Abstract
ESCRT-III (endosomal sorting complexes required for transport-III) subunits cycle between two states: soluble monomers and higher-order assemblies that bind and remodel membranes during endosomal vesicle formation, midbody abscission and enveloped virus budding. Here, we show that the N-terminal core domains of IST1 (increased sodium tolerance-1) and CHMP3 (charged multivesicularbody protein-3) form equivalent four-helix bundles, revealing that IST1 is a previously unrecognized ESCRT-III family member. IST1 and its ESCRT-III binding partner, CHMP1B, both form higher-order helical structures in vitro, and IST1-CHMP1 interactions are required for abscission. The IST1 and CHMP3 structures also reveal that equivalent downstream α5 helices can fold back against the core domains. Mutations within the CHMP3 core-α5 interface stimulate the protein’s in vitro assembly and HIV inhibition activities, indicating that dissociation of the autoinhibitory α5 helix from the core activates ESCRT-III proteins for assembly at membranes.
The ESCRT (endosomal sorting complexes required for transport) pathway functions in a series of important membrane remodeling processes, including multivesicular body vesicle formation at endosomal membranes, enveloped virus budding from the plasma membrane, and midbody abscission during cytokinesis (reviewed in 1–4). In performing these functions, ESCRT components cycle between two different states: an inactive state, in which they are dispersed throughout the cytoplasm, and an active state in which they are recruited to specific membranes by upstream adaptor proteins that induce tight membrane binding and component assembly. ESCRT recruitment and assembly ultimately lead to membrane fission, producing endosomal vesicles, budded viruses, or daughter cells. The ESCRT pathway is therefore apparently a mobile machine that can be recruited to different membranes to help mediate membrane remodeling and fission.
Most of the ~30 proteins of the human ESCRT pathway function as components of one of five different complexes (termed the ESCRT-0-III and VPS4 complexes). These complexes are recruited sequentially to sites of action, with each successive complex recruiting and activating downstream complexes for membrane association and assembly1–4. Recent studies have revealed that subunits of the late acting ESCRT-III complex can form rings, helical filaments, and tubes in vitro and in vivo. Thus, ESCRT-III complexes may form “collars” within the cytoplasm-filled necks of membrane vesicles and tubules that can constrict the neck and promote membrane fission5–10. Variants of this general model suggest that one specific subset of ESCRT-III proteins, those of the CHMP4/Snf7p family, may create an encircling ring or spiral, and the other ESCRT-III proteins serve primarily to nucleate and cap the CHMP4/Snf7p filament8–10. ESCRT-III proteins also recruit VPS4 ATPases and induce them to assemble into active complexes. Enzyme recruitment and activation are further promoted by the accessory factors IST1 (Increased Sodium Tolerance-1)11–14 and Vta1p/LIP51–4, both of which can bind ESCRT-III and VPS4 proteins. Once assembled, VPS4 uses the energy of ATP hydrolysis to release the assembled ESCRT-III machinery back into the cytoplasm, thereby allowing multiple rounds of membrane deformation and fission10.
The eleven known human ESCRT-III-like proteins can be divided into seven different families, six of which (CHMP1-6) correspond to the six ESCRT-III-like proteins of S. cerevisiae. Most ESCRT-III proteins can form both homo- and hetero-oligomers, and can pair preferentially at membrane sites of action (e.g., CHMP4-CHMP6 and CHMP2-CHMP3)6,15. ESCRT-III proteins vary significantly in primary sequence, but all share common N-terminal core domains of ~150 residues. The core domain of one ESCRT-III protein, CHMP38–183, has been crystallized and shown to form a four-helix bundle16. The longer first two helices form an extended hairpin, and the shorter two helices pack at the open end of the hairpin. Sequences located beyond the helical core exhibit autoinhibitory activities, and there is now considerable biochemical and genetic evidence that these downstream sequences prevent premature membrane binding and assembly6,16–21. Downstream sequence elements also bind a variety of different late-acting ESCRT factors, including ALIX22 and a series of MIT domain-containing proteins and enzymes such as ubiquitin hydrolases (e.g., AMSH and UBPY)23,24, proteases (calpain 7)25,26, ATPases (Spastin and VPS4 proteins)21,27–30, and ATPase activators (Vta1p/LIP5)31–33. Different ESCRT-III subunits exhibit distinct binding partner specificities, which helps rationalize the need for so many different ESCRT-III proteins.
IST1 is a recently identified, late-acting ESCRT pathway factor that binds LIP5/Vta1p, VPS4 and CHMP1/Did2p11–14. The precise function(s) of IST1 are not yet well understood and, indeed, IST1 can exert both positive11–14 and negative12 effects on ESCRT pathway functions. Yeast Ist1p exhibits synthetic interactions with the Vta1p-Vps60p (LIP5-CHMP5) complex in endosomal protein sorting reactions, implying that Ist1p functions in MVB vesicle formation, albeit in a non-essential role12,13. Human IST1 localizes to midbodies, and its depletion inhibits abscission but not HIV budding11,14. IST1 proteins therefore appear to play important roles in cytokinesis, auxiliary roles in endosomal protein sorting, and no essential role in HIV budding. Biochemical and secondary structure analyses indicate that IST1 is composed of a helical N-terminal domain (residues 1–189, here termed IST1NTD, but also called the “ELYC” domain after a conserved tetrapeptide sequence12), C-terminal MIT Interacting Motifs (MIM) that can bind cooperatively to the MIT domains of VPS4 enzymes (321–366)11,14, and a central linker region that is predicted to lack persistent secondary structure (~190–320)11,14.
Extensive structural and biochemical analyses have revealed many of the relevant molecular interactions that allow different ESCRT components to associate with one another, with PI(3)P-containing membranes, and with ubiquitylated protein cargoes1–4. In contrast, the mechanisms that regulate ESCRT pathway assembly and disassembly are much less well understood. Here, we have determined crystal structures of human IST1 and CHMP3 proteins, and performed complementary biochemical analyses that characterize the functional importance of the IST1-CHMP1 binding interaction in cytokinesis and suggest how ESCRT-III proteins can switch between their inactive, soluble states and their active, membrane-bound assemblies.
RESULTS
Characterization of recombinant CHMP3 and IST1 proteins
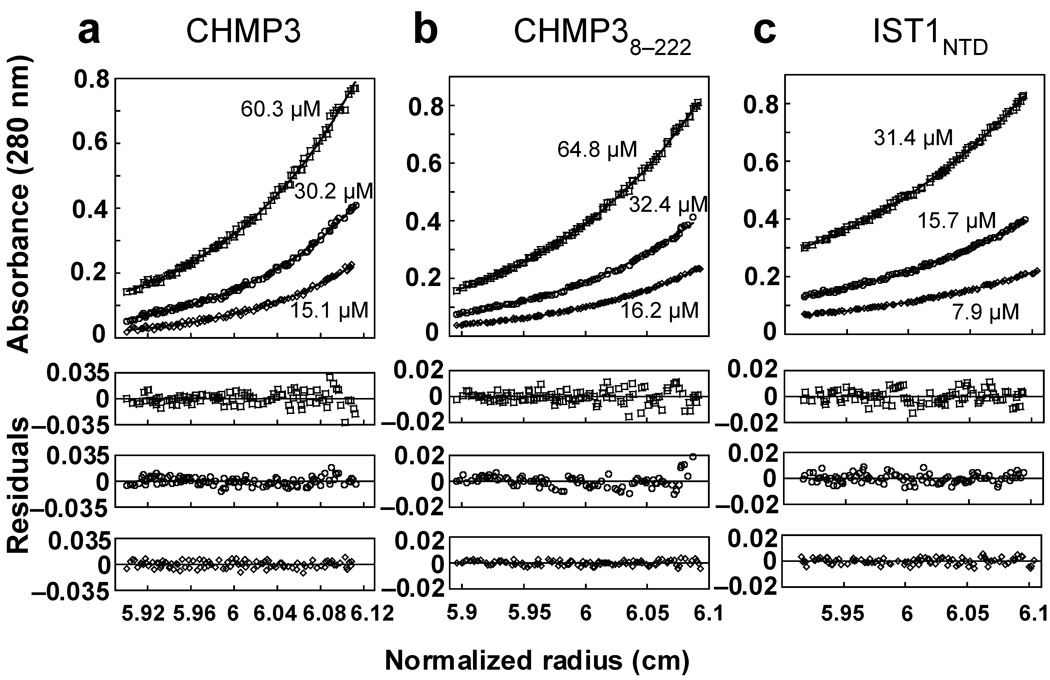
We developed methods for expressing and purifying recombinant human IST1 and CHMP3 proteins for structural and biochemical studies. Full length CHMP3 and an N-terminally truncated construct (CHMP38–222) were both monomeric in solution as analyzed by equilibrium sedimentation (Fig. 1), in good agreement with a previous report16. Full length IST1 was also monomeric under high ionic strength conditions (>300 mM NaCl)11, but aggregated at lower ionic strength. The purified N-terminal domain of IST1 (IST1NTD, residues 1–189) behaved like the full-length protein in that it was also monomeric under high ionic strength conditions (Fig. 1c) and insoluble in lower ionic strength solutions. This construct was particularly amenable for structural and biochemical studies, presumably because it lacked the proline-rich linker region present in the full-length protein.
