Abstract
Summary
The genetic contribution to age-related bone loss is not well understood. We estimated that genes accounted for 25–45% of variation in 5-year change in bone mineral density in men and women. An autosome-wide linkage scan yielded no significant evidence for chromosal regions implicated in bone loss.
Introduction
The contribution of genetics to acquisition of peak bone mass is well documented, but little is know about the influence of genes on subsequent bone loss with age. We therefore measured 5-year change in bone mineral density (BMD) in 300 Mexican Americans (>45 years of age) from the San Antonio Family Osteoporosis Study to identify genetic factors influencing bone loss.
Methods
Annualized change in BMD was calculated from measurements taken 5.5 years apart. Heritability (h2) of BMD change was estimated using variance components methods and autosome-wide linkage analysis was carried out using 460 microsatellite markers at a mean 7.6 cM interval density.
Results
Rate of BMD change was heritable at the forearm (h2=0.31, p=0.021), hip (h2 =0.44, p=0.017), spine (h2=0.42, p=0.005), but not whole body (h2=0.18, p=0.123). Covariates associated with rapid bone loss (advanced age, baseline BMD, female sex, low baseline weight, postmenopausal status, and interim weight loss) accounted for 10% to 28% of trait variation. No significant evidence of linkage was observed at any skeletal site.
Conclusions
This is one of the first studies to report significant heritability of BMD change for weight-bearing and non-weight-bearing bones in an unselected population and the first linkage scan for change in BMD.
Keywords: Bone Loss, Bone Mineral Density, Heritability, Osteoporosis, Quantitative trait locus (QTL)
Introduction
Osteoporosis is a skeletal condition of major public health significance, contributing toward the risk of fragility fracture in women and men of all populations. The degenerative disorder and associated fragility fractures have devastating effects on health, resulting in substantial morbidity, and increased mortality for hip and vertebral fractures [1]. Bone mineral density (BMD), as the foremost determinant of bone strength and major predictor of future fractures, has been extensively studied to help identify the environmental [2–4] and genetic factors [5–8] influencing risk for osteoporosis.
While the contribution of genetics to variation in BMD is widely acknowledged, the mechanisms by which genetic factors affect BMD are not well understood. Bone is dynamically maintained through the cycle of bone formation and resorption, with changes in bone mass resulting from an imbalance of bone turnover processes. In general, bone turnover yields a net increase in BMD during adolescence and young adulthood leading to peak bone mass attainment, followed by a net decrease in BMD resulting in the subsequent loss of bone with advanced age. Cross-sectional study designs cannot sufficiently distinguish between processes leading to peak BMD acquisition versus loss with age, and this has been a persistent limitation of cross-sectional epidemiological and genetic studies of BMD, particularly those carried out in older individuals since such studies cannot allow for variation in rates of change in BMD during aging. Longitudinal studies have shown that weight, interim change in weight, alcohol consumption, smoking, sex, estrogen replacement therapy, menopausal status, exercise, calcium intake, and serum vitamin D level may affect change in BMD over time, and that rates of BMD change may differ among skeletal sites [9–13]. Rate of bone loss as a risk factor for fracture independent of bone mass has recently been reported in a cohort of postmenopausal women (mean age of 62 years) [14], reinforcing the clinical importance of change in BMD for bone health.
The degree to which genes affect the rate of BMD change over time remains largely unresolved [15]. To date, very few studies have explicitly addressed the role of genetics for change in bone traits over time. One investigation of 25 monozygotic (MZ) and 21 dizygotic (DZ) pairs of twins reported no evidence of heritability for decline in radial bone mass over 16 years [16], while a second study reported greater similarity between 21 MZ than 19 DZ twin pairs for annualized percent change in lumbar spine and Ward’s triangle over 1 to 5 years [17]. A larger study of 177 monozygotic and 185 dizygotic female twin pairs (ages 45 to 82), revealed evidence for genetic effects on 5-year change in BMD at the lumbar spine, whole body, and forearm, but not hip [18]. Finally, a study of premenopausal sisters from 178 sibships demonstrated significant heritability (h2=0.29 to 0.35) of 6-year change in femoral neck bone mineral content (BMC) and BMD [19]. Although there is some consistency among the results of these studies, there are also differences; for example, two studies report significant genetic effects on change in hip BMD [17, 19], and one does not [18]. These differences may be due to the small sample sizes of some studies, length of time elapsed, age range of subjects, and study design (twins versus siblings). Thus, additional research is needed because of the inconsistency of results regarding the heritability of BMD change at all high-risk fracture sites (i.e., spine, hip, and forearm) in postmenopausal women. Furthermore no studies have clearly demonstrated heritability of BMD change in men of any age, or performed linkage analysis to find QTLs influencing BMD change in any population.
The San Antonio Family Osteoporosis Study (SAFOS) was designed to investigate the influence of genes and environmental factors on BMD and change in BMD over time in Mexican Americans. In the current report, we assessed the cumulative effects of genes (i.e., additive heritability) and performed autosome-wide linkage analyses on approximately 5-year longitudinal change in BMD (ΔBMD) at several skeletal sites among 300 men and women (>45 years of age) in 32 extended pedigrees.
