Abstract
Apicomplexan parasites are characterised by the presence of specialised organelles, such as rhoptries, located at the apical end of invasive forms that play an important role in invasion of the host cell and formation of the parasitophorous vacuole. In this study, we have characterised a novel Plasmodium falciparum rhoptry protein, Pf34, encoded by a single exon gene located on chromosome 4 and expressed as a 34 kDa protein in mature asexual stage parasites. Pf34 is expressed later in the life cycle than the previously described rhoptry protein, Rhoptry Associated Membrane Antigen (RAMA). Orthologues of Pf34 are present in other Plasmodium species and a potential orthologue has also been identified in Toxoplasma gondii. Indirect immunofluorescence assays show that Pf34 is located at the merozoite apex and localises to the rhoptry neck. Pf34, previously demonstrated to be glutathione-S-transferase (GPI)-anchored (Gilson et al., 2006, Identification and stoichiometry of GPI-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics, 1286–1299), is associated with parasite-derived detergent-resistant microdomains (DRMs). Pf34 is carried into the newly invaded ring, consistent with a role for Pf34 in the formation of the parasitophorous vacuole. Pf34 is exposed to the human immune system during infection and is recognised by human immune sera collected from residents of malaria endemic areas of Vietnam and Papua New Guinea.
Keywords: Plasmodium falciparum, Rhoptry, Pf34, GPI-anchored, Detergent resistant microdomain
1. Introduction
Invasion of red blood cells (RBCs) by invasive asexual forms of Plasmodium falciparum (merozoites) represents an attractive target in the parasite life cycle for the development of anti-invasion therapies. Invasion is an active process that is comprised of a series of complex and well coordinated events. Invasion commences with an initial low affinity interaction between proteins of the merozoite coat surface and RBC receptors and is followed by reorientation of the merozoite, host cell entry and establishment of the parasitophorous vacuole (PV). These latter steps are facilitated by the actin-myosin motor and proteins localised within specialised apical secretory organelles - the rhoptries, micronemes and dense granules. Studies over the last 25 years have attempted to fully characterise the contents of these organelles, particularly the rhoptries. In P. falciparum at least 20 rhoptry proteins have been characterised in detail, while proteomic studies in Toxoplasma indicate that there are likely to be a substantially greater number of rhoptry proteins (Bradley et al., 2005). These proteins are involved both in binding to the exterior of the RBC during the later stages of invasion and in formation of the PV (see Kats et. al., 2006 for recent review). Known rhoptry-resident proteins appear to be predominantly secreted with soluble proteins present in the rhoptry neck and lumen, but a smaller number of proteins are membrane-associated either by glycosylphosphatidylinositol (GPI) anchors, for example Rhoptry Associated Membrane Antigen (RAMA), or integral membrane anchors such as Rhop148 (Lobo et al., 2003; Topolska et al., 2004a).
As invasion is an essential step in the parasite life cycle, it may be possible to block parasite replication in vivo by designing inhibitors that interfere with one or more steps of the invasion process. In addition, merozoites are briefly exposed to the host immune system during RBC egress and invasion. As such, proteins on the surface of merozoites or those that are secreted during invasion are potential targets for neutralising antibodies and potential candidates for vaccine development (Cowman et al., 2002; Cowman and Crabb, 2006; Kats et al., 2006). GPI-anchored proteins have emerged as important candidates for inclusion in a blood-stage subunit vaccine. At least nine GPI-anchored merozoite proteins have been described (Merozoite Surface Protein (MSP)-1, -2, -4, -5, -8, -10, Pf92, Pf38 and Pf12) and two others (RAMA and ASP) have been localised to apical organelles (Holder and Freeman, 1982; Smythe et al., 1988; Marshall et al., 1997, 1998; Black et al., 2001, 2003; Topolska et al., 2004a; O’Keeffe et al., 2005; Sanders et al., 2005; Gilson et al., 2006;). GPI-anchored proteins are preferentially incorporated into detergent resistant microdomains (DRMs) where they are proposed to function as distinct components within lipid bilayers and are thought to participate in receptor signaling, signal transduction and membrane sorting in a variety of eukaryotic cells. In P. falciparum-infected erythrocytes, DRMs are rich in cholesterol and sphingolipids and contain a sub-population of parasite-derived membrane proteins. (Salzer and Prohaska, 2001; Salzer et al., 2002; Hiller et al., 2003; Wang et al., 2003b; Sanders et al., 2005).
In contrast to peripheral MSPs and other apical proteins, most GPI-anchored proteins are refractory to genetic deletion (Sanders et al., 2006). MSP-1, MSP-2, MSP-4 and MSP-5 are currently being developed for clinical trials (see Malkin et al., 2006 for recent review), whereas others such as RAMA are implicated as targets of host protective immunity (Topolska et al., 2004b; Nixon et al., 2005). Gilson and co-workers (Gilson et al., 2006) recently identified Pf34 as a blood-stage GPI-anchored protein. Here, we show that Pf34 is localised to rhoptries and is indeed recruited into DRMs. Pf34 is expressed in mature intra-erythrocytic stages and is carried across by the invading merozoite to the ring stage parasite, suggesting a possible role in the invasion process perhaps at the stage of formation of the PV.
2. Materials and methods
2.1. Plasmodium falciparum parasites
Plasmodium falciparum 3D7 parasites were cultured in vitro using routine culture techniques as previously described (Trager and Jensen, 1976). Parasite extracts (Black et al., 2001) and synchronised cultures (Lambros and Vanderberg, 1979) were prepared as described and sampled at various time points for analysis of stage-specific expression.
