ model) necessarily results in greater entropic restriction, and thus in more unfavorable entropy. This hypothesis is fine as far as it goes, but it neglects entirely the role of water, solvation, and ordering/disordering of water near surfaces. Understanding EEC is, in our opinion, one of the most important challenges at the core of molecular recognition (Krishnamurthy et al. in press).
What has changed?
Given these many conceptual and practical difficulties, what has changed in the last decade to suggest that now might, finally, be the time when it will be possible to make progress in understanding molecular recognition in water? We list seven relevant developments:
(i) ITC directly measures the heat released upon titration of protein by ligand (Wiseman et al. 1989; Turnbull & Daranas, 2003). It thus makes it possible, for the first time, to measure ΔG and ΔH, and to estimate TΔS, all accurately, for association of proteins and ligands of many kinds. Calorimetry remains a limited technique in protein biochemistry, because it requires substantial amounts of material (∼0·5 mg for a 30 kDa protein). Nonetheless, with appropriately chosen model proteins, there is, for the first time, the prospect of having good thermochemical data.
(ii) Recombinant DNA technology, model over-expression systems (e.g. insect cells), and protein purification technology (e.g. Ni2+ chromatography for the purification of His6-tagged proteins) have revolutionized the production of proteins, and the preparation of site-specific mutants. These proteins will be crucial in testing hypotheses of structure–activity relationships in rational ligand design.
(iii) Biostructural analyses – especially X-ray crystallography – have been developed to a level where it is now routine to get accurate structures of a protein with a family of ligands, and of ligands with a family of mutants of a common protein. These types of data provide the basis for any analyses of relationships between structure and binding.
(iv) Hydrogen/deuterium (H/D) exchange experiments reveal the amino-acid residues of a protein that either interact directly with the ligand or that are in regions of the protein that are tightened or loosened as a result of ligand binding. This technique gives information regarding protein–ligand complex dynamics, and thus is complementary to static X-ray crystal structures of these complexes.
(v) Mass spectrometry (particularly, electrospray ionization) has proven useful as a detection method for techniques such as H/D exchange. It has also been used to evaluate and discover high-affinity ligands for proteins.
(vi) Computer simulations are, finally, beginning to give useful information concerning binding, and even concerning protein mobility and solvation. These simulations are still much too slow to give accurate values of thermodynamic parameters, but they are useful in guiding intuition.
(vii) High-throughput assays have allowed the determination of moderate affinity (∼μm) ligands for many proteins from structurally diverse families of ligands. These lead ligands can then be further optimized and studied using ITC to determine the structural basis for the different affinities.
Carbonic anhydrase (CA) as a model
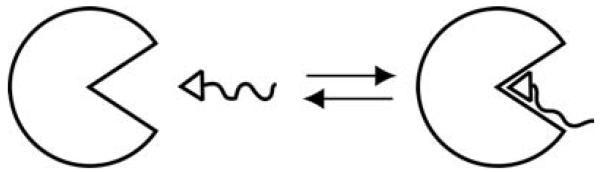
In our studies, we have used the system of bovine carbonic anhydrase II (BCA) and p-substituted benzenesulfonamides as a model to study protein–ligand binding and understand rational ligand design (A. R. Urbach & G. M. Whitesides, unpublished observations). CA has been well-defined structurally using X-ray crystallography. These structures have revealed that CA binds most p-substituted benzenesulfonamides with the same geometry (the ionized sulfonamide nitrogen, ArSO2NH−, binds to the Zn2+ co-factor, and the phenyl ring interacts directly with a hydrophobic pocket of the enzyme); this geometry is independent of the nature of the para substituent. This consistent mode of binding allows us to consider the interaction of the para substituent with the surface of BCA with high confidence that we know how the phenyl ring (and thus, the substituent) is positioned in the active site (Fig. 1). Thus, this system perturbs a known interaction, rather than probing an undefined and/or variable one. Perturbation approaches are often the simplest ones to use in working on complicated problems.
Fig. 1.
Association of a benzenesulfonamide with tail (shown as wavy line) to carbonic anhydrase. The tail can make favorable contacts with the surface of the protein.
