Abstract
Fecal occult blood testing (FOBT) is proven an efficient way of reducing mortality from colorectal cancer but has a relatively low positive predictive value (PPV). This study evaluated the ability to detect K-ras mutations in stool DNA from FOBT cards and to improve the PPV of the screening process. 205 consecutive positive FOBT cards and an arbitrary sample of 38 negative cards from a population-based screening program were included. K-ras mutations in FOBT card stool were sought using allele-specific hybridization. DNA was successfully amplified from 87.2% of cards. In 130 cases with positive FOBT and amplifiable DNA 23 malignancies and 25 adenomas were detected. In 34.8% of the malignancies, a mutation in K-ras was detected. The PPV for malignancies increased from 17.7% (all positive cards) to 60.0% if cards with 4 or more fields were positive and K-ras was positive (RR=2.66,95%CI:1.2–6.1). Testing for k-ras mutations in DNA extracted from stool from positive FOBT cards is feasible. Sequential detection of cancer-associated genetic markers from FOBT-based stool samples may potentially help separate true from false positives in a FOBT-based screening process.
Introduction
Three randomized controlled trials have been conducted over the last 30 years to evaluate the contribution of periodic screening with fecal occult blood tests (FOBT) to the reduction in mortality of colorectal cancer in the studied population [1–5}. The results of these studies demonstrate significant reduction in mortality with biennial as well as annual testing after 8–18 years of follow-up. Yet, despite the cost effectiveness of this screening tool, these tests carry a high false positive rate resulting in the need for many unnecessary colonoscopies.
To improve upon FOBT, stool based tests detecting common colorectal neoplasia-associated DNA mutations were developed by Sidransky et al [6], refined by Jen et al [7] and subsequently validated in a prospective screening clinical trial [8]. The DNA-based diagnostic panel enhanced diagnostic sensitivity for colorectal adenocarcinoma but not for colorectal adenoma [8]. If applied sequentially, stool DNA testing might improve the differentiation of true positive from false positive FOBT results, thus increasing the positive predictive value of the FOB test.
This study was designed to test the feasibility of extracting, of amplifying, and of detecting normal and mutated K-ras DNA from stool samples that were submitted on guaiac-positive and guaiac-negative FOBT cards.
Materials and Methods
Study population
The National FOBT Screening Program for Colorectal Cancer of Clalit Health Services in Israel (CHS, an non-for-profit HMO-type organization covering more than 70% of the Israeli older population), served as the source of FOBT cards for this study. The Screening Program invites all women and men, ages 50–74, members of CHS to have an annual, free-of-charge, FOBT. Of 5000 cards screened, 205 consecutive positive (traces or any positive field) FOBT cards and 38 arbitrarily assigned negative FOBT cards were included in this study. The study was approved by the IRB Committee of Carmel Medical Center.
FOBT screening process
Following suggested diet and medication restriction, subjects applied feces to two windows on a Hemoccult Sensa test card (Beckman Coulter, Inc) on each of three consecutive days for a total of six test windows). The card was then mailed to the CHS National Cancer Control Center laboratories in a pre-paid, pre-addressed envelope supplied as part of the FOBT kit to each subject. In order to eliminate peroxidases from dietary vegetables and fruits known to be responsible for false positive test results [9], the cards were not processed earlier than a week from first stool application. The test was analyzed by applying the supplied developing solution (a stabilized mixture of less than 4.2% hydrogen peroxide and 80% denatured ethyl alcohol and enhancer in an aqueous solution) in a single laboratory. A test was called positive if at least one of the six tested fields showed the typical color change following the guaiac reaction.
Laboratory methods
DNA extraction
After development for the guiac reaction, FOBT cards were kept dry at −20°C until DNA isolation. Stool samples were then excised from all six windows of the FOBT cards and agitated overnight in 5 ml preservation solution containing 1.2 M Guanidium Thiocyanate, 100 mM sodium acetate pH 5.2, 0.5% Triton X100. The next morning, the samples in the preservation solution were extracted briefly with 3 ml of chloroform:isopropanol (94/6; v/v) and then centrifuged at 8000 × g for 10 min at room temperature. The top 4 ml were then diluted with an equal amount of 6 M guanidium hydrochloride. DNA purification resin (Promega) was added and the mix was applied onto a vacuum column. The resin was washed twice with total 6 ml Wash-1 solution (40% Isopropanol, 3 M Guanidium Hydrochloride, 50 mM NaOAc pH 5.2) and then twice with a total of 6 ml Wash-2 solution (60% Ethanol, 100mM sodium acetate pH 5.2). The DNA was eluted with 100 μl of 10 mM Tris hydrochloride pH 7.6. Approximately 200 mg of stool were normally available from the six card-windows of stool, with a varing yield of between 100–1000 ng of apparently mixed bacterial and human DNA.