2.2. Genomic sequences, molecular cloning and analysis
The Pf34 nucleotide sequence encoded by PFD0955w was sourced from the Sanger Institute P. falciparum chromosome 4 sequence (Genbank accession AL844503). Pf34 orthologues were identified in genomic sequence data for Plasmodium vivax (TIGR contig 6950, Genbank AAKM01000006), Plasmodium knowlesi, Plasmodium chabaudi and Plasmodium berghei (http://www.wehi.edu.au/MalDB-www/genome.htm). Gene annotation was completed by the Victorian Bioinformatics Consortium (http://www.wehi.edu.au/MalDB-www/genome.htm). Further sequence analysis was carried out using the various databases and tools available at the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/), PlasmoDB (http://plasmodb.org), ToxoDB (http://toxodb.org), ApiDB (http://apidb.org) and ExPASy (http://ca.expasy.org, including Pfam and Prosite databases).
Three non-overlapping fragments of Pf34 were amplified by PCR from P. falciparum 3D7 genomic DNA and ligated as BamHI/HindIII fragments into the pGEX-KG expression vector (Guan and Dixon, 1991) for expression of glutathione-S-transferase (GST) fusions. Fragment Pf34-A comprised amino acid residues 24–110 of the full-length Pf34 amino acid sequence (amplified using forward primer 5′-acgcggatccAATAATATAAAATTAAAC-3′ and reverse primer 5′-tccgaagcttATCTGCTTTTAAACTTTCAG-3′); fragment Pf34-B comprised residues 111–256 (forward primer 5′-acgcggatccCTTAAGATATTAAATAATG-3′, reverse primer 5′-tcccaagcttATAAATTTTTTTTTTTTTC-3′); fragment Pf34-C comprised residues 260–307 (forward primer 5′-acgcggatccTATGATAATCAAGATAATAG-3′, reverse primer 5′-tcccaagcttAAGTAGAGGAACCACTATG-3′) (Fig. 1A, 1B). The underlined sequences encode restriction endonuclease sites and the capitalised sequence is Pf34 gene-specific. DNA cloning and characterisation procedures were performed as previously described (Black et al., 2001).
Fig. 1.
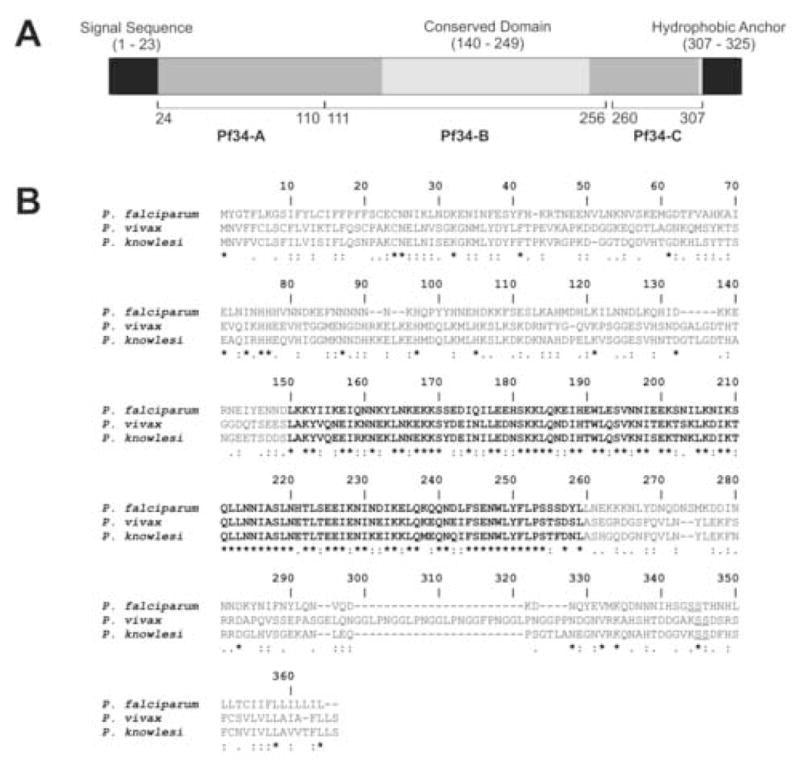
Structure, features and conservation of Pf34. (A) Schematic representation of full-length Pf34, a 325 residue protein. The N-terminus contains a predicted signal peptide (residues 1 – 23), whereas the C-terminus contains a probable glycosylphosphatidylinositol (GPI) attachment site (residue 306), followed by a hydrophobic anchor sequence (residues 307 – 325). Pf34 also contains a central domain (residues 140 - 248) that is highly conserved in orthologues identified in Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi (shown in bold in Fig. 1B). Fragments corresponding to residues 24 – 110 (Pf34-A), 110 – 256 (Pf34-B) and 260 – 307 (Pf34-C) were produced as glutathione-S-transferase fusion proteins. (B) Sequence similarity between P. falciparum Pf34 and orthologues in P. vivax and P. knowlesi. Identical (*), highly similar (:) and similar (.) residues are indicated. The central highly conserved region is shown in bold type. Probable GPI attachment sites are indicated by the di-serine motif (underlined).
2.3. Recombinant protein expression and production of polyclonal antisera
Recombinant Pf34-GST fragments were expressed in Escherichia coli BL21 (DE3) (Invitrogen) and purified as described previously (Black et al., 2001). Polyclonal antisera were raised in 6–8-weeks-old female BALB/C mice (Monash Animal Services, Clayton Vic, Australia). Immunisations were conducted intraperitoneally using 25 μg of purified recombinant protein emulsified in Complete Freund’s adjuvant (Difco Laboratories), with subsequent monthly boosts of 25 μg recombinant protein in Incomplete Freund’s Adjuvant (Difco Laboratories). Experiments involving animals were carried out with appropriate ethics approval from the Monash University Animal Use Committee (BAM/M/2003/14). Work involving handling of transgenic organisms was carried out in an OGTR PC2 certified facility and approved by the Monash University Biosafety Committee.