The protein is available commercially in high purity. This availability allows the use of ITC to determine the thermodynamic parameters for the binding of ligands to it.
Perturbation studies: benzenesulfonamides with flexible ‘tails’
We have used p-substituted benzenesulfonamides in which the para substituents were oligomeric tails (e.g. alkyl, fluoroalkyl, oligoethylene glycol, oligoglycine, and oligosarcosine) (Jain et al. 1994a,b; Gao et al. 1995; Krishnamurthy et al. 2006). Our results for the binding of benzenesulfonamides with alkyl and fluoroalkyl tails to BCA revealed that affinity increased (the dissociation constant decreased) as the length of the tail increased, and reached a plateau when the tail was hexyl or heptyl (Jain et al. 1994b; Gao et al. 1995). [Our results were in good agreement with those reported by King and Burgen for human carbonic anhydrase II (HCA) (King & Burgen, 1976)]. We interpreted this result as originating from a favorable contribution due to the hydrophobic effect that increased in magnitude with increasing tail length (due to increasing buried surface area between the tail and the surface of the protein) until the tail was longer than the depth of the catalytic cleft of BCA. The decrease of the dissociation constant with increasing tail length was greater for the binding of benzenesulfonamides with fluoroalkyl tails to BCA than of benzenesulfonamides with alkyl tails. This variation disappeared, however, when the decrease of dissociation constants with increasing molecular surface area (using the van der Waals radii of fluorine and hydrogen) of the ligands was examined. This result suggested that the greater ‘hydrophobic bonding’ contributed by fluorine than hydrogen was due to its greater size (Gao et al. 1995).
Our results for the binding of benzenesulfonamides with alkyl and fluoroalkyl tails to BCA contrasted with results we obtained for the binding of benzenesulfonamides with oligoethylene glycol and oligoglycine tails (Jain et al. 1994a). For the latter two series of ligands, we found an entirely unexpected result: the values of dissociation constant were approximately independent of tail length for both series, and did not decrease with increasing tail length as the benzenesulfonamides with alkyl and fluoroalkyl tails had done. This insensitivity of dissociation constant to tail length was particularly difficult to rationalize because the interaction of the tails for benzenesulfonamides with oligoglycine tails with the protein was sufficiently strong to decrease the NMR T2 relaxation times of the α (or methylene) protons of the first three residues of these tails to <25 ms (the value for these residues when free in solution was 230 ms) (Jain et al. 1994a). The tails were also sufficiently ordered that we were able to locate the first three residues of the tails (for benzenesulfonamides with both oligoglycine and oligoethylene glycol tails) in contact with a hydrophobic patch (the so-called ‘hydrophobic wall’) of CA in X-ray structures of the protein–ligand complexes (Cappalonga Bunn et al. 1994; Boriack et al. 1995). The principal inference from these studies was that the first three residues of the tails for these two series interacted in a similar fashion (apparently through hydrophobic contacts) with the hydrophobic wall of CA, but that – counter to our expectations – this interaction had no effect on the value of the dissociation constant.
Our initial explanation for these results was a form of EEC: we hypothesized that longer tails interacted more favorably enthalpically with the surface of BCA (due to greater van der Waals contacts, etc.) than shorter ones, but that this interaction was disfavored entropically due to the larger number of degrees of conformational freedom that were restricted to allow such an interaction to occur (Cappalonga Bunn et al. 1994; Jain et al. 1994a). We found it astonishing that perfect compensation was working with two very different types of tails, especially in light of the aforementioned experimental observation that increasing the length of the tail for p-substituted benzenesulfonamides with alkyl tails decreased the dissociation constant.