K-ras mutation analysis
After extracting DNA from the stool samples the first exon of K-ras was amplified by PCR. Primers used were:
Ki-ras 11 primer aggaattcatgactgaatataaacttgt
Ki-ras 12 primer atcgaattcctctattgttggatcatatcc.
5–10 μl of DNA purified from feces (10–100 ng) was added to 50 μl of PCR mix containing PCR-buffer, 200 μM dNTP, 1 μM of Ki-ras11 and Ki-ras12 primers. The mix was heated up to 80°C for 1 min. 1.5 units of Taq polymerase were added. The tubes were sealed and reaction continued for 60 cycles as follows: 56°C for 30 sec, 72°C for 15 seconds, 94°C for 15 seconds. Samples were spotted on nylon membrane (Hybond-N, Amersham or Gene Screen, NEN). 30μl of PCR product was diluted with 150 μl of 0.15N sodium hydroxide and allowed to melt for 20 minutes at room temperature. 200 μl of 0.5M phosphate buffer (pH 7) were added into each well of a dot apparatus, and 12–15 μl of a DNA sample in alkali were transferred into each of the wells before the vacuum was applied. DNA was fixed with UV crosslinking (1200 mW/cm2).
Probe labeling for point mutation detection were labeled by a primer extension approach. A short template was designed that complements the 9 last nucleotides of the 3′-end of the allele specific oligodendronucleotide (ASO) probe detailed below, plus 3 extending thymidine triphosphate nucleotides at its 5′-end, 12 nucleotides in total. After annealing, Klenow DNA polymerase is added in the presence of 32P-deoxyadenine triphosphate, and the ASO probe is labeled by extension upon the template. In this way the final product is the ASO probe containing 3 labeled dAs at its 3′-end. Dotted nylon sheets had been incubated in 20–30 ml of hybridization solution containing 100 mg/ml of herring sperm DNA and 200 pmole of normal sequence competitor oligo for 1hour before the labeled probe was added. Blots were routinely hybridized overnight at 56–59°C. Hybridized filters were briefly washed 2–3 times with a washing solution containing 3XSSC and 0.2% SDS at room temperature, and washed 3–4 times (each time for 15 minutes) with a washing solution prewarmed to 50°C. The washed filters were briefly air dried and exposed to the Phosphoimager plate for 1–3 hours.
Mutations were detected by ASO hybridization. Two copies of each membrane were prepared: One hybridized with a general probe from outside the mutated region detecting any amplified K-ras DNA. The second membrane was hybridized with an ASO probe. Since most of the mutations in codons 12 and 13 in the Israeli population are found in 4 out of 12 possible mutations [10], we used only 4 different ASO probes, representing 86% of the available mutations, as follows:
codon12, AGT cctacgccactagctcca
codon12, GAT cctacgccatcagctcca
codon12, GTT cctacgccaacagctcca
codon13, GAC cctacgtcaccagctcca
competitor oligo, cctacgccaccagctcca
tailer, ttttggagctgg
Signal evaluation
Due to some background expected with normal sequence, a strong signal may be obtained from either the presence of mutated sequences, or the presence of high amount of normal sequences. Also, the efficiency of the different probes was variable. Accordingly, the signals on a given membrane were divided by eye into 3 levels of intensity: background (the majority of signals), weak (slightly above background), and strong (significantly above weak). A signal was considered positive if it was in the strong class of signals, and if it was not highly strong with the general probe.
