SUMMARY
We describe a broad mechanistic framework for the transcriptional induction of mammalian primary response genes by Toll-like receptors and other stimuli. One major class of primary response genes is characterized by CpG-island promoters, which facilitate promiscuous induction from constitutively active chromatin without a requirement for SWI/SNF nucleosome remodeling complexes. The low nucleosome occupancy at promoters in this class can be attributed to the assembly of CpG islands into unstable nucleosomes, which may lead to SWI/SNF independence. Another major class consists of non-CpG-island promoters that assemble into stable nucleosomes, resulting in SWI/SNF dependence and a requirement for transcription factors that promote selective nucleosome remodeling. Some stimuli, including serum and tumor necrosis factor-α, exhibit a strong bias toward activation of SWI/SNF-independent CpG-island genes. In contrast, interferon-β is strongly biased toward SWI/SNF-dependent non-CpG-island genes. By activating a diverse set of transcription factors, Toll-like-receptors induce both classes and others for an optimal response to microbial pathogens.
INTRODUCTION
The availability of complete genome sequences for numerous species has enhanced interest in the organization and regulation of promoters, enhancers, and other DNA regions that control gene transcription in a physiological context. In mammals, promoters can be divided at their most basic level into the approximately 70% that contain CpG islands and the remaining 30% that lack CpG islands (Davuluri et al. 2001; Saxonov et al. 2006). CpG-island promoters are associated with most “housekeeping” genes and many regulated genes. Although CpG dinucleotides are substrates for DNA methyltransferases, most CpG islands are constitutively unmethylated in normal cells (Suzuki and Bird, 2008).
Another common property of promoters in mammals and other eukaryotes appears to be low nucleosome occupancy. In yeast, approximately 95% of promoters exhibit nucleosome deficits (Yuan et al., 2005; Mavrich et al., 2008b). Bioinformatic analyses suggest that reduced nucleosome stability due to a prevalence of rigid poly (dA:dT) sequences is responsible for this deficit (Iyer and Struhl, 1995; Anderson and Widom, 2001; Sekinger et al., 2005; Mavrich et al., 2008b), with regions flanking the promoters enriched in periodic AA/TT dinucleotides that favor stable nucleosome formation (Drew and Travers, 1985; Satchwell et al. 1986; Segal et al., 2006; Mavrich et al., 2008b). Yeast promoters that possess higher nucleosome occupancy are generally found in genes that exhibit greater plasticity of expression (Tirosh and Barkai, 2008; Mavrich et al., 2008b). Genome-wide studies have suggested that Drosophila and human promoters also exhibit reduced nucleosome occupancy (Heintzman et al., 2007; Ozsolak et al., 2007; Mavrich et al., 2008a; Schones et al., 2008), but the relevance of the nucleosome deficit in these organisms has not been examined.
A third common property of promoters is the pre-association of RNA polymerase II with inactive genes. Initial evidence of pre-association emerged from studies of Drosophila heat-shock promoters, the HIV-1 long terminal repeat, and the c-Myc promoter (Gilmour and Lis, 1986; Kao et al., 1987; Krumm et al., 1992). More recent studies have suggested that polymerase molecules are associated with a high percentage of genes that are generally considered to be inactive (Guenther et al., 2007).
Although some inducible promoters are associated with RNA polymerase prior to activation, other inducible model promoters assemble into stable nucleosomes. For example, at the S. cerevisiae PHO5 promoter, activation requires remodeling of promoter-associated nucleosomes by ATP-dependent remodeling complexes (Williams and Tyler, 2007; Boeger et al., 2008 and references therein). At the human IFNB promoter, the SWI/SNF remodeling complex catalyzes the sliding of a nucleosome spanning the TATA box and start site to a location further downstream, allowing pre-initiation complex assembly and transcription (Agalioti et al., 2000). At the inducible Il12b promoter, SWI/SNF-dependent remodeling coincides with increased accessibility of the promoter DNA, although a positioned nucleosome at the promoter does not slide and does not appear to be evicted (Weinmann et al. 1999; Ramirez-Carrozzi et al. 2006).
Although studies of model genes have revealed diverse mechanisms by which inducible transcription can be regulated in a chromatin context, general principles have remained elusive. For example, it is not known why CpG islands are found at some regulated genes, but more generally are associated with constitutively expressed genes. Moreover, the mechanistic and biological distinctions between inducible genes containing a pre-associated polymerase versus those assembled into stable nucleosomes prior to activation have not been established.