Methods
Subjects and data collection
Recruitment and data collection for the baseline phase of the SAFOS has been previously described in detail [8]. The families included for study were selected without regard for health outcomes and represent a relatively random sample of low income families from urban San Antonio. In brief, 34 probands of Mexican American descent aged 40 to 60 were identified, and all first, second, and third degree relatives of probands and their respective spouses were invited to participate in the study. The only criterion for inclusion of probands was that they have large families (>6 members) in the San Antonio area. Anthropometric, medical, and body composition data were collected during medical examinations at baseline between 1997 and 2000. Body composition data and select covariates were reassessed during a follow-up examination (2002–2006) occurring 3 to 8 years later (mean=5.5 years). Lifestyle, medical history, and reproductive history data were collected via questionnaire. Of the original sample of 895 individuals from 34 families considered in this study (ages 18 to 98), 724 (80.9%) have currently been re-enrolled for follow-up.
The present analyses were confined to the subsample of individuals aged >45 years at baseline (n=370 at baseline, n=300 at follow-up). This cohort comprises age-eligible members of 32 families, among whom are 173 sibling pairs, 126 first-cousin pairs, and 189 other relative pairs. Two of the initial 34 families did not include any relative pairs in this age range and were thus excluded from these analyses. The heritability and linkage analyses carried out in this study exclude individuals younger than 45 years at baseline because differences in the biological processes that influence bone turnover between younger and older individuals may be due, in part, to different genes contributing to bone turnover in younger and older individuals. This speculation is supported by (unpublished) genetic modeling in our sample, showing unequal genetic variances between individuals younger and older than 45 years for change in forearm (p=0.04) and hip BMD (p=0.02).
Measurements of BMD of the total hip, total lumbar spine (L1-L4), ultradistal radius, 33% ulna (measured at one-third its total length from the distal end), and whole body were obtained by dual-energy x-ray absorptiometry (DXA) at both baseline and follow-up. Ultradistal radius and 33% ulna sites were analyzed separately because these sites are composed of differing proportions of cortical and trabecular bone: 33% ulna, like total hip, is largely comprised of cortical bone, whereas ultradistal radius, like total spine, is primarily trabecular bone. During the interim between baseline and follow-up clinic visits, DXA equipment was upgraded from a pencil beam Hologic 1500 W to a fan beam Hologic 4500 W absoptiometer (Hologic, Inc., Bedford, MA). A software upgrade was also included for compatibility of scoring algorithms between the two machines. Cross-calibration of absorptiometry equipment used at baseline and follow-up indicated excellent agreement between measurements taken from Hologic 1500 W and 4500 W scanners (e.g., r2>0.99 for measurements taken at the hip and spine from the two scanners on the same 10 subjects); thus the effect of this equipment change on statistical analyses and results should be minimal (see Appendix).
For quality control, the same technician operated all equipment, and phantom measurements were taken daily to guard against measurement drift. Patient positioning was performed according to the Hologic positioning protocol; baseline and follow-up scans were all compared by the same reviewer, and when necessary, scans were re-analyzed to prevent non-overlapping regions of interest.
Covariates considered in this study include sex, age, age2, age × sex, age2 × sex, site-specific baseline BMD, baseline weight, annualized change in weight, annualized change in height, and baseline and follow-up measures of menopausal status (defined by surgical menopause or 1 or more elapsed years without menstrual period). Measurements of all baseline covariates were previously described in Mitchell et al. [8]; follow-up measurements of covariates were assessed identically to the baseline measures. Annualized change in BMD (ΔBMD), height, and weight was calculated as the difference between follow-up and baseline measurements divided by the exact elapsed time between clinic visits.
Genotyping
Genotyping for SAFOS was carried out as previously described in detail [7] for the San Antonio Family Heart Study (SAFHS, the parent project to SAFOS) January 2007 genetic map build: 460 highly polymorphic microsatellite markers across all chromosomes were genotyped, and genetic maps were assembled via the program CRI-MAP [20] and confirmed by deCODE (deCODE genetics, Reykjavik, Iceland). Mean inter-marker distance was 7.6 cM, ranging from <0.1 cM to 15.7 cM (Haldane).
Statistical analyses
The two goals for analyses presented herein were (1) to determine the extent to which genetic and measured environmental factors contribute to the phenotypic variation in ΔBMD at different skeletal sites over time, and (2) to perform an autosome-wide linkage scan for regions affecting ΔBMD. Prior to analyses, the distributions of ΔBMD phenotypes and covariates were assessed, and data points greater than four standard deviations from trait and covariate means were excluded (0 to 3 observations removed per trait or covariate).