Follow up
All screened positive cases were referred to primary care physicians and appropriate subspecialists. Recommended evaluation of a positive case was either a full colonoscopy or a flexible sigmoidoscopy with a double contrast barium enema. Post-evaluation findings were coded as: a. no neoplasia (e.g. hemorrhoid, diverticulosis, inflammatory bowel disease, gastritis), b. hyperplastic polyp (non-neoplastic), c. adenomas, of all types (tubular, tubulo-villous, villous), d. invasive neoplasm. The rest of the cards were summarized as no finding (or false positives). Evaluation of the upper GI tract was not mandatory in cases with positive occult blood and no findings from colonoscopy.
Statistical analysis
Positive predictive value (and 95% confidence intervals) of the addition of K-ras results to a positive FOBT was calculated by dividing the number of cases with a positive FOBT result in which a pathological finding was found in follow-up by the total number of positive FOBT. This was done separately for each type of pathologic finding in follow-up. The analysis was performed separately for a) any FOBT positivity and b) FOB finding of 4 or more positive fields (out of 6). The later was shown to carry a much higher probability of cancer [11]. No measures of sensitivity or specificity were calculated because this pilot study evaluated only K-ras performance as a sequential adjunct to the guiaiac test results. The K-ras outcome is not intended to be interpreted independently.
Results
Feasibility
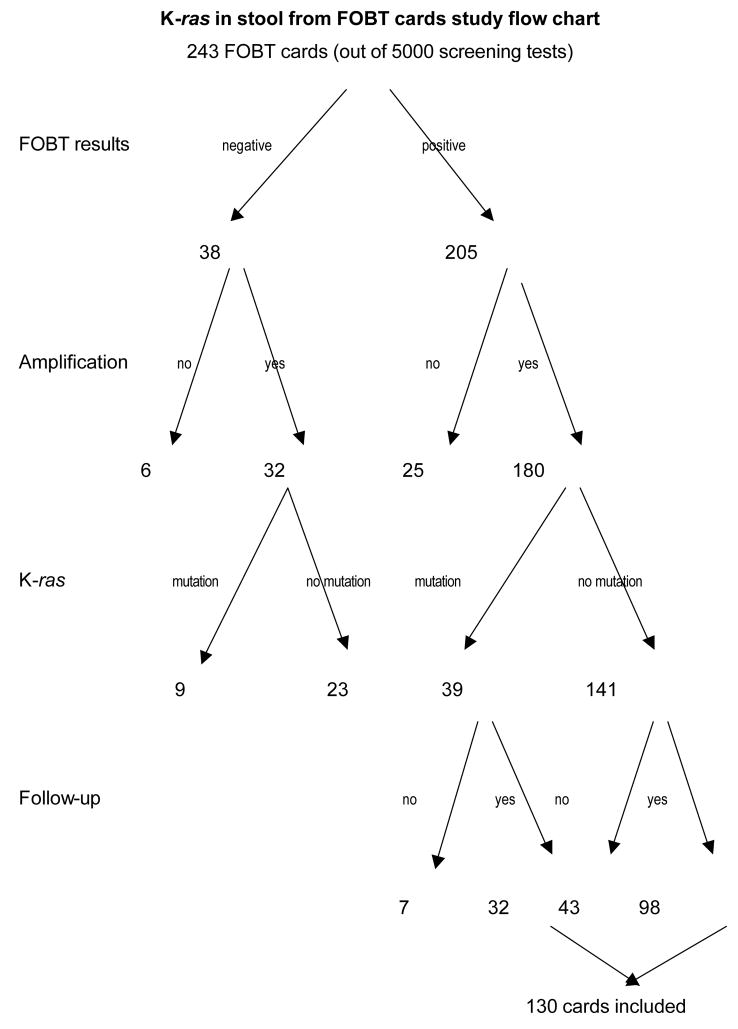
Of the 243 FOBT cards included in this study, 205 (84.4%) tested positive and 38 (15.6%) tested negative. 212 of the 243 (180 positive and 32 negative) samples (87.2%) were successfully amplified. Most of the unamplified samples had insufficient stool mass resulting in insufficient DNA recovery for amplification. Amplification failures were not dependent upon the FOBT reaction (non-amplified, negative FOBT cards (6/38, 15.8%); non-amplified positive FOBT cards (25/205, 12.2%) p=0.6 (Figure 1). Mutations in K-ras were found in 48 amplified samples; 39 among 180 positive FOBT amplified samples (21.7%) and 9 among 32 negative FOBT amplified samples (28.1%) p=0.42.