Figure 1.
CHMP3, CHMP38–222 and IST1NTD are monomers in solution. Equilibrium sedimentation distributions of recombinant CHMP3 (a), CHMP38–222 (b), and IST1NTD (c) (upper panels), and residual differences (lower panels), with data points shown in open symbols and the single species models shown as solid lines. Rotor speeds were 20,000 RPM and initial subunit protein concentrations are shown. Data sets were also collected at 24,000 RPM (not shown) and all of the data were globally fit to single species models in which the molecular weights were allowed to float during the refinement. Estimated molecular weights were: CHMP3, 25,840 Da (MWmonomer = 25,267 Da, Mobs/Mcalc = 1.02); CHMP38–222, 24,390 Da (MWmonomer = 24,663 Da, Mobs/Mcalc = 0.99); IST1NTD, 20,520 Da (MWmonomer = 21,791 Da, Mobs/Mcalc = 0.94).
Crystal structures of IST1NTD
We crystallized IST1NTD under high ionic strength conditions, and the structure was determined in space groups P21 (1.8 Å resolution) and P43212 (2.6 Å, Table 1 and Supplementary Fig. 1a,b). The structures were similar, and the higher resolution P21 structure is described here. IST1NTD is composed of eight helices, which pack to create a flat, elongated structure, with overall dimensions of 21 × 30 × 60 Å (Fig. 2a). The two longest helices, α1 and α2, form a helical hairpin that packs against the smaller α3 and α4 helices to create an asymmetric four-helix bundle (termed the “core”). Beyond the core, the segment between helices α4 and α5 forms a flap that tracks along the length of the molecule, packing against the α2/α3 edge. The flap contains two short helices, which are designated αA and αB in order to maintain a consistent numbering scheme for conserved ESCRT-III helices. α5 packs perpendicularly across α1 and α2 near the closed end of the hairpin and is termed the “autoinhibitory helix”. A final turn takes the polypeptide across the hairpin loop and into α6, which projects away from the core. This helix was only seen in the P21 crystal form where it is stabilized by lattice contacts. As discussed below, the IST1NTD structure appears to represent a closed/autoinhibited ESCRT-III conformation in which the flap and autoinhibitory helix fold against the core to prevent higher order interactions.
Table 1.
IST1NTD and CHMP3 Crystallographic Statistics
| IST1NTD – Native 1 | IST1NTD – Peak 1 | IST1NTD – Inflection 1 | IST1NTD – Remote 1 | IST1NTD – Native 2 | CHMP38–222 | CHMP31–150 | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P43212 | P43212 | P43212 | P43212 | P21 | P21 | P61 |
| Cell dimensions | |||||||
| a, b, c (Å) | 57.2, 57.2, 157.2 | 57.6, 57.6, 157.1 | 57.6, 57.6, 157.2 | 57.7, 57.7, 157.3 | 29.3, 92.4, 33.25 | 36.5, 131.5, 48.5 | 111.5, 111.5, 30.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 97.0, 90.0 | 90.0, 108.1, 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 40 – 2.6 (2.7 – 2.6) | 40 – 3.4 (3.5 – 3.4) | 40 – 3.4 (3.5 – 3.4) | 40 – 3.4 (3.5 – 3.4) | 50 – 1.80 (1.86 –1.80) | 50 – 4.0 (4.14 – 4.0) | 25 – 3.7 (3.83-3.7) |
| Rsym | 6.8 (34) | 7.5 (16) | 5.9 (15) | 6.5 (14) | 5.8 (15) | 0.16 (15) | 0.063 (55) |
| I / σI | 13.5 (1.7) | 23.9 (5.0) | 24.3 (4.4) | 24.4 (3.8) | 19.2 (4.6) | 9.3 (2.9) | 18.4 (2.2) |
| Completeness (%) | 97.7 (88.3) | 88.5 (56.5) | 87.7 (55.4) | 83.4 (42.8) | 97.2 (79.1) | 78.7 (43.9) | 99.2 (96.8) |
| Redundancy | 3.8 (2.8) | 6.7 (5.8) | 3.9 (3.6) | 3.9 (4.0) | 3.1 (1.8) | 4.0 (2.8) | 3.2 (3.1) |
| Refinement | |||||||
| Resolution (Å) | 40 – 2.6 | 50 – 1.80 | 38 – 4.0 | 24 – 3.7 | |||
| No. reflections | 7968 | 15213 | 2936 | 2449 | |||
| Rwork / Rfree (%) | 25.9 / 29.8 | 19.5 / 24.7 | 37 | 41 | |||
| No. atoms | |||||||
| Protein | 1438 | 1558 | 2274 | 973 | |||
| Water | 35 | 163 | 0 | 0 | |||
| B-factors (Å2) | |||||||
| Protein | 87.2 | 26.0 | n/a1 | n/a1 | |||
| Water | 69.0 | 39.4 | n/a1 | n/a1 | |||
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.0007 | 0.011 | n/a1 | n/a1 | |||
| Bond angles (°) | 1.2 | 1.3 | n/a1 | n/a1 | |||
Bfactors and individual atoms were not refined and are unchanged from the molecular replacement model.
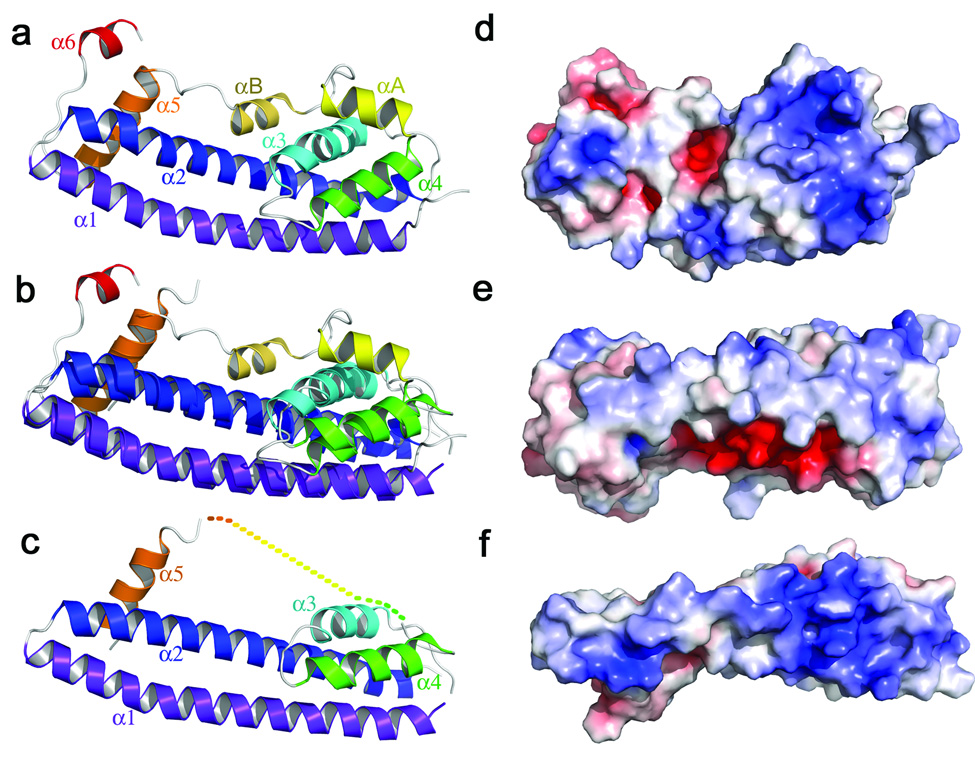
Figure 2.
Structures of IST1NTD and CHMP3. (a) Ribbon diagram and helix labeling scheme for IST1NTD. (b) Overlay of the ordered regions of IST1NTD and CHMP38–183. (c) Ribbon diagram of CHMP38–222. (d) Space filling model of IST1NTD, color coded to show the surface charge distribution (blue, basic; red, acidic; ± 7kV)36,37. The molecule is shown in the same orientation as in a. (e) Same as d. The view is toward α1, and was generated from d by rotation about the horizontal so that the bottom edge of d faces the viewer. (f) Space filling model of CHMP38–222 shown in an equivalent orientation to the view of IST1NTD shown in e, emphasizing the basicity of the α1 surface of CHMP316.
IST1NTD bears an unexpected resemblance to the published CHMP38–183 structure16 (Figs. 2a–c), and the four helices of the IST1NTD core superimpose with the first four helices of CHMP3 with an RMSD of 2.4 Å (128 Cα positions). Muziol et al. noted that the α1 edge of CHMP3 is distinctly cationic and proposed that this surface binds membranes16. The equivalent IST1 surface is also basic, although the α3/α4 face is even more so (Figs. 2d–f).