As previously described in detail [8, 21], heritability of ΔBMD was estimated using variance decomposition methods, which model phenotypic variation in ΔBMD at each bone site as a function of effects attributable to the measured covariates, additive genetic (based on expected allele sharing between pairs of relatives), and unmeasured error components. This model takes the general form where yi is the annualized ΔBMD for the ith individual, μ is the sample mean ΔBMD, Xij is the jth covariate for the ith individual, βj is the corresponding regression coefficient, gi is the additive genetic effect, and ei is the residual error effect (which includes unmeasured environmental and non-additive genetic components). Pedigree-based maximum likelihood methods were used to estimate these parameters, from which residual narrow-sense heritability (h2r, the proportion of total trait variance due to the additive genetic component after adjusting for environmental covariates) was determined. The likelihood ratio test was used to assess the significance of model parameters by comparing the full model (all covariates and additive genetic effects) with a nested model lacking a specific component. The test statistic asymptotically follows the chi-squared distribution with one degree of freedom for testing covariates, and follows a 50:50 mixture of chi-squared distribution with one degree of freedom and a point mass at zero for testing heritability. Power was 94%, 84%, and 66% to detect true heritability of 0.45, 0.35, and 0.25, respectively at α=0.05. The analysis of each ΔBMD phenotype was limited to individuals with observed data for all retained covariates. The proportion of total variance attributable to covariates was estimated by comparing the estimated variance in the model excluding all significant covariates to that of the model including significant covariates.
Multipoint linkage scans were performed using the variance components method, which extends the above model by also including the effect of a presumed QTL, σ2QTL, as a component of ΔBMD genetic variance. Maximum likelihood methods were used to estimate σ2QTL based on the expected covariance of relatives due to their identity-by-descent (IBD) at a given marker (two-point analyses) or at an arbitrary chromosomal location (multipoint analyses) in tight linkage with the presumed QTL. A Markov Chain Monte Carlo algorithm, as implemented in Loki, was used to calculate multipoint IBD probabilities using data from all genotyped individuals [22]. The likelihood ratio test was used to compare the linkage model to the polygenic (i.e., no linkage, σ2QTL =0) model, and findings were reported in LOD scores (i.e., log10 of the likelihood ratio). To remedy the potential consequence of phenotype distribution on calculated LOD scores, 10,000 simulations of an unlinked marker were performed and linkage analyses carried out on each to determine the empirical LOD score distribution for each phenotype. Linear LOD score adjustments according to the empirical distribution were then applied to our findings [23]. Power to detect linkage was low: 20% and 33%, respectively, to detect a QTL describing 35% of phenotype variance at a LOD threshold of 2.0 and 1.5. Genetic analyses were performed using the sequential oligogenetic linkage analysis routines (SOLAR) software [24]. For illustrative purposes, histograms and LOD score plots were created in R (R Foundation for Statistical Computing, Vienna, Austria).
Results
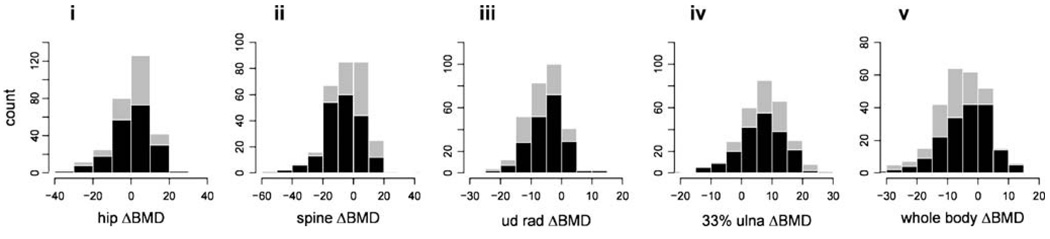
Population characteristics of the 300 individuals in our study are summarized in Table 1. Mean length of follow-up was 5.5 years and ranged from 3.2 to 8.0 years. The prevalence of obesity in the sample was high, as indicated by mean body mass index (BMI) for men and women of 31.1 and 32.9 kg/m2, respectively. Distributions of annualized ΔBMD for different skeletal sites, calculated from measurements taken at baseline and follow-up, are depicted in Fig. 1. Negative values indicate average yearly decline in BMD per year, whereas positive values indicate average yearly gain. BMD declined, on average, for lumbar spine, ultradistal radius, and whole body, but not 33% ulna or total hip. The differences in magnitude and direction of ΔBMD across skeletal sites reflects the site-specific consequence of aging on bone; indeed, correlations for ΔBMD among skeletal sites are low (r=0.15 to 0.41). Also, mean ΔBMD of the spine differed markedly between men and women (p<0.001), possibly due to the high prevalence of osteoarthritis and aortic calcification in men, leading to inflated densitometry values with increasing age [25–27].
Table 1.