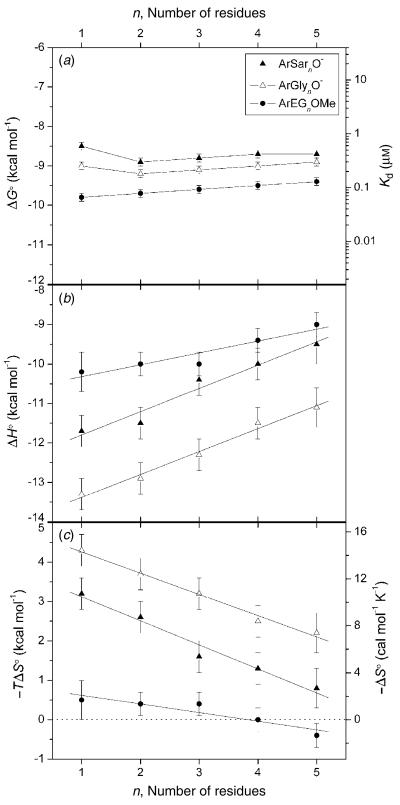
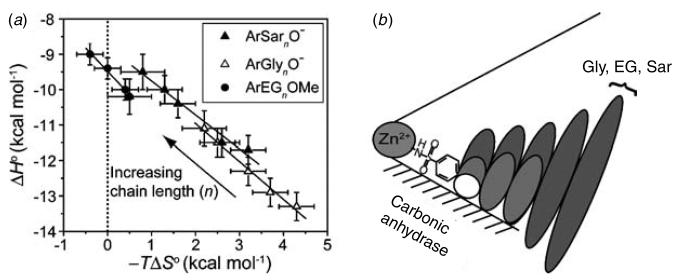
To test our explanation of EEC, we recently undertook a calorimetric investigation of the binding of benzenesulfonamides with oligoglycine, oligoethylene glycol, and oligosarcosine tails to BCA to separate the dissociation constants into their enthalpic and entropic components (Krishnamurthy et al. 2006). Our intent was to characterize the interaction of these tails with the surface of BCA, in order to further our understanding of the interaction between these classes of tails and proteins. These calorimetric data clearly demonstrate that our initial explanation was completely incorrect: the enthalpy of binding became less favorable as the length of the tail increased, while the entropy of binding became less unfavorable for all of the series studied (Fig. 2). The changes in enthalpy and entropy of binding with tail length (total variation of 1–2 kcal mol−1, or 10–15%) perfectly compensated one another (making the dissociation constant insensitive to tail length) for all three series of ligands studied (Fig. 3a). On the basis of these data, and of measurements of heat capacities, we now rationalize the behavior of this system using a model in which the interface between the ligand and the hydrophobic wall of the protein becomes less intimate (less ‘tight’) as the length of the tail increases. In this model, the decreasing tightness of the interface arises because residues of the tail farther from the phenyl ring of the ligand destabilize the binding of residues of the tail closer to the phenyl ring (Fig. 3b). The decreasing tightness of the interface (with increasing length of the tail) results in increasing mobility of the tail of the ligand in the complex, and in decreasing magnitude of the unfavorable entropy of binding. In parallel, the decreasing tightness of the interface results in fewer van der Waals contacts between the ligand and protein, and in decreasing exothermicity of binding.
Fig. 2.
Variation in (a) free energy of binding and dissociation constant, (b) enthalpy of binding, and (c) entropy of binding with the number of residues in the tail for benzenesulfonamides with oligoethylene glycol (ArEGnOMe), oligoglycine (ArGlynO−), and oligosarcosine (ArSarnO−) tails. Linear fits to the data in (b) and (c) are shown. The horizontal dashed line in (c) separates favorable (−TΔS° <0) from unfavorable (−TΔS° >0) entropy of binding. (Reprinted with permission from Krishnamurthy et al. 2006. Copyright 2006 American Chemical Society.)
Fig. 3.