2.4. SDS-PAGE and Immunoblotting
Recombinant proteins and parasite lysate were resolved by SDS-PAGE on 12% (w/v) polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes (NEN) for immunoblotting as previously described (Black et al., 2001), with all samples resolved under denaturing conditions. Polyclonal antisera raised in mice (against Pf34 fragments, diluted to 1:500) and rabbits (RAMA, 1:1000; HSP70, 1:10,000 and Glucose-Regulated Protein GRP (BiP), 1:1,000) were used as primary antibodies. Human hyperimmune sera from malaria endemic areas of Papua New Guinea (PNG) (Marshall et al., 1997) and Vietnam (Wang et al., 2001) and pooled sera from individuals in Melbourne, Australia, who have never been exposed to malaria were used to assess reactivity of Pf34 (Wang et al., 2003a). The primary antibodies were detected using anti-mouse, anti-rabbit or anti-human immunoglobulin conjugated to horseradish peroxidase (Silenus) and developed as previously described (Black et al., 2001).
2.5. Indirect Immunofluorescence Assays (IFA)
Plasmodium falciparum blood-stage parasites were cultured to approximately 8% parasitemia as previously described (Wang et al., 1999). Blood smears were taken when the majority of the parasites were at late trophozoite and schizont stages. IFA was performed as previously described (Black et al., 2001). Polyclonal mouse anti-Pf34, rabbit anti-RAMA diluted 1:500 (provided by Dr. A. Topolska) and rabbit anti-AMA-1 diluted 1:500 (Apical Membrane Antigen 1, provided by Dr. A. Hodder), were used as primary antibodies. Reactivity was detected using Alexa Fluor 488-conjugated anti-mouse immunoglobulin and Alexa Fluor 568-conjugated anti-rabbit immunoglobulin (diluted 1:3,000; Molecular Probes Inc.) as secondary antibodies. Smears from asynchronous cultures were examined by widefield fluorescent microscopy and selected samples were also examined by confocal fluorescent microscopy.
2.6. Isolation of detergent resistant microdomains (DRMs)
Asynchronous P. falciparum blood-stage parasites were cultured to approximately 6% parasitaemia as described previously (Wang et al., 1999). The parasites were harvested using 0.15% (w/v) saponin, washed with PBS and stored in 100 μL aliquots. To test the detergent solubility of Pf34, three separate aliquots of parasites were each resuspended in 1 mL of ice-cold TNET buffer (1% (w/v) TX-100, 25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5) containing a cocktail of protease inhibitors (Roche Diagnostics). Separate aliquots were incubated as follows: (i) on ice for 30 min and centrifuged at 10,000 × g, 4°C for 10 min; (ii) at 37°C for 30 min and centrifuged at 10,000 × g, room temperature for 10 min; (iii) on ice for 30 min and centrifuged at 10,000 × g, 4°C for 10 min. For sample (iii), the resulting soluble and pellet fractions were collected. The insoluble pellet was further resuspended in 1 mL of ice-cold TNET, incubated at 37°C for 30 min and centrifuged at 10,000 × g at room temperature for 10 min. All samples were resolved by SDS-PAGE and immunoblotting. To confirm that Pf34 was present in DRMs, parasite samples were fractionated by sucrose density flotation as previously described (Wang et al., 2003b).
3. Results
3.1. Pf34 identification, gene structure and orthologues
Global microarray analyses of asexual parasites of P. falciparum have revealed that most known invasion-related genes are coordinately and maximally transcribed in schizont stage parasites (Bozdech et al., 2003; Le Roch et al., 2003). We analysed previously uncharacterised genes that showed coordinated transcription profiles with known invasion genes. One of these genes, PFD0955w, is a single exon gene located on chromosome 4. PFD0955w encodes for a 325 amino acid protein with a predicted N-terminal signal sequence and a C-terminal hydrophobic sequence preceded by a di-serine motif that is often found in sequences of Plasmodium GPI-attachment sites (Fig. 1). The encoded protein was named Pf34 from its apparent molecular mass on SDS-PAGE. It was shown to be radiolabelled following biosynthetic labelling with glucosamine, indicating the presence of a GPI-anchor (Gilson et al., 2006). No identifiable motifs other than the GPI-attachment motif or domains of known function were identified in the Pf34 sequence.
Pf34 orthologues are present in other Plasmodium species. Alignment of the Pf34 amino acid sequence with those of predicted orthologues in P. vivax and P. knowlesi show an overall identity of 25.7% and similarity of 62% (Fig. 1B). Upon alignment, a central region corresponding to residues 140-249 of the Pf34 sequence shows markedly higher cross-species conservation (64% identity, 89% similarity) and may represent a conserved, functional domain. Genomic synteny of the PFD0955w locus was found to be conserved in the P. vivax and P. knowlesi genomes. Analysis of other available Plasmodium genomic sequences indicated that Pf34 orthologues are present in the murine parasites P. chabaudi and P. berghei. Although the full sequence of these orthologues is currently incomplete, the available sequences show a high degree of similarity to Pf34 and include putative GPI-anchor attachment sites preceding a hydrophobic terminus (data not shown).
Efforts to characterise the Toxoplasma gondii rhoptry proteome identified 12 proteins with orthologues in P. falciparum (Bradley et al., 2005), including a possible orthologue of Pf34 encoded by TGG_99455w (227295-226243) showing 50% amino acid similarity to Pf34. No other matches were found amongst the available genomic sequences from other apicomplexan parasites and further searching failed to identify any homologous sequences outside the phylum Apicomplexa.