Mean population characteristics (SD) [95% CI]
| total sample | women | men | ||||
|---|---|---|---|---|---|---|
| trait | n=300 | n=197 | n=103 | |||
| length of follow-up (years) | 5.5 | (0.6) | 5.5 | (0.6) | 5.4 | (0.6) |
| demographic | ||||||
| age (years) | 56.7 | (8.8) | 56.3 | (8.7) | 57.6 | (9.0) |
| lifestyle | ||||||
| alcohol consumption (%) | 27.0 | - | 15.7 | - | 48.5 | - |
| smoking history (%) | 17.3 | - | 9.6 | - | 32.0 | - |
| medical | ||||||
| diabetes (%) | 28.3 | - | 28.4 | - | 28.2 | - |
| reproductive | ||||||
| post-menopausal (%) | 41.2 | - | 62.8 | - | - | |
| anthropometric | ||||||
| height (cm) | 160.0 | (8.9) | 155.2 | (5.4) | 169.1 | (6.9) |
| weight (kg) | 82.7 | (18.8) | 79.3 | (17.4) | 89.3 | (19.7) |
| BMI (kg/m2) | 32.3 | (6.6) | 32.9 | (6.9) | 31.1 | (6.0) |
| change in height (cm/year) | −0.12 | (0.30) | −0.14 | (0.28) | −0.07 | (0.32) |
| change in weight (kg/year) | 0.00 | (1.37) | 0.09 | (1.19) | −0.19 | (1.66) |
| change in BMI (kg/m2/year) | 0.05 | (0.53) | 0.10 | (0.50) | −0.04 | (0.58) |
| BMD (g/cm2) | ||||||
| total hip | 0.96 | (0.16) | 0.93 | (0.15) | 1.02 | (0.15) |
| total spine | 1.01 | (0.17) | 0.97 | (0.16) | 1.07 | (0.17) |
| ultradistal radius | 0.47 | (0.08) | 0.44 | (0.07) | 0.52 | (0.07) |
| 33% radius | 0.67 | (0.11) | 0.62 | (0.07) | 0.77 | (0.08) |
| whole body | 1.10 | (0.13) | 1.05 | (0.12) | 1.19 | (0.10) |
| annual BMD change (mg/cm2/year) | ||||||
| total hip | 0.3 | [−0.09, 1.5] | −0.2 | [−1.8, 1.3] | 1.2 | [−0.6, 3.1] |
| total spine | −4.6 | [−6.0, −3.2] | −6.9 | [−8.6, −5.2] | −0.2 | [−2.3, 1.9] |
| ultradistal radius | −5.8 | [−6.5, −5.2] | −5.2 | [−6.0, −4.8] | −7.0 | [−8.1, −5.9] |
| 33% radius | 7.0 | [6.2, 7.8] | 6.3 | [5.5, 7.2] | 8.3 | [7.0, 9.6] |
| whole body | −5.6 | [−6.5, −4.6] | −4.3 | [−5.6, −3.1] | −7.9 | [−9.3, −6.6] |
Figure 1.
Distributions of annual change in BMD (mg/cm2/yr) for (i) total hip, (ii) total spine, (iii) midpoint radius, (iv) ultradistal radius, and (v) whole body. Gray bars represent the total sample and black bars represent women.
Table 2 shows for each bone site the residual heritability of ΔBMD after adjusting for sex, age, age2, age × sex, age2 × sex, baseline BMD, baseline weight, menopausal status, interim change in weight, and (for spine and whole body ΔBMD only) interim change in height. In light of the well-documented bone loss following menopause, we adjusted ΔBMD for baseline post-menopausal status to correct for women having already undergone transient menopause-related bone loss, as well as interim incidence of menopause to correct for women undergoing rapid menopause-related bone loss during the years of follow-up. After incorporating covariates, we observed significant residual heritability for ΔBMD of the total hip, total spine, and 33% ulna (h2r=0:31 to 0.44; p<0.03 for all). Additionally, we observed modest residual heritability for ultradistal radius ΔBMD (h2r=0:25, p=0.07), but not whole body ΔBMD (h2r=0:18, p=0.123). Approximately 10% to 30% of total variation in ΔBMD was attributable to covariates. Heritability of unadjusted ΔBMD (not shown in table) was similar to heritability after covariate adjustment for all bone sites.
Table 2.
Residual heritability of BMD change
| BMD site | n | h2r | SE | p | R2 |
|---|---|---|---|---|---|
| total hip | 272 | 0.44 | 0.24 | 0.017 | 0.19 |
| total spine | 272 | 0.42 | 0.18 | 0.005 | 0.28 |
| ultradistal radius | 277 | 0.25 | 0.18 | 0.064 | 0.10 |
| 33% radius | 277 | 0.31 | 0.17 | 0.021 | 0.11 |
| whole body | 253 | 0.18 | 0.17 | 0.123 | 0.16 |
h2r=residual heritability
R2=proportion of variation attributable to covariates: sex, age, age2, age × sex, age2 × sex, baseline BMD, baseline weight, baseline postmenopausal status, interim menopause, interim change in weight, interim change in height (included only for spine and whole body ΔBMD)
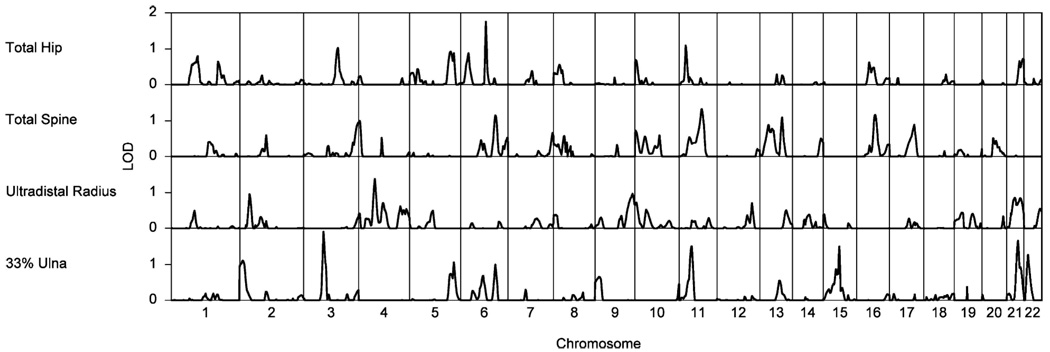
Whole genome multipoint linkage scans were performed for ΔBMD at total hip, total spine, radius, and ulna (Fig. 2), but not whole body, because whole body ΔBMD was not heritable. No evidence of linkage was detected at genome-wide significance; the greatest linkage signal was observed for 33% ulna ΔBMD with a LOD score of 1.90 at 81 cM on chromosome 3p (unadjusted p=0.0018). This region has previously been implicated in lumbar spine BMD in unselected twins and extremely discordant or concordant sib pairs [28]. A novel signal for hip ΔBMD with a LOD score of 1.75 at 103 cM on chromosome 6q was also observed (unadjusted p=0.0008).