The association of para-substituted benzenesulfonamides with oligoethylene glycol (ArEGnOMe), oligoglycine (ArGlynO−), and oligosarcosine (ArSarnO−) tails with bovine carbonic anhydrase II. (a) EEC plot for the association. The solid lines are linear fits to the datasets and give values for the compensation (from the slopes) as follows: −0·96±0·08 (ArSarnO−), −1·07±0·07 (ArGlynO−), and −1·32±0·09 (ArEGnOMe); these observations demonstrate perfect compensation between enthalpy and entropy. The dotted vertical line separates favorable (−TΔS°<0) from unfavorable (−TΔS°>0) entropy of binding. (b) A schematic diagram for the association. This schematic diagram represents the catalytic cleft of the enzyme as a cone with the Zn2+ co-factor at the apex. The bottom surface (shaded) of the cleft is the ‘hydrophobic wall’ of the enzyme. Ellipses depict the residues of the ligand; the sizes of the ellipses are roughly proportional to the mobility of the individual residues. Benzenesulfonamide ligands with one (white), three (light gray), and five (dark gray) residues in the tail are shown. (Modified with permission from Krishnamurthy et al. 2006. Copyright 2006 American Chemical Society.)
Conclusions
Rational ligand design has been exceptionally challenging because we still do not understand non-covalent intermolecular interactions in water. The crucial issues that we must understand to be able to frame the problem include: the relative importance of enthalpy and entropy for molecular recognition in water, the origin of the hydrophobic effect, the attraction and repulsion of point charges in water, the effect of protein plasticity on ligand binding, the importance (if any) of shape and charge complementarity between the ligand and active site of the protein (specifically, how can small molecules, which do not fill the active site of a protein, be good ligands for a protein?), and the surprising interplay between enthalpy and entropy for association of systematically varied ligands with a protein (EEC).
Many technological advances have been made in recent years and promise to allow for more careful examination and understanding of these different issues. Particularly notable has been the advent of commercially available microcalorimeters, which allow the measurement of the free energy, enthalpy, and entropy of binding. Dissecting the free energy of binding into enthalpy and entropy is, we believe, crucial to addressing the problem of rational ligand design.
The examination of model protein–ligand systems, where structural parameters of the ligand can be varied in controllable ways, offers a means to begin to understand the problem of rational drug design. Appropriate model systems require extensive biophysical characterization to be able to assign increases and decreases in affinity (that is, function) with variation of ligand structure to distinct, interacting components in the protein–ligand complex.
So: in an area that is vitally important to understanding ‘life’ in molecular terms, what can we say after 50 years of work by a number of superb, thoughtful scientists? What do we know about why ligands bind to proteins, and what can we say about how to move from empiricism to design? We can say, we believe, that we have begun to understand the borders of the problem. We know much more than we knew 50 years ago: most importantly, we know (perhaps, or at least partly) what we do not know. We can begin to define the problem with some accuracy. We have begun to understand some of the subtleties of water. We understand better how little we understand about entropy. We understand some of the surprising complexities of ‘simple’ electrostatic interactions. We understand something of the complexity of the dynamic structure of proteins. We have begun to develop experimental and analytical tools that display – and in some cases solve – specific problems.
We are certainly not close to the end of the problem, but we are at least at the beginning.
Acknowledgements
Research on rational ligand design in our laboratory has been supported by the National Institutes of Health (GM51559 and GM30367).
References
- Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- Boriack PA, Christianson DW, Kingery-Wood J, Whitesides GM. Secondary interactions significantly removed from the sulfonamide binding pocket of carbonic anhydrase II influence binding constants. Journal of Medicinal Chemistry. 1995;38:2286–2291. doi: 10.1021/jm00013a004. [DOI] [PubMed] [Google Scholar]
- Cappalonga Bunn AM, Alexander RS, Christianson DW. Mapping protein-peptide affinity: binding of peptidylsulfonamide inhibitors to human carbonic anhydrase II. Journal of the American Chemical Society. 1994;116:5063–5068. [Google Scholar]