In addition to their similar core structures, the autoinhibitory α5 helices of CHMP38–183 and IST1NTD sit in equivalent positions (α5, orange). Thus, all of the ordered helices in the CHMP38–183 structure have analogs in IST1NTD. However, the assigned connectivity of the autoinhibitory helices differ between the two structures because this helix is connected in “cis” to the IST1NTD core via the flap segment, whereas it was assigned to an adjacent molecule in the CHMP38–183 lattice16. Muziol et al. noted, however, that the CHMP38–183 connectivity could not be defined unambiguously owing to a lack of visible electron density for the flap region16. We therefore speculate that the CHMP38–183 connectivity may actually match that of IST1NTD and that the CHMP38–183 structure may also represent a closed conformation.
Crystal structures of CHMP3
The CHMP38–183 construct lacked the final 39 CHMP3 residues, which may have favored an open conformation16. We therefore attempted to remove this ambiguity by crystallizing both the full-length monomeric CHMP31–222 protein and a slightly truncated CHMP38–222 protein. Both proteins crystallized isomorphously in space group P21, but CHMP38–222 diffracted to slightly higher resolution (4.0Å), and this structure is therefore reported (Fig. 2c and Table 1). The CHMP38–222 structure was determined by molecular replacement using the core of CHMP38–183 as a search model and then subjected to rigid body refinement. Discernible features beyond the search model were limited, but the structure solution is supported by the molecular replacement statistics (Methods) and by unbiased electron density maps (Supplementary Fig. 2). In addition to the four core helices, density for the autoinhibitory helix was evident in unbiased electron density maps, particularly for one of the two independent molecules in the asymmetric unit (Supplementary Fig. 2c,d). Although the connectivity between α4 and α5 was not defined by experimental electron density, α5 appears to sit in the same position in both the CHMP38–183 and CHMP38–222 structures despite different crystal packing interactions in the two lattices. We have connected α4 and α5 in cis rather than in trans to create a structure with the same topology as IST1NTD, and this assignment is consistent with biochemical analyses described below.
Finally, we reasoned that a construct corresponding to the CHMP3 core alone could not adopt an autoinhibited conformation and might therefore reveal authentic protein-protein interactions used for ESCRT-III assembly. The CHMP3 core construct (CHMP31–150) was crystallized in space group P61 and the structure solved by molecular replacement (3.7 Å resolution, Table 1 and Supplementary Fig. 2b). As expected, the CHMP31–150 core was very similar to the cores of the other CHMP3 and IST1NTD structures, although, as described below, the lattice interactions were unique.
Crystal packing interactions
We analyzed lattice contacts in the different IST1 and CHMP3 crystal structures to determine whether they were strong candidates for authentic interactions used in ESCRT-III protein assembly (see Supplementary Fig. 3 for a complete catalog of the different lattice interactions). Overall, there are not compelling reasons to believe that any of the various crystal contacts represent authentic assembly interfaces. Moreover, both of our CHMP3 structures lacked the “side-to-side” dimer interface seen in the CHMP38–183 structure16 and postulated as a possible ESCRT-III assembly interface6,7.
Two of the new CHMP3 interfaces merit comment because they are mediated by a conserved, exposed hydrophobic surface on the closed “tip” of the α1/α2 hairpin. In the CHMP8–222 structure, two adjacent molecules associate through a small (359 Å2) two-fold symmetric “tip-to-tip” dimer interface (Fig. 3a,b). A similar tip-to-tip interface was also present in the CHMP38–183 lattice16, and this is the only interface that has been seen in more than one crystal form.
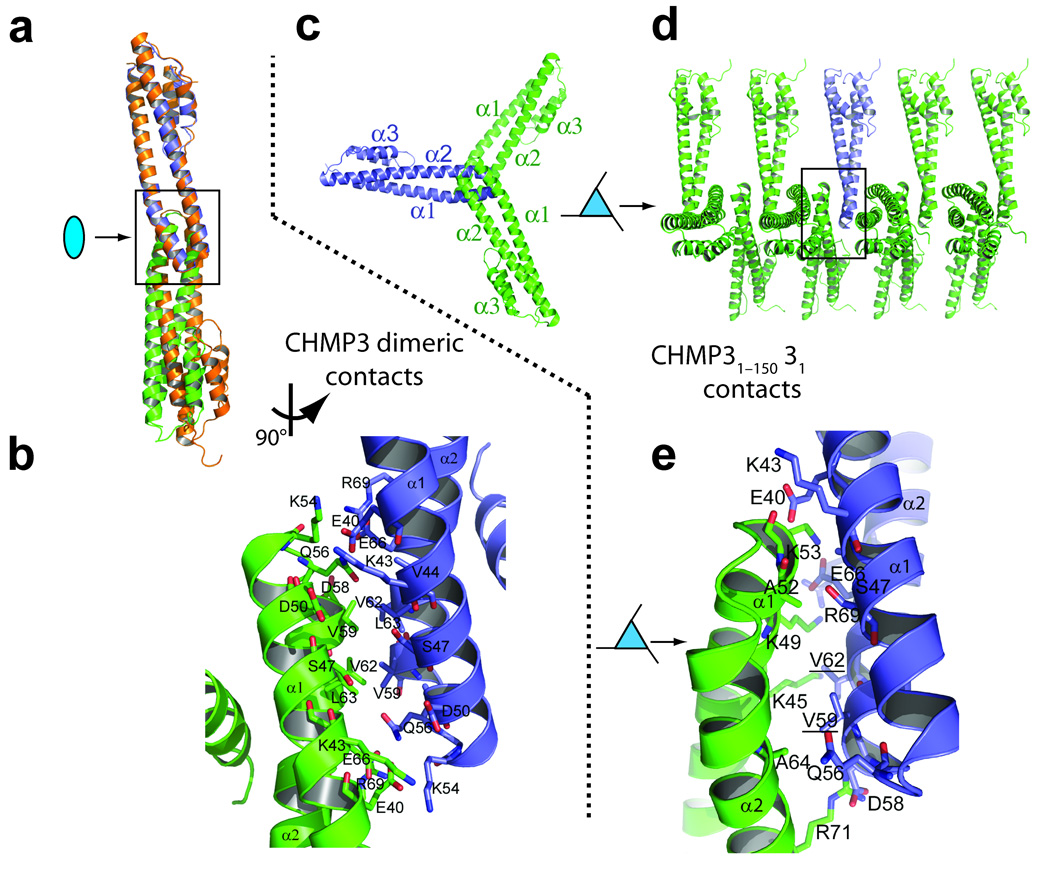
Figure 3.
CHMP3 crystal packing interactions. (a) Overlay of the cores of the “tip-to-tip” dimers in the crystal lattices of CHMP38–183 (orange)16 and CHMP38–222 (blue and green). The upper subunits were aligned, and the lower subunits diverge owing to small differences in their tip-to-tip interfaces. (b) Detailed expansion of the boxed region of CHMP38–222 shown in a. Both here and in e, most side chains conformations are shown as defined in the CHMP38–183 search model, with some minor adjustments to avoid clashes. Precise description of these side chain conformations is not possible at current resolutions. (c) Subunit packing down the 31 screw axis of the CHMP31–150 crystal lattice, with a single CHMP31–150 molecule highlighted in blue. (d) Same assembly as in c, but viewed perpendicularly to the 31 screw axis. (e) Detailed expansion of the boxed area shown in d.
A different type of tip-to-tip interaction is present in the CHMP31–150 lattice. In this case, the interaction lacks two-fold rotational symmetry and instead follows a crystallographic 31 screw axis that creates an infinite CHMP31–150 filament with interfaces on both sides of the tip (Fig. 3c–e). The yeast homolog of CHMP3 (Vps24p) can assemble into three-stranded filaments in vitro7, although the relationship between the Vps24p filaments and the crystallographic CHMP3 filaments remains to be determined. In summary, the crystal packing interactions in our IST1 and CHMP3 structures do not obviously correspond to authentic biological interfaces, but do reinforce the idea that the exposed hydrophobic surface located at the tip of the helix 1/2 loop is a preferred site for protein-protein interactions6,7,16.
IST1NTD binds CHMP1 proteins
Many ESCRT-III proteins bind one another, and previous studies have shown that the N-terminal domain of IST1 binds preferentially to the C-terminal region of the CHMP1 subset of ESCRT-III proteins11–14. To map the IST1 binding surface more precisely, we created an ensemble of IST1 proteins with point mutations at conserved, surface-exposed residues and tested them for CHMP1A and CHMP1B interactions using: 1) immunoprecipitation (IP) reactions in 293T cells (not shown), 2) GST-pulldown experiments with recombinant proteins (not shown), and 3) biosensor binding experiments in which pure recombinant CHMP1B bound immobilized GST-IST1NTD (Fig. 4).