Figure 2.
Multipoint LOD scores for change in hip BMD (mg/cm2/yr) on chromosome 6
Discussion
Numerous family and sibling studies of peak BMD have been performed in a variety of populations, and these studies have universally shown the high heritability of peak BMD [3, 8, 21, 29, 30]. Many whole genome linkage scans have also been performed, with QTLs reported at a number of chromosomal regions, though specific QTLs have rarely been replicated across studies [5–7, 28, 31–36], probably due, in part, to genetic heterogeneity among the populations studied. Analyses of BMD data from these previous studies, however, cannot adequately distinguish between loci affecting loss of BMD with age and those affecting the acquisition of peak bone mass occurring in young adulthood. Moreover, the models of BMD variation in these cross-sectional family studies generally assume universal rates of change with age (i.e., by adjusting BMD for age and/or age2) thereby further reducing their ability to find genes that influence individual rates of change. In the current study, we have directly calculated ΔBMD from longitudinal measurements, to better investigate the role of genes on ΔBMD.
Our study provides support that the rate of bone loss in middle-to-older-aged Mexican Americans is heritable. Specifically, we estimated that genes account for 25% to 44% of residual variation in ΔBMD for three bone sites (hip, spine, and forearm) at high risk of fracture. The confidence intervals surrounding these estimates, however, are wide—a feature that is partly a consequence of our sample size (n=300) of adults >45 years of age from large multigenerational families, and partly due to the measurement uncertainty for ΔBMD. We have reported heritability estimates for the combined cohort of men and women aged >45 years; heritability estimates for ΔBMD of similar magnitude were obtained when we restricted analysis to women only (n=197), with the exception that heritability of ΔBMD at the ulna was reduced and did not differ significantly from zero (results not shown). Sample size limitations precluded performing genetic modeling of ΔBMD in men alone. In general, heritability estimates of ΔBMD in our sample (both overall, and in the women-only subset) are similar in magnitude to recently published estimates obtained in both pre-menopausal [19] and peri- and post-menopausal women [18], with the exception that, unlike in our study, significant genetic effects on bone loss of the hip were not observed for the latter [18].
Though significantly heritable, our genome-wide linkage scan for ΔBMD revealed no strong evidence for QTLs (including in the women-only subset; results not shown). However, power in our sample was very low to detect QTLs having relatively modest effects (e.g., at a LOD threshold of 2.0 we have 20% power to detect a locus accounting for 35% of residual phenotypic variance), and it is likely that QTLs of modest effect sizes were missed. While the cumulative influence of genes on ΔBMD is large, the contribution of environmental factors is also important; measured covariates accounted for 10% to 28% of phenotypic variation. Associations with BMD of several of the environmental correlates identified in this study, including age, female sex, postmenopausal status, low body weight, and weight loss, have been detected in previous studies [11–13, 37]. Interestingly, we observed differences in magnitude and direction for ΔBMD across skeletal sites, as has been observed by others, although not always for the same sites [9–11]. Unfortunately, there are insufficient reports of ΔBMD across multiple age ranges and ethnic groups to develop hypotheses regarding potential mechanisms for these differences at this time. However, this result, combined with the observed differences in heritability and variation attributable to environmental correlates among bone sites, suggests that factors regulating 5-year ΔBMD may vary across the skeleton.
A major strength of this study is the use of extended families and longitudinal measurements of BMD to investigate the genetics of bone loss. Previous linkage scans for BMD, of which there have been many, have been ill-suited to find genes affecting change, and all previous attempts to estimate the heritability of ΔBMD have been carried out exclusively in twins or siblings [16–19]. By using information from many types of relative pairs (cousins, avuncular pairs, etc.) in addition to siblings, our estimates of heritability more accurately reflect the truly genetic factors affecting ΔBMD by reducing the contribution of familial non-genetic factors such as household effects to our heritability estimation. Other strengths of this study are the inclusion of both axial (weight-bearing) and peripheral (non-weight-bearing) skeletal sites, and the incorporation into our models of important covariates previously shown to have potential effects on ΔBMD. Furthermore, this is one of the first studies to consider ΔBMD in a population of Mexican descent and, to our knowledge is the only study, to date, reporting a linkage scan for a change in bone phenotype.