Figure 1.
K-ras in stool from FOBT cards study flow chart
Follow-up evaluation
Follow up on cases with positive FOBT was complete in 130 of the 180 positive amplified samples (72.2%). Among these, 23 malignancies in the colon and rectum, 25 adenomatous polyps and 14 hyperplastic polyps were detected. Among the 50 amplified samples with positive FOBT who did not have an evaluation procedure, 31 cases (62.0%) had one positive FOBT field and 11 cases (22.0%) had two positive fields. The main reasons for incomplete follow-up were, in equal proportions, patient refusal and primary care physician decision not to procede. This was most commonly the case when only one positive field was found. After a median of 6 years followup, no colorectal cancer cases were detected among hemoccult negative subjects. Four more cancers were detected three to five years after the index positive FOBT in cases with either no findings or adenomas only in colonoscopy.
The proportion of K-ras positive tests differed between cases with positive and negative FOBT, and by the type of finding in follow up (p=0.07) (Table 1). No differences were noted in the distribution of the specific K-ras mutations between the different diagnostic groups. AGT was the most commonly identified K-ras mutation and was noted in 50% of the mutated stools of cases with or without tumor findings.
Table 1.
Proportion of K-ras positivity in stool from positive FOBT cards with amplified DNA, by type of finding in the colon
| Finding in colon | Number of cases with positive FOBT and DNA amplification | Number of cases with K-ras mutation | Proportion of K-ras positivity |
|---|---|---|---|
| Carcinoma+ Adenoma | 48 | 16 | 33.0% |
| Adenocarcinoma | 23 | 8 | 34.8% |
| Adenoma | 25 | 8 | 32.0% |
| All other* | 82 | 16 | 19.5% |
| No follow up | 50 | 7 | 14.0% |
| Total positive FOBT | 180 | 39 | 21.7% |
Includes non-adenomatous polyps, diverticuli, IBD, hemorrhoids and upper GI findings
Impact of adding K-Ras Detection upon FOBT Detection of Colorectal Neoplasia
The positive predictive value for cancer diagnosis of the combined test (FOBT+K-ras) approximately doubled that of a single FOBT measure. For adenomas, K-ras improved FOBT’s positive predictive value when the FOBT tested positive in less then four of the six test slides. Such a situation is common because adenomas usually bleed less. The positive predictive value of FOBT in four or more fields reached 60% (positive/negative cards ratio=2.66, 95% CI:1.2–6.1). K-Ras enhanced the positive predictive value of FOBT in four or more fields to 80% for all neoplastic lesions; cancer + adenoma (ratio=1.46, 95% CI:0.9–2.3). K-ras reduced the FOBT positive predictive value (from 35.4% to 10.0%) for non-neoplastic conditions (e.g. diverticulosis, hemmorhoids, ulcers) (Table 2).
Table 2.
Positive predictive values of FOBT alone or in combination with stool K-ras results, for the detection of malignancies, adenomas, any tumors or benign non-neoplastic findings in the colon
| Findings in colon | Any FOBT+ | FOBT+ >4 fields | Any K-ras+ In FOBT+ | FOBT>4 + K-ras+ |
|---|---|---|---|---|
| N | 130 | 41 | 32 | 10 |
| Adenocarcinoma | 23 | 13 | 8 | 6 |
| PPV | 17.7% (2.1–33.3) | 31.7% (12.7–50.7) | 25.0% (7.3–42.7) | 60.0% (40.0–80.0) |
| Adenoma | 25 | 9 | 8 | 2 |
| PPV | 19.2% (3.8–34.7) | 22.0% (5.7–38.2) | 25.0% (8.0–42.0) | 20.0% (4.3–35.7) |
| All Neoplasms* | 62 | 25 | 17 | 8 |
| PPV | 47.7% (35.3–60.1) | 61.0% (48.8–73.1) | 53.1% (40.7–65.5) | 80.0% (70.0–90.0) |
| No-neoplastic GI disease** | 46 | 13 | 9 | 1 |
| PPV | 35.4% (21.6–49.2) | 31.7% (18.3–45.2) | 28.1% (15.1–41.1) | 10.0% (1.3–18.7) |
| Normal colonoscopy*** | 22 | 3 | 6 | 1 |
| PPV | 16.9% | 7.3% | 18.8% | 10.0% |
Includes adenocarcinomas, adenomas and polyps NOS
Includes diverticulosis, hemorrhoids, IBD.