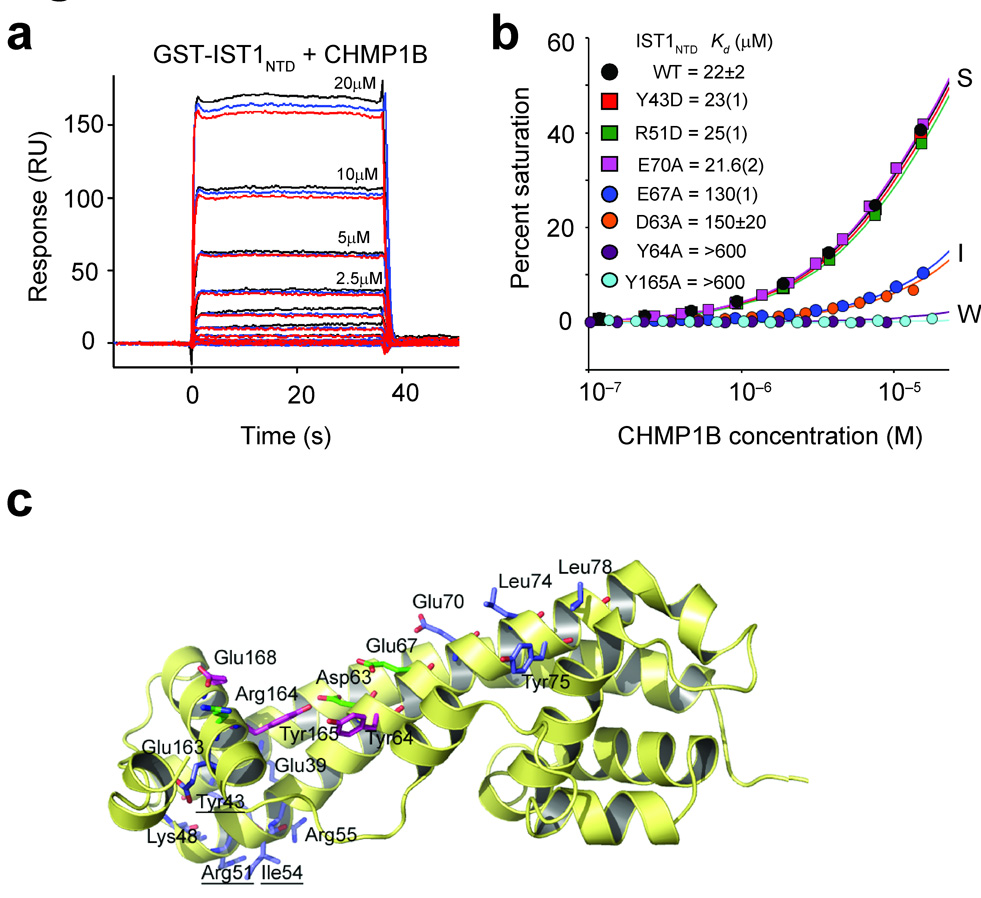
Figure 4.
Mutational analyses of IST1-CHMP1B interactions. (a) Sensorgrams showing different concentrations of purified CHMP1B binding to immobilized GST-IST1NTD. Triplicate measurements in response units (RU) are shown for each CHMP1B concentration. (b) Representative biosensor binding isotherms showing CHMP1B binding to wild type (WT) and mutant GST-IST1NTD proteins with strong (S), intermediate (I) and weak (W) binding affinities. IST1NTD mutations and estimated dissociation constants are given in the inset. Errors represent either standard deviations from multiple independent measurements (n≥3), or standard deviations derived from single isotherms measured in triplicate (values in parentheses report the standard deviations in the final digit of the measurement).(c) Ribbon diagram showing the location of all IST1NTD mutations tested for CHMP1 binding. Mutated residues are shown explicitly, and the binding affinities of the mutant proteins are color-coded as follows: blue, strong (S) binding (binding affinities within 1.5-fold of wild type IST1NTD); green, intermediate (I) binding (binding affinity reduced 1.5–8 fold); magenta, weak (W) binding (binding affinity reduced ≥8-fold).
As shown in Fig. 4a,b, CHMP1B bound wild type IST1NTD with an estimated dissociation constant of 22±2 µM. CHMP1B binding was unaltered by most mutations tested, including mutations within the closed tip of the α1/α2 hairpin (e.g., E39A, Y43D, K48D, R51D, I54D and R55D), and in the IST1NTD surface that corresponded to the “side-to-side” dimer interface seen in the crystal structure of CHMP38–183 and included the signature 73ELYCEL78 motif (e.g., E70A, L74R, Y75A, and L78R). However, one mutation along helix 2 (E67A) reduced CHMP1B binding in the IP and pulldown experiments and lowered the CHMP1B binding affinity six-fold in the biosensor binding assay, suggesting that this residue contributed to the CHMP1B binding site. Mutations in adjacent surface exposed residues were therefore tested to identify the CHMP1B binding site more precisely, and these analyses revealed three IST1 mutants that bound CHMP1B weakly (>8-fold affinity reductions, IST1Y64D, IST1Y165A and IST1E168R), three IST1 mutants that bound CHMP1B with intermediate affinities (2- to 8-fold reductions, IST1D63A, IST1E67A and IST1R164D), and one mutation that still bound strongly to CHMP1B (IST1E163R, data not shown).
Given the proximity of the apparent CHMP1B binding site to the IST1 autoinhibitory helix α5, we also tested the effects of a series of IST1 mutations predicted to destabilize the α5-core interface (V56D, I60D, L166D, and I169D). All of these IST1 mutations blocked CHMP1B binding entirely (not shown), indicating that CHMP1B binds the closed conformation of IST1NTD seen in the crystal structures. As summarized in Fig. 4c, mutations that inhibited CHMP1B binding clustered within or near a groove created by packing of autoinhibitory helix α5 against core helix α2. The groove features a pair of exposed Tyr residues (Tyr64 and Tyr165), which likely contribute to CHMP1B binding.
The IST1-CHMP1B interactions function in cytokinesis
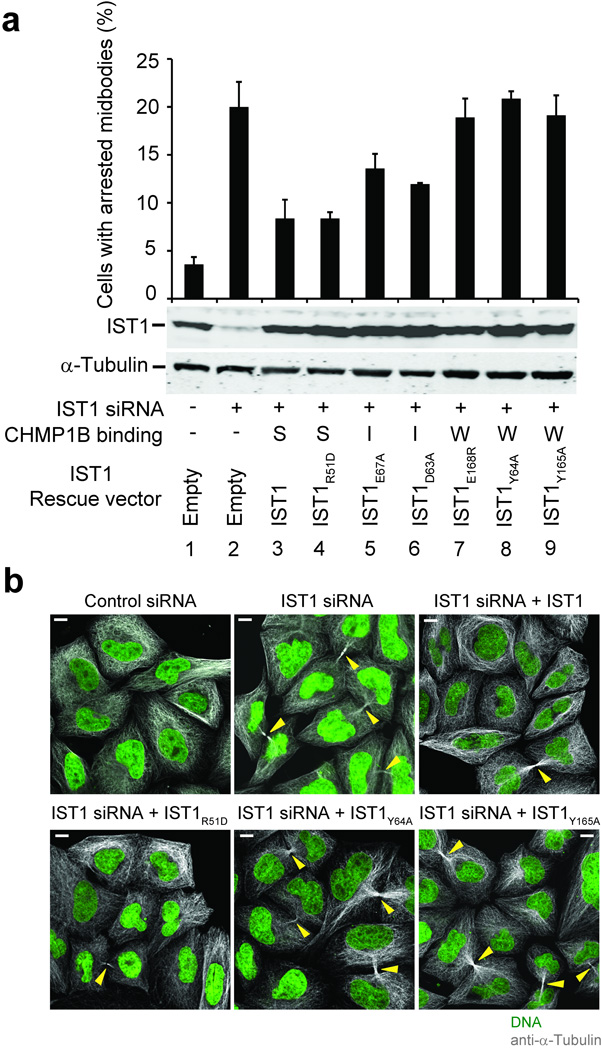
To determine whether CHMP1 binding was necessary for IST1 cytokinesis function(s), we tested whether IST1 mutations that impaired CHMP1B binding also inhibited abscission. Depletion of endogenous IST1 increased the percentage of HeLa cells with visible midbodies from 4±1% to 20±3% (Fig. 5a, compare lanes 1 and 2 and Fig. 5b, panels 1 and 2), and this midbody arrest defect was largely (though not entirely) rescued by re-expression of a wild type, siRNA-resistant IST1 construct (8±2%, lane/panel 3)11. Midbody arrest was also corrected by an IST1 protein with a mutation in the tip of the α1/α2 hairpin that retained strong (S) CHMP1B binding (IST1R51D, 8±1%, lane/panel 4). In contrast, three weak-binding IST1 mutants failed to rescue midbody arrest (18–21%, Fig. 5a, lanes 7–9 and Fig. 5b, panels 5 and 6). Point mutations within the α5-core interface that blocked CHMP1B binding also blocked rescue of the midbody arrest defect. However, these mutant proteins partitioned almost exclusively into the insoluble/membrane-bound cellular fractions, and their failure to support abscission may therefore reflect either aberrant CHMP1 binding or IST1 protein mislocalization (see Supplementary Fig. 4 and the Supplementary Discussion). Finally, two IST1 mutants with intermediate (I) CHMP1B binding affinities rescued midbody arrest to an intermediate extent (12–13%, Fig. 5a lanes 5 and 6). Hence, there was an excellent correlation between the ability of different IST1 constructs to bind CHMP1B in vitro and to function in abscission, indicating that CHMP1 binding is required for IST1 abscission function(s).