3.2. Expression of Pf34 recombinant fragments and reactivity to human immune sera
Three non-overlapping fragments spanning the length of the predicted mature Pf34 polypeptide, designated Pf34-A, Pf34-B and Pf34-C, (Fig. 1A) were produced as recombinant GST fusion proteins in E. coli (Fig. 2A). In order to investigate the ability of Pf34 to elicit an immune response in human infection, Pf34 fusion fragments were resolved alongside a GST control and immunoblotted with immune sera from long-term residents from endemic regions of PNG and Vietnam. All three recombinant proteins were recognised by PNG sera (Fig. 2A). The Vietnam sera recognised Pf34-A with weak reactivity to the other two fragments suggesting the immunodominant region of this protein is localised to the N-terminus. GST alone was not recognised by any sera, and no reactivity was observed when all proteins were probed with sera from naïve, non-exposed residents of Australia (Fig. 2A).
Fig. 2.
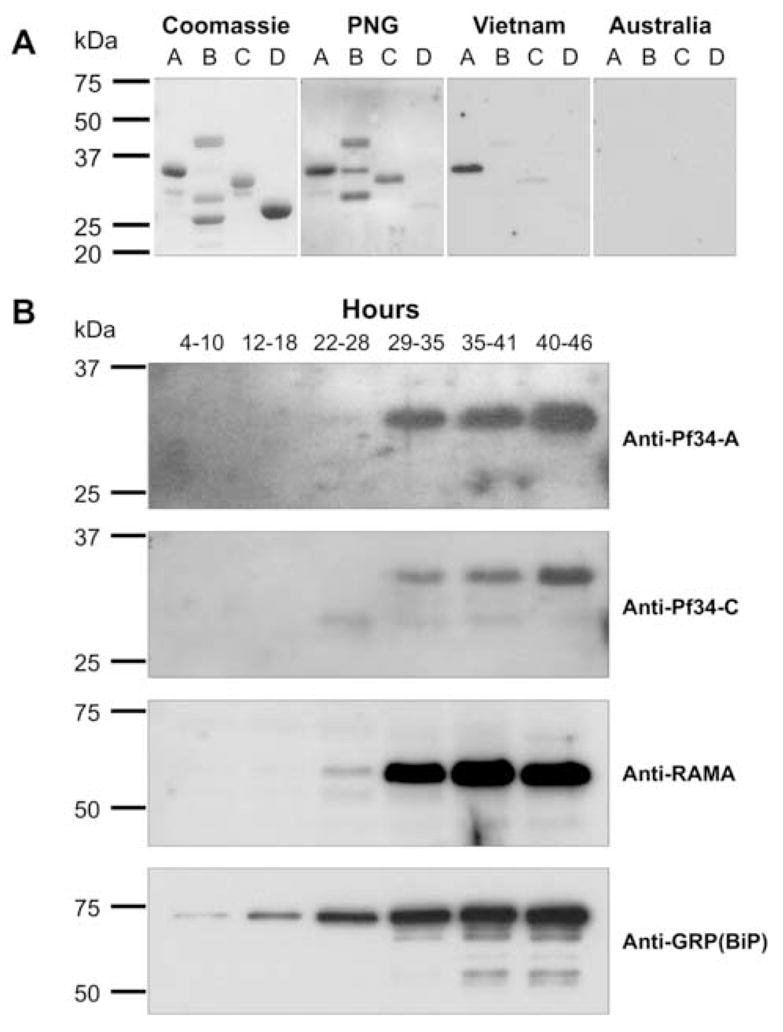
Reactivity of human immune sera and stage-specific expression of Pf34. (A) The recombinant proteins Pf34-A (lane A), Pf34-B (lane B) and Pf34-C (lane C) and glutathione-S-transferase alone (lane D) were resolved under denaturing conditions. Shown are Coomassie-stained samples and those immunoblotted with human immune sera from endemic areas of Vietnam and Papua New Guinea, and non-immune sera from Australia. (B) Stage-specific expression of Pf34. Synchronised parasite culture was sampled at various time points (indicated in hours) and lysates resolved by SDS-PAGE. All samples were immunoblotted with antisera raised against Pf34 fragments A, C and RAMA. Anti-GRP(BiP) sera was used as a loading control. Pf34 expression is not convincingly demonstrable at 22–28 h.
3.3. Stage-specific expression of Pf34
Antisera to all three recombinant Pf34 proteins raised in mice reacted with the corresponding immunised recombinant protein (data not shown). The anti-Pf34 sera were used to probe time-course samples from a highly synchronised parasite culture (Fig. 2B). Consistent with previous observations, all three antisera recognised a protein with a molecular mass of 34 kDa (Gilson et al., 2006). This data suggests that the protein does not undergo proteolytic processing during the asexual life cycle. Full-length Pf34 was first detected in late trophozoite samples (29–35 h post invasion) and was present until schizont rupture, consistent with the reported maximal transcription profiles for Pf34 (Bozdech et al., 2003; Le Roch et al., 2003). The samples were also probed with RAMA-specific antisera which were able to detect RAMA from 22–28 h, consistent with previously published data (Topolska et al., 2004a) (Fig. 2B). This suggests that although RAMA and Pf34 expression are both detected from the late trophozoite stage, RAMA expression appears to begin earlier than that for Pf34. Probing the samples with GRP(BiP) (Kumar et al., 1991) antisera demonstrated the increase in parasite material over the time course. GRP(BiP) expression was detected at all time points (Fig. 2B).