does not preclude upper GI findings
Discussion
This study demonstrated that DNA extracted from a Hemoccult Sensa card is of sufficient quality and quantity to detect common mutations associated with colorectal carcinogenesis progression. K-ras was chosen as the test gene for this new method of DNA isolation and amplification from stool guiac cards due to its high mutational frequency in the earlier phases of the colonic epithelial transformation process [12]. Among the histologically verified malignancies, K-ras was found in 32%. While we did not perform a concordance validation with tissue samples from the sample patients’ stool samples we amplified, the percentage of positive K-ras tests in the subjects who had subsequent neoplastic disease proven corresponds well with the available literature [12–15].
K-ras mutations occurred in 28% of the stool samples of negative FOBT cards. Other groups have also demonstrated a similar rate of “false positive” results for K-ras in urine [16] and in stool [14]. Ito et al [14] reported that 3 out of 6 stools found positive for K-ras not to have K-ras in the corresponding tumor tissue. Jen et al [17] showed 25% of hyperplastic polyps to be K-ras positive. Others, such as Puig et al [18] did not find any false positive samples for K-ras. The source of these positive tests without known tissue-based mutation is unclear [14, 16, 18].
Many somatic events have been reported to be part of the colorectal neoplastic progression process [10,12]. Testing for K-ras can only represent a partial approach towards colorectal cancer detection from stool samples. It is conceivable that probing expression and mutations of other commonly detected genes on the same DNA samples may improve the detection process further [19, 20]. Other such markers [21, 22] may identify K-ras negative malignancies, thus further increasing the efficacy of the screening process. This process is similar in concept to several recently published studies of single or multiplexed, colorectal cancer associated gene mutations or gene silencing events on stool samples [8, 13, 23–27] and on colon tissue [28].
The 205 positive cards in this study are the result of screening 5,000 asymptomatic people (for a positive test rate of 4.1%). A positive predictive value of 5.5% for cancer and 26.6% for all tumors was formerly reported for this screening program [11]. Of the 130 positive FOBT tests which amplified and for which a followed up was available, 23 or 17.7% were later diagnosed with malignancy.
In this pilot, proof of principle, study we demonstrated the ability to detect genetic events in the stool from FOBT cards and attempted to use these events to separate true from false positive FOBTs. On the basis of these preliminary data, we are pursuing further validation by assaying stool samples and matched colonic neoplastic and flat mucosa obtained multi-center, case control trial. This work will allow us to more carefully evaluate the value of using K-ras and potentially other commonly mutated genes as adjuncts to FOBT testing prior to endoscopic evaluation.