Despite these strengths, several limitations of this study must be acknowledged. Foremost is the fact that different absorptiometers were used to collect baseline and follow-up BMD measurements. However, the BMD scoring algorithms were standardized to the degree possible and our cross-calibration experiment (albeit in a limited number of subjects) confirmed outstanding agreement in measurements between the two scanners (see Methods and Appendix). Other limitations and sources of error for this study include the reduced sample size available for follow-up (86.5%) and the possibility of non-random loss to follow-up with respect to bone health. Also, power to detect significant linkage was limited, and our sample size and methods preclude direct detection of gene-by-environment interactions (in particular, gene-by-sex and gene-by-menopausal status interactions). Analyses could not be performed separately in pre- and postmenopausal women (again, due to sample size); however, the similarity of our results with those in both pre- and postmenopausal women in other studies [18, 19] suggests that the impact of this limitation may be not be critical. Finally, there are inherent drawbacks to using DXA to assess BMD, notably the estimation of bone mineral content from a two-dimensional projection, which fails to precisely account for the size (depth) of bone [38]. These limitations, however, would reduce our chances of detecting heritable effects or QTLs, but would not inflate them.
In conclusion, this study provides evidence that ΔBMD is heritable for several skeletal sites in middle-aged men and women. Our results corroborate recent findings for significant heritability of change in femoral neck BMD in premenopausal women [19], as well as other previous twin studies [16, 17] and racial comparisons [39, 40]. Moreover, we report on the genetics of bone loss in Mexican Americans, a population that is under-represented in the osteoporosis literature. While evidence for the heritability of age-related ΔBMD is mounting, the localization of QTLs, and identification of specific genetic factors contributing to variation in bone loss has yet to be realized. Such genes represent a novel area for investigation into the risk factors for osteoporosis.
Acknowledgments
We are deeply grateful for the cooperation of the families participating in the SAFOS. This work was supported by research grants R01-AR43351 and P01-HL45522 awarded by the National Institutes of Health. Support for the Frederic C. Bartter General Clinical Research Center was made available by Clinical National Institutes of Health Grant M01-RR-01346. Development of SOLAR is supported by R01-HG59490. We would also like to express our gratitude to three anonymous reviewers for their thoughtful consideration of this work.
Appendix
DXA measurement and cross-validation
Due to an upgrade in equipment, densitometry was carried out on a pencil-beam Hologic Model 1500 at baseline examination (1997–2000) and a fan-beam Model 4500 W at follow-up examination (2002–2006). For both densitometers, areal BMD was calculated by manufacturer’s software as per current recommendations by dividing bone mineral content (g) by the projected area of the region scanned (cm2). Precision of pencil-beam DXA was 0.009 g/cm2 for spine, 0.007 g/cm2 for total hip, and 0.002 g/cm2 for the manufacturer’s spine phantom. Precision of fan-beam DXA was 0.006 g/cm2 for spine, 0.007 g/cm2 for hip, and 0.002 g/cm2 for radius. Based on the precision of our equipment, least significant change (at 95% confidence) was 3.2 mg/cm2/year for hip, 3.1 mg/cm2/year for spine, and 1.1 mg/cm2/year for forearm DXA measurements.
To address the comparability in our study of measurements taken from Hologic 1500 and 4500 W scanners, we performed cross-calibration of absorptiometry equipment used at baseline and follow-up on 10 participants. Measurements obtained from the two scanners showed near-perfect agreement (R2 values = 99.95%, 99.81%, and 99.87% for spine, total hip, and femoral neck sites, respectively; p<10−13 for all). Moreover, regression slopes (0.99, 0.99, and 1.01, respectively, for spine, total hip, and femoral neck; p<10−13 for all) and paired T-tests (p>0.1, for all) revealed no evidence of systematic or mean differences between absorptiometers. This evaluation of equipment indicates that measurements from Hologic 1500 and 4500 W scanners used in this study are comparable, and that 5-year change in BMD can be adequately calculated. Cross-calibration was not performed for forearm BMD, although we expect that measurements at this site are equally comparable.
Robustness of methods to measurement error
Though we found no evidence to suggest that systematic differences exist in the measurements between scanners, we have nonetheless employed statistical methods that are robust to potential differences. If present, such bias (e.g., systematic measurement error) would decrease our power to detect covariate effects and attenuate our estimate of heritability and linkage, but should not otherwise affect our results. That is, machine differences leading to biased estimates of BMD change could prevent us from assessing environmental and genetic factors affecting BMD change, but would not produce false positive results or lead to overestimation of effects sizes. Our findings, therefore, are conservative. Furthermore, we recognize that deviations from absolute agreement between machines would not adversely affect the genetic analyses as long as the relative BMD between individuals as measured by each scanner is accurate (i.e., variances are comparable). In other words, as long as BMD measured for an individual at baseline is accurate relative to the baseline measurements of the rest of the study sample, and the same holds for measurements at follow-up, the estimation of heritability will be unaffected by inter-machine differences. This is because the genetic modeling used to assess heritability decomposes the trait variance into genetic and environmental components irrespective of the trait mean.