3.4. Localisation of Pf34 to the rhoptries
IFA was performed using antisera raised against Pf34 to determine its sub-cellular location in P. falciparum parasites. The antisera were tested in dual labelling IFAs with either anti-RAMA and anti-RhopH3 (rhoptry markers) or anti-AMA-1 (microneme marker) antibodies (Hodder et al., 1996; Topolska et al., 2004a) (Fig. 3A). In segmented schizonts, Pf34 antisera produced a punctuate pattern of fluorescence. Each of the developing merozoites has a single small punctate Pf34-positive structure located in the apical end. Pf34 showed close localisation with the rhoptry markers RAMA and RhopH3. The partial overlapping of the small, punctate Pf34- or RAMA-positive areas with the larger and more diffuse AMA-1-positive area suggests that Pf34 was not a micronemal protein (Fig. 3A).
Fig. 3.
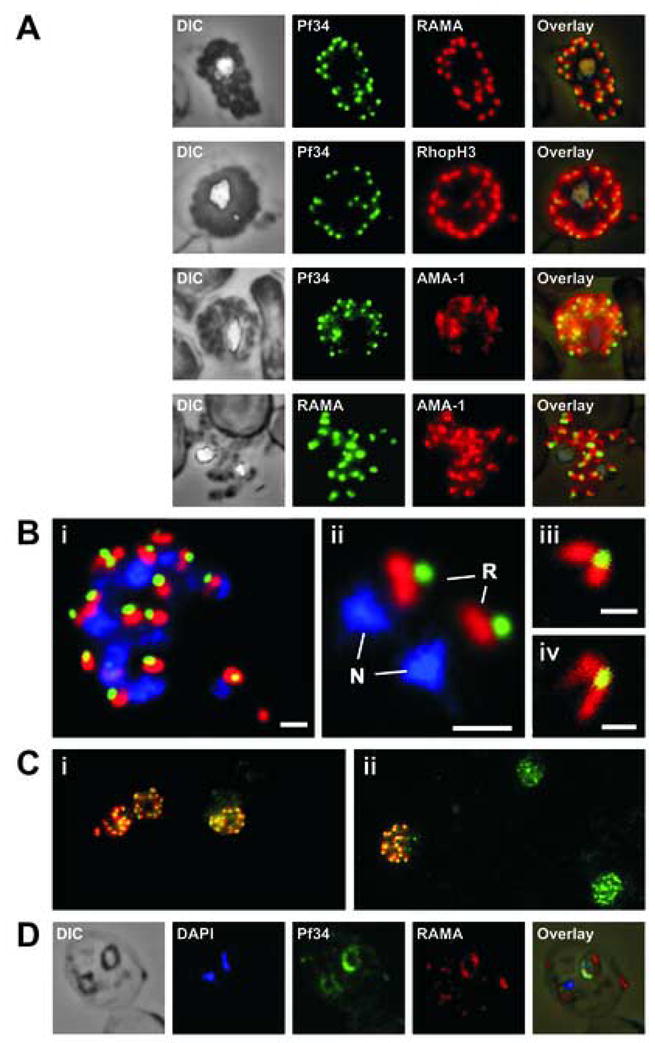
Localisation of Pf34 to the rhoptries. (A) Rhoptry localisation was demonstrated in late stage parasites by immunostaining infected blood smears with anti-Pf34 and co-localising with either anti-RAMA or anti-RhopH3 (both rhoptry markers) or anti-AMA-1 (microneme marker) antibodies. (B) A mature schizont (i) and two extracellular merozoites (ii) showing the close apposition of Pf34-positive (green) and RAMA-positive (red) structures in the apex of the merozoite. Panels (iii) and (iv) show the apex of merozoites and the two rhoptries connected to the Pf34-positive structure at the anterior when visualised by confocal microscopy. N – nucleus; R – rhoptry. Bars represent 1 μm in (i) and (ii) and 500 nm in (iii) and (iv). (C) Late stage parasites were stained with anti-Pf34 (green) and anti-RAMA (red) (i) or anti-Pf34 (green) and anti-AMA-1 (red) (ii) antibodies, highlighting the differential expression of the rhoptry proteins (Pf34 and RAMA) when compared with AMA-1 (microneme protein). (D) Ring stage expression of Pf34 was shown using anti-Pf34 co-localised with anti-RAMA.
Closer examination of the mature schizont (Fig. 3B, panel i) and extracellular merozoites (Fig. 3B, panel ii) revealed that there was limited co-localisation of RAMA and Pf34, with the Pf34-positive area close to, but slightly anterior to, the RAMA-positive area. Confocal microscopy of mature schizonts confirmed these observations (not shown). Confocal imaging of extracellular merozoites resolved the two bulbous RAMA-positive areas of the rhoptries with apparent overlapping of the Pf34-positive area toward the anterior (Fig. 3B, panels iii and iv). Combined, this data suggests that Pf34 is compartmentalised to the rhoptry neck and absent from the rhoptry bulb. This type of staining pattern has been previously demonstrated in the T. gondii rhoptry neck proteins (Bradley et al., 2005). In schizonts, Pf34 was consistently co-expressed with the rhoptry markers RAMA and RhopH3. However, in cells double-labelled with Pf34 and AMA-1, many of the developing schizonts exhibiting the punctate Pf34-positive staining were unstained with AMA-1 (Fig. 3C). Only a sub-population of schizonts was positive for both Pf34 and AMA-1. Taken together, these results suggest that Pf34 is not a micronemal protein, is indeed localised to the rhoptry neck and that it is expressed at an earlier stage than AMA-1.
Pf34 staining was also detected in young rings as a rim of fluorescence surrounding the parasite (Fig. 3D). Ring stage Pf34 staining shows co-localisation with RAMA, a protein that is known to be transferred to the PV during invasion (Topolska et al., 2004a).