Figure 5.
Requirement for IST1-CHMP1 interactions during abscission. (a) The upper panel shows quantified abscission defects as reflected in the percentages of HeLa M cells with visible midbodies following siRNA treatment to deplete endogenous IST1 (lanes 2–9) and rescue with an empty vector control (lane 2, negative control) or with vectors expressing wild type IST1 (lane 3, positive control) or the designated IST1 mutants (lanes 4–9). Untreated cells are shown in lane 1. Error bars show standard deviations from three independent repetitions of the experiment. The middle panel is a western blot (anti-IST1) showing levels of soluble endogenous IST1 (lanes 1 and 2) or exogenously expressed IST1 proteins (lanes 3–9). The bottom panel is a western blot (anti-α-Tubulin) showing expression levels of endogenous α-Tubulin (loading control). CHMP1B binding phenotypes of the different IST1 proteins are shown below: strong (S), intermediate (I), or weak (W). (b) Immunofluorescence images showing the midbody phenotypes of cells from a designated subset of the experiments from a. Microtubules (anti-α-Tubulin, grey) and nuclei (SYTOX green) were stained for reference, and yellow arrowheads highlight midbodies. Scale bars are 10µm.
IST1NTD and CHMP1B form homopolymeric helices in vitro
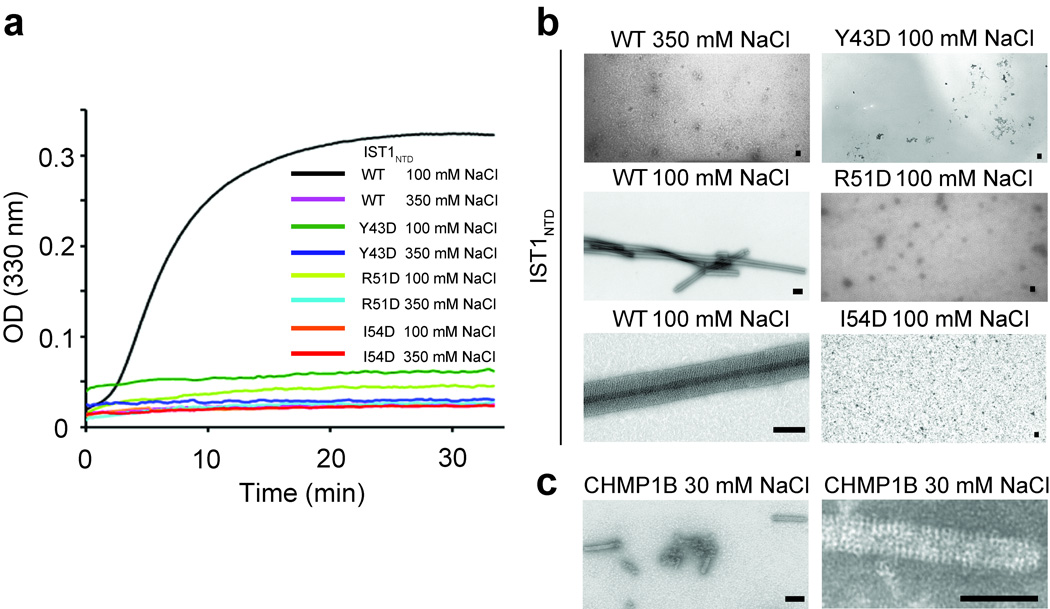
CHMP3 can co-polymerize with truncated CHMP2A proteins to form 50 nm diameter helical tubes in vitro6. As noted above, both IST1 and IST1NTD became insoluble under low ionic strength conditions, and we therefore tested whether these proteins assembled into regular structures. Assembly conditions were initially surveyed using light scattering to follow complex formation. As shown in Fig. 6a, monomeric IST1NTD spontaneously polymerized into high molecular weight complexes that strongly scattered light at 330 nm when the protein was diluted from a high salt buffer (350 mM NaCl) into a low salt buffer (100 mM NaCl). In contrast, IST1NTD did not scatter light when diluted into an otherwise equivalent high salt buffer (350 mM NaCl).
Figure 6.
IST1NTD and CHMP1B tube assembly. (a) Assembly of wild type (WT) IST1NTD or three different IST1NTD mutants with the designated amino acid substitutions at the tip of the α1/α2 hairpin (mutated residues are underlined in Fig. 4c). IST1NTD proteins were diluted from concentrated protein stocks in high salt buffers to final concentrations of 62 µM. Salt concentrations in the assembly buffers are provided in the inset key and protein assembly was followed by light scattering at 330 nm. (b) Transmission electron microscopic images of a subset of the wild type and mutant IST1NTD assembly reactions from a. Note that IST1NTD only assembled under low salt conditions (compare upper and middle left panels) and that IST1NTD proteins with different point mutations at the tip of the α1/α2 hairpin did not assemble in either high salt (not shown) or low salt conditions (right panels). An expanded view of a single IST1NTD tube is shown in the lower left panel. Scale bars are 500 nm. (c) Transmission electron microscopic images of CHMP1B tubes assembled under low salt conditions. A field of tubes is shown in the left panel and an expanded view of a single CHMP1B tube is shown in the right panel. Scale bars are 500 nm.
EM analyses revealed that the light scattering IST1NTD assemblies were large tubular structures with diameters of ~700 nm (Fig. 6b, lower left panels). The tubes appeared to be open along one edge and may therefore be curled sheets rather than fully closed helices. Lattice lines were evident in some assemblies, and the clearest lattice spacing was ~440 Å, which presumably represents a supermolecular spacing because the maximal length of a single IST1NTD molecule is only ~60 Å. As noted above, CHMP3 interactions can be mediated by various different kinds of tip-to-tip interactions (refs. 6,7 and Fig. 3). A series of three different point mutations in the analogous IST1 surface (Y43D, R51D and I54D) also inhibited IST1NTD polymerization (Fig. 6a,b. right column), and see Supplementary Fig. 5 for ESCRT-III protein alignments). IST1NTD assembly therefore likely also requires some type of tip-to-tip interaction.
We also attempted to test whether IST1NTD and CHMP1B could co-polymerize like the CHMP2A9–161-CHMP3 pair6. However, controls for these experiments revealed that pure recombinant CHMP1B alone polymerized into helical tubes when diluted into low salt buffers, even in the absence of IST1NTD (Fig. 6c). CHMP1B tubes were of intermediate size (~230 nm in diameter), appeared to be fully closed, and had visible lattice spacing of ~420 Å, which were similar to the lattice spacing in the IST1NTD tubes. Thus, both IST1NTD and CHMP1B, like CHMP2A-CHMP36, form tubular structures in vitro, and the ability to assemble into helical tubes therefore appears to be a property common to many ESCRT-III core domains. However, the IST1 and CHMP1B tubes were homopolymeric, and were significantly larger than those formed by CHMP2A-CHMP3 heteropolymers6.