To demonstrate this point, we removed the possibility of any inter-machine effects by first standardizing (mean=0, SD=1) the BMD measurements in our sample separately at baseline and follow-up, and then analyzing yearly change in standardized values. This process retains the information of individuals’ measurements relative to the sample, but not of the absolute magnitude of measurements. In doing so, potential unknown machine differences that could invalidate direct inter-machine comparisons are avoided. Results (not shown) of change in standardized BMD are very similar to the absolute change reported herein. Likewise, results (not shown) of percent change in BMD were also similar. Attenuation of heritability due to measurement error The precision of DXA, which is excellent when looking at cross-sectional BMD measurements, is poor when looking at change over time, leading to considerable uncertainty of actual rates of change. Therefore, rates of change assessed in this study are notably crude, with a large percentage of observations being less than our measurement uncertainty (i.e., no measurable change). Since noise constitutes a substantial portion of the variation in observed rates of change, our results represent a considerable underestimation of the heritability of observed BMD change compared to that of true change (free of such measurement error) [19]. Noise due to our crude assessment of BMD change effectively drowns the heritability signal, and diminishes our ability to detect linkage.
References
- 1.Dennison E, Cole Z, Cooper C. Diagnosis and epidemiology of osteoporosis. Curr Opin Rheumatol. 2005;17:456–461. doi: 10.1097/01.bor.0000166384.80777.0d. [DOI] [PubMed] [Google Scholar]
- 2.Cummings SR, Kelsey JL, Nevitt MC, et al. Epidemiology of osteoporosis and osteoporotic fractures. Epidemiol Rev. 1985;7:178–208. doi: 10.1093/oxfordjournals.epirev.a036281. [DOI] [PubMed] [Google Scholar]
- 3.Krall EA, Dawson-Hughes B. Heritable and life-style determinants of bone mineral density. J Bone Miner Res. 1993;8:1–9. doi: 10.1002/jbmr.5650080102. [DOI] [PubMed] [Google Scholar]
- 4.Wilkins CH, Birge SJ. Prevention of osteoporotic fractures in the elderly. Am J Med. 2005;118:1190–1195. doi: 10.1016/j.amjmed.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 5.Eisman JA. Genetics of osteoporosis. Endocr Rev. 1999;20:788–804. doi: 10.1210/edrv.20.6.0384. [DOI] [PubMed] [Google Scholar]
- 6.Huang QY, Kung AW. Genetics of osteoporosis. Mol Genet Metab. 2006;88:295–306. doi: 10.1016/j.ymgme.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Kammerer CM, Schneider JL, Cole SA, et al. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18:2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell BD, Kammerer CM, Schneider JL, et al. Genetic and environmental determinants of bone mineral density in Mexican Americans: results fromthe San Antonio Family Osteoporosis Study. Bone. 2003;33:839–846. doi: 10.1016/s8756-3282(03)00246-1. [DOI] [PubMed] [Google Scholar]
- 9.Dirschl DR, Henderson RC, Oakley WC. Accelerated bone mineral loss following a hip fracture: a prospective longitudinal study. Bone. 1997;21:79–82. doi: 10.1016/s8756-3282(97)00082-3. [DOI] [PubMed] [Google Scholar]
- 10.Engelke K, Kemmler W, Lauber D, et al. Exercise maintains bone density at spine and hip EFOPS: a 3-year longitudinal study in early postmenopausal women. Osteoporos Int. 2006;17:133–142. doi: 10.1007/s00198-005-1938-9. [DOI] [PubMed] [Google Scholar]
- 11.Hannan MT, Felson DT, Dawson-Hughes B, et al. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:710–720. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 12.Warming L, Hassager C, Christiansen C. Changes in bone mineral density with age in men and women: a longitudinal study. Osteoporos Int. 2002;13:105–112. doi: 10.1007/s001980200001. [DOI] [PubMed] [Google Scholar]
- 13.Knoke JD, Barrett-Connor E. Weight loss: a determinant of hip bone loss in older men and women. The Rancho Bernardo Study. Am J Epidemiol. 2003;158:1132–1138. doi: 10.1093/aje/kwg265. [DOI] [PubMed] [Google Scholar]
- 14.Sornay-Rendu E, Munoz F, Duboeuf F, et al. Rate of forearm bone loss is associated with an increased risk of fracture independently of bone mass in postmenopausal women: the OFELY study. J Bone Miner Res. 2005;20:1929–1935. doi: 10.1359/JBMR.050704. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Shen H, Jiang H, et al. On genetic studies of bone loss. J Bone Miner Res. 2006;21:1676–1677. doi: 10.1359/JBMR.060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christian JC, Yu PL, Slemenda CW, et al. Heritability of bone mass: a longitudinal study in aging male twins. Am J Hum Genet. 1989;44:429–433. [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly PJ, Nguyen T, Hopper J, et al. Changes in axial bone density with age: a twin study. J Bone Miner Res. 1993;8:11–17. doi: 10.1002/jbmr.5650080103. [DOI] [PubMed] [Google Scholar]