- Carlson HA. Protein flexibility and drug design: how to hit a moving target. Current Opinion in Chemical Biology. 2002;6:447–452. doi: 10.1016/s1367-5931(02)00341-1. [DOI] [PubMed] [Google Scholar]
- Chandler D. Two faces of water. Nature. 2002;417:491. doi: 10.1038/417491a. [DOI] [PubMed] [Google Scholar]
- Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Enthalpy-entropy compensation: a phantom phenomenon. Journal of Biosciences. 2002;27:121–126. doi: 10.1007/BF02703768. [DOI] [PubMed] [Google Scholar]
- Dill KA, Bromberg S. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. Garland Science; New York: 2003. [Google Scholar]
- Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chemistry & Biology. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- Gao J, Qiao S, Whitesides GM. Increasing binding constants of ligands to proteins by using ‘greasy tails’. Journal of Medicinal Chemistry. 1995;38:2292–2301. doi: 10.1021/jm00013a005. [DOI] [PubMed] [Google Scholar]
- Gilli P, Gerretti V, Gilli G, Borea PA. Enthalpy-entropy compensation in drug-receptor binding. Journal of Physical Chemistry. 1994;98:1515–1518. [Google Scholar]
- Gitlin I, Carbeck JD, Whitesides GM. Why are proteins charged? Networks of charge-charge interactions in proteins measured by charge ladders and capillary electrophoresis. Angewandte Chemie International Edition. 2006;45:3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Houk KN, Leach AG, Kim SP, Zhang XY. Binding affinities of host-guest, protein–ligand, and protein-transition-state complexes. Angewandte Chemie International Edition. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- Jain A, Huang SG, Whitesides GM. Lack of effect of the length of oligoglycine- and oligo(ethylene glycol)-derived para-substituents on the affinity of benzenesulfonamides for carbonic anhydrase II in solution. Journal of the American Chemical Society. 1994a;116:5057–5062. [Google Scholar]
- Jain A, Whitesides GM, Alexander RS, Christianson DW. Identification of two hydrophobic patches in the active site cavity of human carbonic anhydrase II by solution-phase and solid-state studies and their use in the development of tight-binding inhibitors. Journal of Medicinal Chemistry. 1994b;37:2100–2105. doi: 10.1021/jm00039a023. [DOI] [PubMed] [Google Scholar]
- King RW, Burgen ASV. Kinetic aspects of structure-activity relations: the binding of sulphonamides by carbonic anhydrase. Proceedings of the Royal Society London B. 1976;193:107–125. doi: 10.1098/rspb.1976.0034. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy VM, Bohall BR, Semetey V, Whitesides GM. The paradoxical thermodynamic basis for the interaction of oligoethylene glycol, glycine, and sarcosine chains with bovine carbonic anhydrase. II: An unexpected manifestation of enthalpy/entropy compensation. Journal of the American Chemical Society. 2006;128:5802–5812. doi: 10.1021/ja060070r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy VM, Estroff LA, Whitesides GM. Multivalency in ligand design. In: Jahnke W, Erlanson DA, editors. Fragment-Based Approaches in Drug Discovery. Wiley-VCH; Weinheim: in press. [Google Scholar]
- Lazaridis T. Solvent size vs cohesive energy as the origin of hydrophobicity. Accounts of Chemical Research. 2001;34:931–937. doi: 10.1021/ar010058y. [DOI] [PubMed] [Google Scholar]
- Liu YF, Sturtevant JM. Significant discrepancies between van't Hoff and calorimetric enthalpies. 2. Protein Science. 1995;4:2559–2561. doi: 10.1002/pro.5560041212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Sturtevant JM. Significant discrepancies between van't Hoff and calorimetric enthalpies. 3. Biophysical Chemistry. 1997;64:121–126. doi: 10.1016/s0301-4622(96)02229-6. [DOI] [PubMed] [Google Scholar]
- Naghibi H, Tamura A, Sturtevant JM. Significant discrepancies between van't Hoff and calorimetric enthalpies. Proceedings of the National Academy of Sciences USA. 1995;92:5597–5599. doi: 10.1073/pnas.92.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle MS, Westwell MS, Williams DH. Application of a generalized enthalpy-entropy relationship to binding cooperativity and weak associations in solution. Journal of the Chemical Society – Perkin Transactions. 1995;2:141–151. [Google Scholar]
- Southall NT, Dill KA, Haymet ADJ. A view of the hydrophobic effect. Journal of Physical Chemistry B. 2002;106:521–533. [Google Scholar]
- Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? Journal of the American Chemical Society. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Williams DH, Stephens E, O'Brien DP, Zhou M. Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angewandte Chemie International Edition. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin L-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]