Mutations at the tip of the CHMP3 α1/ α2 hairpin inhibit CHMP2A binding
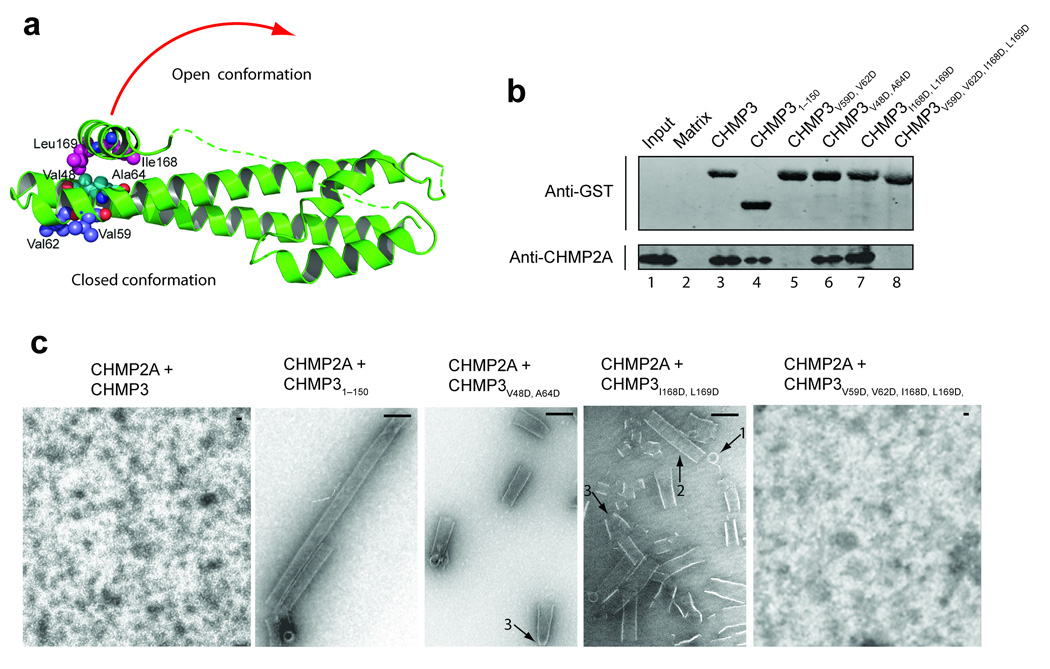
Like the IST1-CHMP1 pair, CHMP2A and CHMP3 bind one another preferentially. To investigate this interaction further, we used GST pulldown assays to test for binary interactions between CHMP2A and GST-CHMP3 (Figs. 7a,b). These experiments were performed at low protein concentrations where higher order assemblies were not detectable (35µM CHMP2A, ~7 µM GST-CHMP3). CHMP2A protein bound GST-CHMP3 (Fig. 7b, lane 3) but did not bind a matrix control (lane 2). CHMP2A also bound the truncated GST-CHMP31–150 protein (lane 4), indicating that CHMP2A interacted with the CHMP3 core and that the binary interaction was not dramatically affected by removal of the terminal CHMP3 autoinhibitory sequences. In contrast, a double point mutation (V59D, V62D) that disrupted the exposed hydrophobic surface on one side of the tip of the CHMP3 α1/ α2 hairpin eliminated CHMP2A binding (lane 5). These data suggest that CHMP2A and CHMP3 interact through a tip-to-tip type interaction, and demonstrate that the binary CHMP2A-CHMP3 and IST1-CHMP1B pairs interact via different binding surfaces and therefore form structurally distinct complexes.
Figure 7.
CHMP3 activation in vitro. (a) Ribbon diagram showing the locations of mutated CHMP3 residues at the tip of the α1/α2 hairpin (purple) and activating mutations on either side of the interface between the core α2 helix (cyan) and the autoinhibitory α5 helix (magenta). The red arrow suggests how the closed CHMP3 conformation might convert into an open conformation by dissociation of the autoinhibitory α5 helix from the core. (b) GST pull-down analyses of the binary CHMP3/CHMP2A interaction. Pure recombinant CHMP2A (lower panel, anti-CHMP2A) was tested for binding to a glutathione sepharose matrix (lane 2, negative control) or to immobilized wild type (lane 3) or mutant GST-CHMP3 proteins (lanes 4–8). Both proteins were detected by Western blotting, and input CHMP2A (0.3%) is shown in lane 1 for reference. (c) EM analyses of helical CHMP3-CHMP2A assembly. Different panels show assemblies formed by 1:1 mixtures CHMP2A with: full length, wild type CHMP3 (panel 1, negative control, no assembly), CHMP31–150 core domain (panel 2, positive control), activated CHMP3V48D,A64D (panel 3), activated CHMP3I168D,L169D (panel 4), and activated and tip mutant CHMP3I168D,L169D,V59D,V62D (panel 5, no assemblies). Arrows highlight rings (1), tubes (2), and cones/tapered tubes (3). Scale bars are 100 nm.
Autoinhibitory helix interface mutations activate CHMP3 for polymerization
Comparative analyses of the IST1NTD, CHMP38–183, and CHMP38–222 structures suggested a model for ESCRT-III autoinhibition in which the α5 helix and flap elements fold back against the protein core to prevent higher order assembly. We tested this idea by introducing destabilizing mutations within the α5-core interface of the full length CHMP3 protein and testing their effects on CHMP2A co-assembly. Although the full length CHMP2A and CHMP3 proteins bind one another, these two proteins do not readily form higher order helical assemblies (Fig. 7c, panel 1, and see ref. 6). In contrast, a CHMP31–150 protein that lacked the flap and autoinhibitory helix readily copolymerized with CHMP2A to form long, regular helical tubes (Fig. 7c, panel 2, positive control). Since CHMP2A forms binary complexes with both CHMP3 and CHMP31–150, these experiments imply that CHMP3 residues 151–222 inhibit additional interaction(s) required for higher order assembly.
To examine the mechanism of CHMP3 autoinhibition, we mixed full length CHMP2A with full-length CHMP3 proteins that contained mutations on either side of the core-α5 interface (CHMP3V48D, A64D and CHMP3I168D, L169D, see Fig. 7a). Both sets of mutations activated CHMP3 for assembly, as reflected in the formation of numerous rings, tubes, and cones (Fig. 7c, panels 3 and 4). The tubes were similar in diameter to those formed in the CHMP2A/CHMP31–150 control reaction (~50 nm) but were typically shorter, possibly because the point mutations activated CHMP3 proteins less fully than removal of the entire autoinhibitory region.
We also tested whether the binary CHMP2A-CHMP3 interface detected at low protein concentrations was required for higher order assembly. As expected, mutations on either side of the CHMP3 autoinhibitory interface did not significantly affect CHMP2A binding to GST-CHMP3 (CHMP3V48D, A64D and CHMP3I168D, L169D, Fig. 7b, lanes 6 and 7). However, a CHMP3 protein carrying mutations in both the tip and autoinhibitory interfaces (CHMP3V59D, V62D, I168D, L169D) could neither bind CHMP2A in the GST pulldown assay (Fig. 7b, lane 8) nor assemble into helical tubes (Fig. 7c, panel 5). These experiments imply that tip-to-tip interactions contribute to CHMP2-CHMP3 co-assembly, in good agreement with a previous study6.
Mutations in the autoinhibitory helix activate CHMP3 in vivo
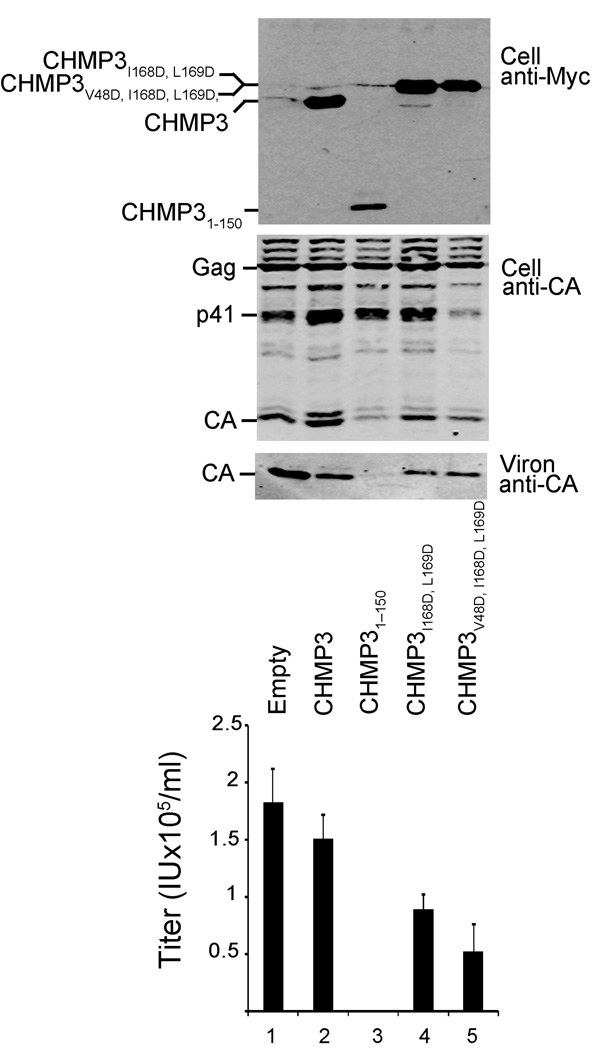
Although co-expression of full length CHMP3 has only minimal effects on HIV-1 budding, truncated CHMP3 constructs lacking C-terminal autoinhibitory elements potently inhibit virus budding, presumably because the activated protein sequesters itself and other ESCRT-III proteins on cellular membranes20. Inhibition of HIV release therefore provides a sensitive assay for CHMP3 activation in vivo. We used this system to examine whether mutations in the α5-core interface also activate CHMP3 in vivo. As shown in Fig. 8, co-overexpression of full length CHMP3 with an HIV-1 vector system had little effect on the release of viral particles, as measured by the levels of virion-associated CA proteins released from 293T cells (Fig. 8, panel 3, compare lanes 1 and 2) or by viral titers (panel 4, compare lanes 1 and 2). In contrast, HIV release and infectivity were potently inhibited by co-expression of a CHMP3 protein that lacked the autoinhibitory region entirely (CHMP31–150, positive control, compare lanes 2 and 4, >250-fold infectivity reduction). In the actual experiment, two different CHMP3 proteins with point mutations within the α5-core interface (CHMP3I168D, L169D and CHMP3V48D, I168D, L169D) reduced HIV vector release and infectivity by 1.7- and 3-fold, respectively (compare lanes 4 and 5 to lane 2). Western blots confirmed that the full-length CHMP3 proteins and viral Gag proteins and proteolytic processing products were expressed at similar levels (panels 1 and 2, respectively). Thus, point mutations in the α5-core interface enhanced the ability of CHMP3 proteins to inhibit HIV-1 budding in vivo, although this enhancement was much weaker than that seen upon deletion of the entire autoinhibitory region. We presume that the weaker effects seen for the point mutants reflects either incomplete activation and/or their ability to interact with VPS4 which will reduce their propensity to become “trapped” on membranes as compared to CHMP31–150 (which lacks a terminal VPS4 binding site).