Acknowledgments
This study was funded by NIH-CA86400, The Great Lakes New England Clinical Epidemiology Center of the Early Detection Research Network (EDRN) of the National Cancer Institute, the General Clinical Research Center, University of Michigan Medical Center (M01-RR00047) and the Israel Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mandel JS, Bond JH, Church TR, et al. Reducing mortality from colorectal cancer by screening for faecal occult blood. N Engl J Med. 1993;328:1365–1371. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 2.Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: Effectiveness of biennial screening for fecal occult blood. J Natl Cancer Inst. 1999;91:434–437. doi: 10.1093/jnci/91.5.434. [DOI] [PubMed] [Google Scholar]
- 3.Hardcastle JD, Chamberlain JO, Robinson MHE, et al. Randomised controlled trial of faecal occult blood screening for colorectal cancer. Lancet. 1996;348:1472–1477. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 4.Kronborg O, Fenger C, Olsen J, Jorgensen OD, Sondergaard O. Randomised study of screening for colorectal cancer with faecal occult blood test. Lancet. 1996;348:1467–1471. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 5.Rennert G. Fecal Occult Blood (FOB) Screening – Trial evidence, Practice and Beyond. Recent Results in Cancer Research. Tumor Prevention and Genetics. 2003;163:248–253. doi: 10.1007/978-3-642-55647-0_22. [DOI] [PubMed] [Google Scholar]
- 6.Sidransky D, Tokino T, Hamilton SR, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–5. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 7.Jen J, Johnson C, Levin B. Molecular approaches to colorectal cancer screening. Eur J Gastro Hepatol. 1998;10:213–217. doi: 10.1097/00042737-199803000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 9.Pignone M, Campbell MK, Carr C, Phillips C. Meta-analysis of dietary restriction during fecal occult blood testing. Eff Clin Pract. 2001;4:150–6. [PubMed] [Google Scholar]
- 10.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 11.Rennert G, Rennert HS, Miron E, Peterburg Y. Population colorectal cancer screening with fecal occult blood test. Cancer Epidemiology, Biomarkers and Prevention. 2001;10:1165–1168. [PubMed] [Google Scholar]
- 12.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 13.Dong SM, Traverso G, Johnson C, et al. Detecting colorectal cancer in stool with the use of multiple genetic targets. J Natl Cancer Inst. 2001;93:858–65. doi: 10.1093/jnci/93.11.858. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y, Kobayashi S, Taniguchi T, Kainuma O, Hara T, Ochiai T. Frequent detection of K-ras mutation in stool samples of colorectal carcinoma patients after improved DNA extraction: comparison with tissue samples. Int J Oncol. 2002;20:1263–8. [PubMed] [Google Scholar]
- 15.Calistri D, Rengucci C, Bocchini R, Saragoni L, Zoli W, Amadori D. Fecal multiple molecular tests to detect colorectal cancer nin stool. Clin Gastroenterol Hepatol. 2003;1:377–83. doi: 10.1053/s1542-3565(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang M, Block TM, Steel L, Brenner DE, Su YH. Preferential isolation of fragmented DNA enhances the detection of circulating mutated K-ras. DNA Clin Chem. 2004;50:211–3. doi: 10.1373/clinchem.2003.026914. [DOI] [PubMed] [Google Scholar]
- 17.Jen J, Powell SM, Papadopoulos N, et al. Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 1994;54:5523–6. [PubMed] [Google Scholar]
- 18.Puig P, Urgell E, Capella G, Sancho FJ, Pujol J, Boadas J, Farre A, Lluis F, Gonzalez-Sastre F, Mora J. A highly sensitive method for K-ras mutation detection is useful in diagnosis of gastrointestinal cancer. Int J Cancer. 2000;85:73–7. doi: 10.1002/(sici)1097-0215(20000101)85:1<73::aid-ijc13>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Brenner DE, Rennert G. Fecal DNA biomarkers for the detection of colorectal neoplasia: attractive, but is it feasible? J Natl Cancer Inst. 2005;97:1107–9. doi: 10.1093/jnci/dji244. [DOI] [PubMed] [Google Scholar]
- 20.Lev Z, Kislitsin D, Rennert G, Lerner A. Utilization of K-ras mutations identified in stool DNA for the early detection of colorectal cancer. J Cell Biochem. 2000;77:35–39. doi: 10.1002/(sici)1097-4644(2000)77:34+<35::aid-jcb8>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 21.Fearon ER. Human cancer syndromes: clues to the origin and nature of cancer. Science. 1997;27:1043–50. doi: 10.1126/science.278.5340.1043. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumrigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 23.Eguchi S, Kohara N, Komuta K, Kanematsu T. Mutations of the p53 gene in the stool of patients with resectable colorectal cancer. Cancer. 1996;77(Suppl):1707–10. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1707::AID-CNCR43>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Tagore KS, Lawson MJ, Yucaitis JA, et al. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer. 2003;3:47–53. doi: 10.3816/CCC.2003.n.011. [DOI] [PubMed] [Google Scholar]
- 25.Traverso G, Shuber A, Levin B, et al. Detection of APC mutations in fecal DNA from patients with colorectal tumors. N Engl J Med. 2002;346:311–20. doi: 10.1056/NEJMoa012294. [DOI] [PubMed] [Google Scholar]
- 26.Muller HM, Oberwalder M, Fiegl H, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–5. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 28.Samuels Y, Wang Z, Bardelli A, et al. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]