3.5. Pf34 is recruited into DRMs
Experimentally, DRMs are defined as being insoluble in cold non-ionic detergents (Brown and London, 1997; Wang et al., 2003b). To determine whether Pf34 is associated with DRMs, detergent-solublised parasite material was probed with anti-Pf34 antisera. Pf34 was partially insoluble in TX-100 at 4°C, but completely soluble at 37°C in TX-100. Furthermore, when insoluble material collected following treatment at 4°C was incubated at 37°C, the majority of Pf34 was re-solublised (Fig. 4A).
Fig. 4.
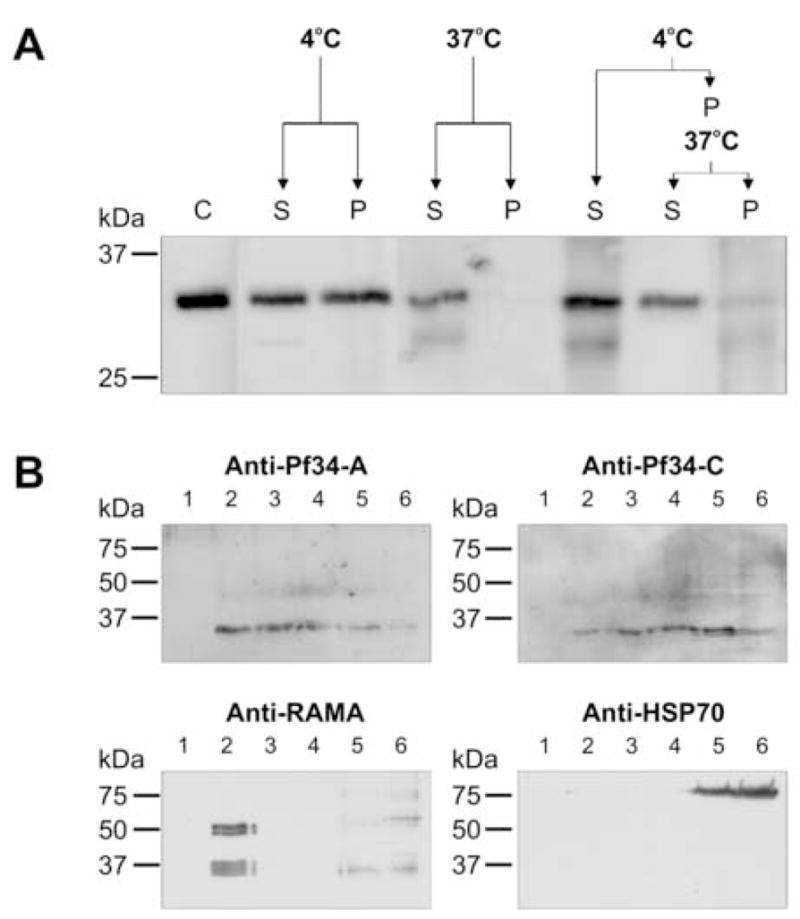
Pf34 is associated with detergent-resistant microdomains (DRMs). (A) Asynchronous parasites were saponin treated (C) and resuspended in TX-100 at 4°C or 37°C. After incubation the soluble (S) and pellet (P) fractions were resolved by SDS-PAGE. Enhanced solubility at 37°C is indicative of proteins associated with DRMs. The samples were immunoblotted with anti-Pf34-A sera. (B) Immunoblot detection of sucrose density gradient samples of Plasmodium falciparum parasites. Asynchronous parasites were extracted in cold TX-100 and subjected to sucrose density gradient flotation. Six gradient fractions were obtained (1–6), where fractions 2–4 contain floating material, and thus, DRMs. Fractions 5 and 6 represent the high density loading fraction. The samples were immunoblotted with anti-RAMA (positive control), anti-HSP70 (negative control) and anti-Pf34-A and C antibodies.
Following solubilisation in cold non-ionic detergent and fractionation by sucrose density gradient flotation, DRMs float within low density sucrose fractions whereas non-DRM components remain in the high density loading fraction (Hiller et al., 2003; Wang et al., 2003b). Following sucrose density gradient flotation, Pf34 was detected in low density fractions (2–4). The same fractions were probed with antisera against the known DRM-associated protein RAMA (Topolska et al., 2004a), which was found in fraction 2 only. Antisera raised against heat shock protein 70 (HSP70), which is not associated with DRMs, showed reactivity to the high density loading fractions (5–6) (Fig. 4B). Taken together, these results indicate that Pf34 associates with DRMs.
4. Discussion
Gilson and colleagues (2006) recently demonstrated that Pf34 is a GPI-anchored protein that is expressed in asexual RBC-stage P. falciparum parasites (Gilson et al., 2006). Using protein-specific antibodies, we have shown that Pf34 is expressed in mature asexual stage parasites and is localised to the rhoptry neck. Repeated attempts to localise Pf34 using immuno-electron microscopy have not been successful. However, these ultrastructural observations did allow evaluation of the anterior structures during merozoite formation. It was observed that the two rhoptries and their ducts formed early in merozoite formation and it was only in the late stages of merozoite formation that a number of micronemes appeared in the apical region of the merozoite (not shown). These observations are consistent with previously published data describing merozoite biogenesis (Margos et al., 2004). The delayed formation of the micronemes relative to the rhoptries is consistent with the appearance of Pf34 and other rhoptry markers in a greater number of schizonts than those positive for the micronemal protein AMA-1 (Fig. 3C). Pf34 is transferred to the PV of newly infected RBCs (Fig. 3D). Despite being visible by immunofluorescence (0–2 h) in newly-invaded rings, we were unable to detect Pf34 by immunoblotting in 4–10 h- old rings, suggesting loss of Pf34, perhaps by proteolytic degradation. This characteristic is common for rhoptry proteins, including RAMA (Kats et al., 2006) and suggests that Pf34 functions earlier either during RBC invasion or the early steps of PV formation.