Figure 8.
CHMP3 activation in vivo. HIV-1 vector expression and release upon co-expression with an empty vector (negative control, lane 1), or with vectors expressing wild type CHMP3 (negative control, lane 2), a CHMP31–150 protein lacking the entire autoinhibitory region (positive control, lane 3), proteins with activating mutations in the α5-core interface: CHMP3I168D, L169D (lane 4), and CHMP3V48D, 168D, L169D (lane 5). Western blots in the first two panels show cellular expression levels of CHMP3-Myc proteins (panel 1, anti-Myc) and of the viral Gag protein and its p41 and CA processing products (panel 2, anti-CA). The western blot in panel 3 shows levels of released, virion-associated CA proteins. The bottom graph shows the infectious titers of HIV-1 vectors released under the different conditions (infectious units ml−1, error bars show standard deviations in multiple titer measurements, n≥4).
DISCUSSION
Our studies have unexpectedly revealed that the N-terminal domain of IST1 contains an asymmetric four-helix bundle that closely resembles the core of the ESCRT-III protein, CHMP3 (Fig. 2). Like other ESCRT-III proteins, IST1 also: 1) localizes to midbodies and functions in cytokinesis11,14 and MVB vesicle sorting12,13, 2) binds preferentially to another subset of ESCRT-III proteins (the CHMP1 proteins)11–14, 3) contains MIM elements near its C-terminus that can bind VPS4 and LIP5 MIT domains11,14, and 4) can self-assemble into tubular structures in vitro (Fig. 6). Thus, despite its larger size and primary sequence divergence, IST1 is an ESCRT-III protein family member.
Despite these similarities, there are also important differences between IST1 and CHMP3, particularly in the binary interactions that they make with their CHMP1 and CHMP2 binding partners. Our mutational analyses mapped the CHMP1 binding site to a groove formed by the autoinhibitory α5 helix and the core α2 helix of IST1NTD (Fig. 4). CHMP1B binding was also inhibited by mutations expected to favor the (hypothetical) open IST1NTD conformation, implying that CHMP1 binds the closed conformation seen in our IST1NTD crystal structures (Fig. 2). IST1 mutations that blocked CHMP1B binding also prevented IST1 from functioning in abscission, implying that these two protein proteins must act together during this final stage of cytokinesis (Fig. 5). In contrast, binary interactions between CHMP2 and CHMP3 were not inhibited by mutations that displaced or even removed the α5 helix entirely (Fig. 7). This interaction was sensitive to mutations at the tip of the α1/α2 hairpin, however, suggesting that unlike IST1 and CHMP1B, CHMP2 and CHMP3 likely interact through some sort of a tip-to-tip interaction (Fig. 7).
In vitro assemblies formed by ESCRT-III proteins include filaments, rings, cones, curled sheets, and tubes (Figs. 6b,c, Fig. 7c and refs. 6,7). At present, we do not know which (if any) of these different in vitro assemblies mimic different ESCRT-III assemblies formed in vivo. As noted previously5–7, however, filaments, rings and/or tubes could correspond to “collars” within the necks of budding particles, and tapered tubes/spiraling cones could provide a mechanism for closing the neck during budding6 (see Fig. 7c, structure types 3). Although the precise molecular interactions that mediate higher order ESCRT-III assembly remain to be determined, both IST1NTD and CHMP2-CHMP3 assemblies are inhibited by point mutations within the conserved, exposed hydrophobic surface located at the tip of the α1/α2 hairpin (Fig. 6b, Fig. 7c), suggesting that some type of tip-mediated interface forms within both assemblies. Two distinct types of homopolymeric ESCRT-III tip-to-tip interactions have now been seen crystallographically (ref. 16 and Fig. 3), and a third type was inferred from cryo-EM studies of helical yeast Vps24/CHMP3 assemblies7. Thus, the conserved hydrophobic surface at the tip of ESCRT-III proteins is a preferred interaction site, although it is not yet clear which, if any, of the tip-to-tip interactions characterized to date represent biologically relevant interactions. We also note that although the IST1 R51D tip mutation inhibits IST1 assembly (Fig. 6a,b) it does not block the ability of IST1 to function in cytokinesis (Fig. 5), implying different requirements for IST1 assembly in vitro and abscission function in vivo. Finally, in addition to tip contacts, at least one more protein-protein interface is required to create two-dimensional surfaces in helices or sheets. These secondary interaction(s) are disfavored by C-terminal autoinhibitory interactions (ref. 6 and this work), and may correspond to one of the several different kinds of higher order packing interactions that have been proposed on the basis of cryo-EM analyses of helical ESCRT-III protein assemblies6,7.
Our IST1NTD structure revealed that the downstream flap and autoinhibitory helix can fold back against the ESCRT-III core, and this topology was unambiguously established by high quality electron density throughout the flap region (Fig. 2 and Supplementary Fig. 1). Analogous models can be built for the CHMP8–183 and CHMP328–222 structures, although in those cases the connectivity has not been established unambiguously. Nevertheless, we suggest that all of these structures likely correspond to the closed, autoinhibited ESCRT-III conformation because: 1) both IST1NTD and CHMP38–222 are monomers in solution under the conditions used for crystallization (although IST1NTD polymerizes under low salt conditions) (Fig. 1), 2) IST1 is redistributed into the insoluble membrane-bound/assembled fractions of cellular extracts by mutations that disrupt core packing of the autoinhibitory elements (Supplementary Fig. 4), 3) CHMP3 and CHMP2A can be activated for co-assembly by deletions that remove the autoinhibitory helices from CHMP2A6 or CHMP3, or by point mutations that destabilize packing of the CHMP3 autoinhibitory helix against the core (Fig. 7), and 4) mutations that remove the autoinhibitory helix or destabilize its core packing interactions render CHMP3 a dominant inhibitor of HIV-1 budding (refs. 17,20 and Fig. 8). Nevertheless, there is also good evidence that additional contacts beyond those visualized in our IST1NTD or CHMP3 structures help to stabilize the closed CHMP3 conformation even further because point mutations in the autoinhibitory helix-core interface do not activate CHMP3 to the same extent as does deletion of the entire autoinhibitory region (Fig. 7c and Fig. 8) and because short C-terminal deletions downstream of the autoinhibitory α5 helix can also enhance ESCRT-III membrane binding and inhibition of HIV-1 budding, albeit to a lesser extent than deletions that include α517,20. These observations indicate that the terminal ESCRT-III helix 6 (and possibly also flap sequences) make energetically important contacts that favor the closed ESCRT-III conformation.
In summary, many ESCRT-III proteins can adopt two distinct conformational states6,16–21, and our studies provide a molecular model for the closed, monomeric state adopted by ESCRT-III proteins when they are distributed throughout the cytoplasm (or loosely associated with non-specific membranes)34. Primary sequence and secondary structure analyses indicate that all ESCRT-III proteins have analogous α5 helices in equivalent positions, and the mechanism of autoinhibition described here is probably quite general (see Supplementary Fig. 5a). Surrounding elements can vary significantly in sequence, length, and secondary structure, however, and likely mediate a variety of different protein-protein interactions33. Interestingly, these different protein-protein interactions can be enhanced, inhibited, or unchanged by mutations that favor the open states of various ESCRT-III proteins (e.g., refs. 21,22,28,33). Thus, changes in protein-protein and protein-membrane interactions that accompany ESCRT-III activation and assembly are likely to reflect the conformational changes described here, coupled with avidity effects created by the assembly of oligomeric ESCRT-III protein arrays.
METHODS
Expression vectors and antibodies
Vectors and antibodies are summarized in Supplementary Table 1 and Supplementary Table 2.
Protein expression and purification
We expressed IST1 and CHMP proteins in E. coli with N-terminal GST affinity tags. Protein expression, purification, and tag removal are described in the Supplemental Methods.