As an initial step toward an understanding of the diverse strategies used to regulate inducible transcription in mammalian cells, we previously used retroviral short hairpin RNAs (shRNA) to simultaneously knock down expression of Brg1 and Brm, the catalytic subunits of mammalian SWI/SNF remodeling complexes (Ramirez-Carrozzi et al., 2006). Brg1/Brm knockdown in murine macrophages followed by stimulation with lipopolysaccharide (LPS) through Toll-like receptor 4 (TLR4) revealed that only a subset of TLR4-induced genes require SWI/SNF complexes for activation. Almost all secondary response genes (i.e. genes requiring new protein synthesis for activation) exhibited strong SWI/SNF dependence, whereas primary response genes (i.e. genes activated in the absence of new protein synthesis) could be divided into SWI/SNF-dependent and -independent classes. The promoters of representative SWI/SNF-independent genes exhibited constitutively high accessibility to nucleases, whereas SWI/SNF-dependent promoters exhibited inducible accessibility and inducible association of Brg1. However, in this initial analysis, we were unable to identify features of the promoters that could explain why a specific subset could be activated in a SWI/SNF-independent manner.
To better understand the distinctions between SWI/SNF-dependent and SWI/SNF-independent inducible genes, we used microarrays to identify and quantitative RT-PCR (qRT-PCR) to validate a much larger set of genes that are strongly induced by TLR4 in murine macrophages. We mainly focused on primary response genes because of the expectation that secondary response genes would be regulated by a more diverse array of mechanisms. By identifying and characterizing defining features of different promoter classes, we obtained insight into the functional and mechanistic distinctions between inducible CpG-island and non-CpG-island promoters, SWI/SNF-independent and SWI/SNF-dependent promoters, and promiscuous and tightly regulated inducible genes. The resulting model explains the variable properties of mammalian genes induced by a wide range of stimuli.
RESULTS
Prevalence of CpG-Island Promoters at SWI/SNF-Independent Primary Response Genes
To understand the distinctions between SWI/SNF-independent and -dependent genes, we used microarrays to expand our set of TLR4-induced genes, with an emphasis on primary response genes. Fifty-five primary response genes were validated using qRT-PCR with mRNA from mouse bone marrow-derived macrophages stimulated with LPS in the presence and absence of the protein synthesis inhibitor cycloheximide (CHX) (Figure 1A and data not shown). Twelve secondary response genes were also included in our analyses. qRT-PCR analyses of these 67 genes rather than microarrays were used for all subsequent expression studies.
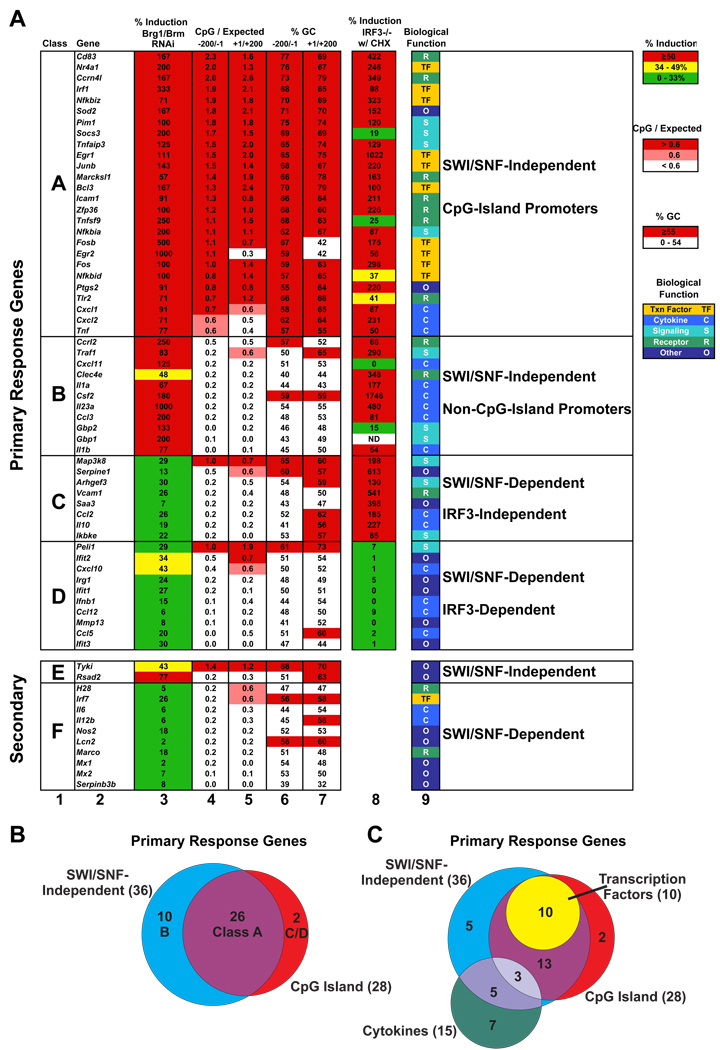
Figure 1. Classification of LPS-Induced Primary- and Secondary-Response Genes.