The data presented here show that Pf34 associates with DRMs. These findings are inconsistent with recent data that failed to detect Pf34 within the P. falciparum DRM proteome (Sanders et al., 2005). The fractionation pattern observed for Pf34 was distinct from that of the previously characterised DRM protein RAMA. This observation supports the possibility of multiple populations of DRMs located in the rhoptries. Distinct populations of RBC DRMs have been demonstrated in other studies (Salzer et al., 2002; Murphy et al., 2004). Localisation to distinct DRM populations might be consistent with the observation that RAMA traffics to the rhoptry body and Pf34 to the rhoptry neck. Residence of both proteins in a single population of DRMs would then require some form of sorting at the rhoptry membrane to partition them to distinct regions of the rhoptry. If the proteins are present in different DRM populations then each population could be trafficked separately to a different portion of the rhoptry. RAMA, the only other GPI-anchored rhoptry protein, is implicated in chaperoning other rhoptry proteins to the rhoptry body (Topolska et al., 2004a). It is not yet known if Pf34 has a similar role with respect to secreted proteins that are sited at the rhoptry neck.
Pf34 is conserved across several Plasmodium species. All Pf34 orthologues identified to date also possess a highly conserved region (Fig. 1B), indicating that this domain is likely to be important to the parasite. The lack of similarity to any known protein domains suggests that Pf34 and its orthologues are functionally unique. To date, no Pf34 orthologue has been identified in P. yoelii, however the synteny of other available P. falciparum and P. yoelii sequences, combined with the high degree of conservation between murine species suggests that it is highly likely that an as yet unidentified orthologue is present in P. yoelii. Some similarity exists between Pf34 and P. berghei reticulocyte binding protein (PbRBP; 41% amino acid similarity). However, the lack of high similarity within the proposed central conserved domain of Pf34 and the lack of a putative GPI attachment site suggests that PbRBP is not a true orthologue of Pf34. It is not clear if Pf34 homologues are present in genera other than Plasmodium. An open reading frame identified in T. gondii shows sequence similarity to Pf34 (Bradley et al., 2005). Together with putative rhoptry location and a GPI-attachment signal, it is possible that the two proteins share a similar function. However, the sequence similarity is scattered and the highly conserved region described here for Plasmodium Pf34 orthologues is not evident in the T. gondii gene. This may reflect the divergence in the respective parasites’ host cell specificity and invasion mechanisms. It remains possible that the T. gondii protein has been mistakenly identified as a Pf34 orthologue.
Pf34 was recognised by sera from Plasmodium-infected human populations but not by non-immune sera from Australia. Interestingly, the PNG and Vietnam sera showed differing results. The PNG sera reacted with epitopes across the entire protein whereas sera from Vietnam only recognised the N-terminal fragment, which appears to harbour an immunodominant epitope in each case. These data indicate that Pf34 is expressed in geographically distinct field isolates during natural infection and is able to elicit an immune response. Differential reactivity to Pf34 may be a useful feature in further epidemiological studies. It would be interesting to examine this differential reactivity with other putative rhoptry proteins.
In summary, we have characterised a novel DRM protein which is localised in the rhoptries of P. falciparum. To date, Pf34 and RAMA are the only identified DRM-associated GPI-anchored proteins localised to the rhoptries. It is therefore possible that Pf34, like RAMA, serves to recruit other proteins into DRMs. Gene knockout studies are currently underway in an attempt to elucidate the function of Pf34 in merozoite biology.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia, the ARC/NHMRC Research Network for Parasitology and the Australian Postgraduate Awards (NIP, LK, BJM). KLW is an NHMRC Howard Florey Centenary Research Fellow. DJPF received a travel grant from the ARC/NHMRC Research Network for Parasitology and an equipment grant from the Wellcome Trust. We would like to thank Robert Huestis of the Victorian Bioinformatics Consortium (VBC) for assistance with the bioinformatic analyses. We also thank Dr. Agnieszka Topolska for providing anti-RAMA antibodies, Dr. A. Hodder for providing anti-AMA-1 antibodies and the Malaria Research and Reference Reagent Resource Center (MR4) for supplying the anti-GRP(BiP) antibodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Black CG, Wu T, Wang L, Hibbs AR, Coppel RL. Merozoite surface protein 8 of Plasmodium falciparum contains two epidermal growth factor-like domains. Mol Biochem Parasitol. 2001;114:217–226. doi: 10.1016/s0166-6851(01)00265-1. [DOI] [PubMed] [Google Scholar]