Equilibrium sedimentation analyses
We centrifuged purified CHMP3, CHMP38–222, and IST1NTD in 10 mM Tris-HCl pH 8.0, 100mM NaCl (CHMP3 proteins) or 50 mM Tris-HCl pH 7.0, 350 mM NaCl, and 1 mM DTT (IST1NTD) at 4°C in an Optima XL-A centrifuge (Beckman). Initial protein concentrations are given in Fig. 1a–c. Data were collected after equilibrium was obtained at two speeds (20,000 and 24,000 rpm) and the molecular masses were determined from the resulting six data sets for each protein by globally fitting to single ideal species models using the nonlinear least-squares algorithms in the Heteroanalysis software38. Protein partial specific volumes and solvent densities were calculated with the program SEDNTERP (version 1.09)39.
Protein crystallization and data collection
We crystallized IST1NTD, CHMP38–222 and CHMP31–150 by vapor diffusion at 13°C. IST1NTD crystals grew from a 12 mg ml−1solution in gel filtration buffer. Crystal form I (diamond-shaped morphology, P43212, one molecule per asymmetric unit (AU)) grew in 1–3 days by simple dehydration of a 2 µl drop. Crystals were cryoprotected by passage through silica oil, suspended in a nylon loop and plunged into liquid nitrogen. Crystal form II (rods/plates, P21, one molecule per AU) grew from a 1:1 v/v mixture of protein and well solution (180 mM KCl, 50 mM HEPES, pH 7.6, 35% pentaethritol propexylate and 7% v/v ethylene glycol). Crystals were cryocooled directly without solution modification. Native data for both crystal forms were collected on a Rigaku Micromax 007HF rotating anode generator equipped with VeriMax-HR optics.
CHMP38–222 crystals (flat plates, P21, two molecules per AU) grew from grew from a 1:2 v:v mixture of protein (30 mg ml−1 in gel filtration buffer) and well solution (10–22% PEG 6000 and 0.1M Na HEPES, pH 7.0–8.0). Data were collected at SSRL beamline 11−1. CHMP31–150 crystals (hexagonal rods, P61) grew from a 1:1 v/v mixture of protein (22mg ml−1 in gel filtration buffer) and well solution (16% v/v PEG 3350, 0.1 M Na HEPES, pH 7.0–8.0, 200 mM proline). Data were collected on a rotating anode generator. Despite extensive effort, we were unable to collect better data at the synchrotron. Data were indexed and scaled with the program HKL200040 (Table 1).
Structure determinations
Three-wavelength MAD data were collected from selenomethionine-substituted IST1NTD at SSRL beamline 9−1 to 3.4 Å. Three of the six potential selenium sites were identified using SOLVE41, and the resulting 4.0 Å phases were solvent flattened and extended to 2.6 Å resolution using the native data and the program DM42. The four core helices (α1–4) were built using O43, and this partial model was used for molecular replacement determination of the higher resolution form II crystal structure using PHASER44. The models were completed with COOT45, refined with REFMAC546, and analyzed using programs within the CCP4 suite47. The similar IST1NTD structures in the two crystal forms overlapped with a root mean square deviation (RMSD) of 1.25 Å on Cα atoms 12–170.
Both CHMP3 structures were determined by molecular replacement using the published CHMP38–183 structure (pdb code 2GD516) as the search model. The correctness of the solutions is supported by PHASER44 Z-scores of 9.1 (CHMP38–222) and 8.6 (CHMP31–150) and by the appearance of electron density when portions of the search model were omitted from the molecular replacement calculation (Supplementary Fig. 2). Both structures are hampered by low resolution and a correspondingly low number of unique reflections. Rfree sets of 5% contain too few reflections (~120) to be statistically useful. An optimal strategy was therefore determined by considering Rfree values from multiple refinement runs, each starting from the same molecular replacement solution and with a different small number (~200) of reflections in the test set. This indicated that the best results were obtained using rigid-body refinement in PHENIX35, with each helix treated as a rigid unit. This approach was therefore applied to the starting molecular replacement solution but using all of the reflections in the calculation (no test set).
Biosensor binding assays
We used BIACORE2000 and 3000 instruments (GE Healthcare) to measure the binding of pure recombinant CHMP1B to immobilized GST-IST1NTD proteins captured from E. coli extracts28 as described in Supplemental Methods.
IST1 depletion and rescue experiments
We transfected HeLa-M cells with IST1 expression vectors (1 µg/well, Lipofectamine 2000, Invitrogen) and siRNA (20nM, Lipofectamin RNAi MAX, Invitrogen) following the time course: t=0, seed cells in DMEM/10% FCS (six well plates, 4 × 105 cells per well (t=0 h); t=24 h, IST1 expression vector; t=32 h, media change + IST1 vector; t=44 h, trypsin treat and reseed on 18 mm glass cover slips for immunofluorescence analyses (3×104 cells per cover slip) and in 6 well plates for western blotting (3×105 cells per well); t=56 h, media change + siRNA; t=68 h, media change + siRNA; t=94 h, fix or harvest cells and analyze.
Direct visualization of cytokinesis defects
We fixed transfected HeLa-M cells with 4% paraformaldehyde and stained with SYTOX-Green (Invitrogen) and with mouse anti-α-Tubulin and a secondary Alexa 594-conjugated goat anti-mouse antibody. We scored midbody arrest manually for ≥ 3 blinded sets of 200 cells by direct visualization using fluorescence microscopy.
ESCRT-III assembly reactions
Assembly conditions for IST1NTD, CHMP1B and CHMP2A/CHMP3 proteins are provided in the Supplemental Methods.
Transmission electron microscopy
We analyzed assembly reactions using TEM with negative staining. Carbon-coated grids were placed on aliquots of each assembly reaction (7µl, 90 sec), washed with 3–4 drops of 0.1M KCl, stained with 3–4 drops of 4% uranyl acetate, air dried, and imaged (Hitachi 7100 TEM).
GST pulldown assays
We tested the binding of pure recombinant CHMP2A proteins to various GST-CHMP3 proteins as described in the Supplemental Methods.
HIV-1 vector release and infectivity
We co-transfected 293T cells with vectors expressing CHMP3-Myc and HIV-1 and harvested supernatants and cells after 24 h, Western blotting was used to analyze cellular proteins and virions pelleted through 20% sucrose cushion and HeLa-M cell transduction to measure vector titers. Full details are provided in Supplemental Methods.
Supplementary Material
Acknowledgements
Portions of this research were carried out at the Stanford Synchrotron Radiation Light Source, (SSRL) a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We thank Markus Babst for helpful discussions. This work was supported by NIH Grants AI051174 (WIS) and GM082545 (WIS and CPH).
Footnotes
Accession Codes. Protein Data Bank: Coordinates and structure factor amplitudes have been deposited for the two IST1NTD crystal forms (3FRR, 3FRS), CHMP8–222 (3FRT), and CHMP31–150 (3FRV).
REFERENCES
- 1.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 3.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajorek M, et al. Biochemical Analyses of Human IST1 and Its Function in Cytokinesis. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates vps4 localization and assembly. Mol Biol Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rue SM, Mattei S, Saksena S, Emr SD. Novel ist1-did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babst M, Katzmann D, Estepa-Sabal E, Meerloo T, Emr S. Escrt-III An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 16.Muziol T, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 18.Lata S, et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:816–825. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 20.Zamborlini A, et al. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieffer C, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 22.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci U S A. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang HT, et al. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Row PE, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Galan O, Galindo A, Hervas-Aguilar A, Arst HN, Penalva MA. Physiological involvement in pH signalling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J Biol Chem. 2008 doi: 10.1074/jbc.M808645200. epublication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yorikawa C, et al. Human calpain 7/PalBH associates with a subset of ESCRT-III-related proteins in its N-terminal region and partly localizes to endocytic membrane compartments. J Biochem. 2008;143:731–745. doi: 10.1093/jb/mvn030. [DOI] [PubMed] [Google Scholar]
- 27.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 29.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 30.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azmi IF, et al. ESCRT-III Family Members Stimulate Vps4 ATPase Activity Directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Xiao J, et al. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shim S, Merrill SA, Hanson PI. Novel Interactions of ESCRT-III with LIP5 and VPS4 and their Implications for ESCRT-III Disassembly. Mol Biol Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welsch S, et al. Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic. 2006;7:1551–1566. doi: 10.1111/j.1600-0854.2006.00489.x. [DOI] [PubMed] [Google Scholar]
- 35.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific LLC; 2008. [Google Scholar]
- 37.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. In: Rowe A, Horton J, editors. Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, England: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 40.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Data Collection and Processing. Warrington, UK: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 41.Terwilliger TC, Berendzen J. Automated structure solution for MIR and MAD. Acta Crystallographica. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 43.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Group CCP. The CCP4 Suite:Programs for Protein Crystallography. Acta Crystallogr D50. 1994 Nov 4;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.