- 18.Makovey J, Nguyen TV, Naganathan V, et al. Genetic effects on bone loss in peri-and postmenopausal women: a longitudinal twin study. J Bone Miner Res. 2007;22:1773–1780. doi: 10.1359/jbmr.070708. [DOI] [PubMed] [Google Scholar]
- 19.Hui SL, Koller DL, Foroud TM, et al. Heritability of changes in bone size and bone mass with age in premenopausal white sisters. J Bone Miner Res. 2006;21:1121–1125. doi: 10.1359/jbmr.060412. [DOI] [PubMed] [Google Scholar]
- 20.Green P, Falls K, Crooks S. Documentation for CRI-MAP, Version 2.4. St. Louis, MO, USA: Washington University School of Medicine; 1990. [Google Scholar]
- 21.Brown LB, Streeten EA, Shapiro JR, et al. Genetic and environmental influences on bone mineral density in pre-and post-menopausal women. Osteoporos Int. 2005;16:1849–1856. doi: 10.1007/s00198-005-1948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blangero J, Williams JT, Almasy L. Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol. 2000;19(Suppl 1):S8–S14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger H, van Daele PL, Odding E, et al. Association of radiographically evident osteoarthritis with higher bone mineral density and increased bone loss with age. The Rotterdam Study. Arthritis Rheum. 1996;39:81–86. doi: 10.1002/art.1780390111. [DOI] [PubMed] [Google Scholar]
- 26.Smith JA, Vento JA, Spencer RP, et al. Aortic calcification contributing to bone densitometry measurement. J Clin Densitom. 1999;2:181–183. doi: 10.1385/jcd:2:2:181. [DOI] [PubMed] [Google Scholar]
- 27.Zmuda JM, Cauley JA, Glynn NW, et al. Posterior-anterior and lateral dual-energy x-ray absorptiometry for the assessment of vertebral osteoporosis and bone loss among older men. J Bone Miner Res. 2000;15:1417–1424. doi: 10.1359/jbmr.2000.15.7.1417. [DOI] [PubMed] [Google Scholar]
- 28.Wilson SG, Reed PW, Bansal A, et al. Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet. 2003;72:144–155. doi: 10.1086/345819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown LB, Streeten EA, Shuldiner AR, et al. Assessment of sex-specific genetic and environmental effects on bone mineral density. Genet Epidemiol. 2004;27:153–161. doi: 10.1002/gepi.20009. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Kammerer CM, Wheeler VW, et al. Genetic and environmental determinants of volumetric and areal BMD in multi-generational families of African ancestry: the Tobago Family Health Study. J Bone Miner Res. 2007;22:527–536. doi: 10.1359/jbmr.070106. [DOI] [PubMed] [Google Scholar]
- 31.Devoto M, Spotila LD, Stabley DL, et al. Univariate and bivariate variance component linkage analysis of a whole-genome scan for loci contributing to bone mineral density. Eur J Hum Genet. 2005;13:781–788. doi: 10.1038/sj.ejhg.5201411. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ng MY, Sham PC, et al. Meta-Analysis of Genome Wide Scans Provides Evidence for Gender and Site Specific Regulation of Bone Mass. J Bone Miner Res. 2006;22:173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YJ, Shen H, Xiao P, et al. Molecular genetic studies of gene identification for osteoporosis: a 2004 update. J Bone Miner Res. 2006;21:1511–1535. doi: 10.1359/JBMR.051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peacock M, Koller DL, Hui S, et al. Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int. 2004;15:489–496. doi: 10.1007/s00198-003-1560-7. [DOI] [PubMed] [Google Scholar]
- 35.Streeten EA, McBride DJ, Pollin TI, et al. Quantitative trait loci for BMD identified by autosome-wide linkage scan to chromosomes 7q and 21q in men from the Amish Family Osteoporosis Study. J Bone Miner Res. 2006;21:1433–1442. doi: 10.1359/jbmr.060602. [DOI] [PubMed] [Google Scholar]
- 36.Styrkarsdottir U, Cazier JB, Kong A, et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 2003;1:E69. doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz AV, Sellmeyer DE, Strotmeyer ES, et al. Diabetes and bone loss at the hip in older black and white adults. J Bone Miner Res. 2005;20:596–603. doi: 10.1359/JBMR.041219. [DOI] [PubMed] [Google Scholar]
- 38.Seeman E. Growth in bone mass and size–are racial and gender differences in bone mineral density more apparent than real? J Clin Endocrinol Metab. 1998;83:1414–1419. doi: 10.1210/jcem.83.5.4844. [DOI] [PubMed] [Google Scholar]
- 39.Cauley JA, Lui LY, Stone KL, et al. Longitudinal study of changes in hip bone mineral density in Caucasian and African-American women. J Am Geriatr Soc. 2005;53:183–189. doi: 10.1111/j.1532-5415.2005.53101.x. [DOI] [PubMed] [Google Scholar]
- 40.Tracy JK, Meyer WA, Flores RH. Racial differences in rate of decline in bone mass in older men: the Baltimore men’s osteoporosis study. J Bone Miner Res. 2005;20:1228–1234. doi: 10.1359/JBMR.050310. [DOI] [PubMed] [Google Scholar]