- Black CG, Wang L, Wu T, Coppel RL. Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol. 2003;127:59–68. doi: 10.1016/s0166-6851(02)00308-0. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem Biophys Res Commun. 1997;240:1–7. doi: 10.1006/bbrc.1997.7575. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Baldi DL, Duraisingh M, Healer J, Mills KE, O’Donnell RA, Thompson J, Triglia T, Wickham ME, Crabb BS. Functional analysis of Plasmodium falciparum merozoite antigens: implications for erythrocyte invasion and vaccine development. Philos Trans R Soc Lond B Biol Sci. 2002;357:25–33. doi: 10.1098/rstb.2001.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, Schofield L, Crabb BS. Identification and stoichiometry of GPI-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. 2006;5:1286–1299. doi: 10.1074/mcp.M600035-MCP200. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hiller NL, Akompong T, Morrow JS, Holder AA, Haldar K. Identification of a stomatin orthologue in vacuoles induced in human erythrocytes by malaria parasites. A role for microbial raft proteins in apicomplexan vacuole biogenesis. J Biol Chem. 2003;278:48413–48421. doi: 10.1074/jbc.M307266200. [DOI] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem. 1996;271:29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- Holder AA, Freeman RR. Biosynthesis and processing of a Plasmodium falciparum schizont antigen recognized by immune serum and a monoclonal antibody. J Exp Med. 1982;156:1528–1538. doi: 10.1084/jem.156.5.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats LM, Black CG, Proellocks NI, Coppel RL. Plasmodium rhoptries: how things went pear-shaped. Trends Parasitol. 2006;22:269–276. doi: 10.1016/j.pt.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Lobo CA, Rodriguez M, Hou G, Perkins M, Oskov Y, Lustigman S. Characterization of PfRhop148, a novel rhoptry protein of Plasmodium falciparum. Mol Biochem Parasitol. 2003;128:59–65. doi: 10.1016/s0166-6851(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Malkin E, Dubovsky F, Moree M. Progress towards the development of malaria vaccines. Trends Parasitol. 2006;22:292–295. doi: 10.1016/j.pt.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Margos G, Bannister LH, Dluzewski AR, Hopkins J, Williams IT, Mitchell GH. Correlation of structural development and differential expression of invasion-related molecules in schizonts of Plasmodium falciparum. Parasitology. 2004;129:273–287. doi: 10.1017/s0031182004005657. [DOI] [PubMed] [Google Scholar]
- Marshall VM, Silva A, Foley M, Cranmer S, Wang L, McColl DJ, Kemp DJ, Coppel RL. A second merozoite surface protein (MSP-4) of Plasmodium falciparum that contains an epidermal growth factor-like domain. Infect Immun. 1997;65:4460–4467. doi: 10.1128/iai.65.11.4460-4467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall VM, Tieqiao W, Coppel RL. Close linkage of three merozoite surface protein genes on chromosome 2 of Plasmodium falciparum. Mol Biochem Parasitol. 1998;94:13–25. doi: 10.1016/s0166-6851(98)00045-0. [DOI] [PubMed] [Google Scholar]
- Murphy SC, Samuel BU, Harrison T, Speicher KD, Speicher DW, Reid ME, Prohaska R, Low PS, Tanner MJ, Mohandas N, Haldar K. Erythrocyte detergent-resistant membrane proteins: their characterization and selective uptake during malarial infection. Blood. 2004;103:1920–1928. doi: 10.1182/blood-2003-09-3165. [DOI] [PubMed] [Google Scholar]
- Nixon CP, Friedman J, Treanor K, Knopf PM, Duffy PE, Kurtis JD. Antibodies to Rhoptry-Associated Membrane Antigen Predict Resistance to Plasmodium falciparum. J Infect Dis. 2005;192:861–869. doi: 10.1086/432550. [DOI] [PubMed] [Google Scholar]
- O’Keeffe AH, Green JL, Grainger M, Holder AA. A novel Sushi domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol. 2005;140:61–68. doi: 10.1016/j.molbiopara.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Salzer U, Prohaska R. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood. 2001;97:1141–1143. doi: 10.1182/blood.v97.4.1141. [DOI] [PubMed] [Google Scholar]
- Salzer U, Hinterdorfer P, Hunger U, Borken C, Prohaska R. Ca(++)-dependent vesicle release from erythrocytes involves stomatin-specific lipid rafts, synexin (annexin VII), and sorcin. Blood. 2002;99:2569–2577. doi: 10.1182/blood.v99.7.2569. [DOI] [PubMed] [Google Scholar]
- Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, 3rd, Crabb BS. Distinct protein classes including novel merozoite surface antigens in raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- Sanders PR, Kats LM, Drew DR, O’Donnell RA, O’Neill M, Maier AG, Coppel RL, Crabb BS. A Set of Glycosylphosphatidyl Inositol-Anchored Membrane Proteins of Plasmodium falciparum Is Refractory to Genetic Deletion. Infect Immun. 2006;74:4330–4338. doi: 10.1128/IAI.00054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe JA, Coppel RL, Brown GV, Ramasamy R, Kemp DJ, Anders RF. Identification of two integral membrane proteins of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1988;85:5195–5199. doi: 10.1073/pnas.85.14.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J Biol Chem. 2004a;279:4648–4656. doi: 10.1074/jbc.M307859200. [DOI] [PubMed] [Google Scholar]
- Topolska AE, Richie TL, Nhan DH, Coppel RL. Associations between responses to the rhoptry-associated membrane antigen of Plasmodium falciparum and immunity to malaria infection. Infect Immun. 2004b;72:3325–3330. doi: 10.1128/IAI.72.6.3325-3330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Wang L, Black CG, Marshall VM, Coppel RL. Structural and antigenic properties of merozoite surface protein 4 of Plasmodium falciparum. Infection & Immunity. 1999;67:2193–2200. doi: 10.1128/iai.67.5.2193-2200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Richie TL, Stowers A, Nhan DH, Coppel RL. Naturally acquired antibody responses to Plasmodium falciparum merozoite surface protein 4 in a population living in an area of endemicity in Vietnam. Infect Immun. 2001;69:4390–4397. doi: 10.1128/IAI.69.7.4390-4397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Crouch L, Richie TL, Nhan DH, Coppel RL. Naturally acquired antibody responses to the components of the Plasmodium falciparum merozoite surface protein 1 complex. Parasite Immunol. 2003a;25:403–412. doi: 10.1111/j.1365-3024.2003.00647.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Mohandas N, Thomas A, Coppel RL. Detection of detergent-resistant membranes in asexual blood-stage parasites of Plasmodium falciparum. Mol Biochem Parasitol. 2003b;130:149–153. doi: 10.1016/s0166-6851(03)00165-8. [DOI] [PubMed] [Google Scholar]