(A) 67 genes that are potently induced by LPS in mouse bone marrow-derived macrophages are shown. Classes A–D are primary response genes (resistant to CHX) and Classes E and F are secondary response genes (sensitive to CHX). Column 3 shows the effect of Brg1/Brm knockdown on LPS-induced mRNA levels as a percentage of the mRNA level observed in control cells (set at 100% for each gene), as determined by qRT-PCR. Column 8 shows mRNA levels in IRF3−/− macrophages stimulated with LPS in the presence of CHX as a percentage of mRNA levels in LPS-stimulated wild-type C57BL/6 macrophages, as determined by qRT-PCR. In columns 3 and 8, percentages represent the average of three independent experiments. Columns 4 and 5 show the ratio of the number of observed CpGs to the number expected if CpGs were randomly distributed, for the regions from −200 to −1 (column 4) and +1 to +200 (column 5) relative to the start site indicated in the DBTSS database. Columns 6 and 7 show percentages of GC bps in these same regions. Column 9 shows the established or predicted functions of the 67 genes. Color-coded legends for columns 3 through 9 are shown at the right.
(B) A Venn diagram shows that 26 of 28 primary response genes containing CpG-island promoters are induced in a SWI/SNF-independent manner.
(C) A Venn diagram shows that all 10 primary response genes encoding transcription factors are contained within Class A, whereas only 3 of 15 cytokine genes are found in this class.
The SWI/SNF dependence of each of the 67 genes was determined by simultaneous Brg1/Brm knockdown in LPS-stimulated J774 macrophages as previously described (Ramirez-Carrozzi et al., 2006), using retroviral delivery of an shRNA that targets a conserved region of the Brg1 and Brm mRNAs (see Suppl. Figure 1). qRT-PCR revealed that the effect of Brg1/Brm knockdown on mRNA levels was highly variable (Figure 1A, column 3). mRNA levels of 16 of the 55 primary response genes (29%) were reduced by at least 3-fold (Figure 1A, column 3, green). We refer to these genes as SWI/SNF-dependent. mRNA levels for 36 others (65%) were reduced by less than 2-fold or were increased relative to the control (Figure 1A, column 3, red). We refer to these genes as SWI/SNF-independent. The mRNA levels for the remaining three genes were reduced by more than 2-fold and less than 3-fold (Figure 1A, column 3, yellow). The moderate effects make these genes difficult to classify. Among the 12 secondary response genes, 10 were SWI/SNF-dependent, one was SWI/SNF-independent, and one was in the intermediate group (Figure 1A, Classes E and F).
It is noteworthy that, in our previous study, SWI/SNF-independent genes were generally induced more rapidly than SWI/SNF-dependent genes (Ramirez-Carrozzi et al., 2006). A similar trend was observed with this larger set of genes (see Figure 5A), but we no longer include activation kinetics in our classification scheme because several exceptions were observed and because the precise activation kinetics for some genes varied from experiment to experiment.
Figure 5. Preferential Activation of CpG-Island and Non-CpG-Island Genes by TNFα and IFNβ.
(A) Bone marrow-derived macrophages were left unstimulated or were stimulated for 30 min, 1 hr, or 2 hrs with stimuli for TLR2, TLR3, or TLR4, or with IFNβ or TNFα. mRNA levels for 61 of the 67 genes shown in Figure 1 were monitored by qRT-PCR. mRNA levels are presented as a percentage of the highest level observed at any of the time points by any of the stimuli (set at 100%). Values represent an average of three independent experiments (i.e. independent stimulations of independent macrophage preparations). mRNA levels of at least 15% of the maximum were colored red (> 50%), orange (33–49%), or yellow (15–32%). CpG numbers, Brg1/Brm-dependence, and IRF3-dependence were derived from Figure 1.
(B) A Venn diagram shows that TNFα preferentially induced a high percentage of CpG-island genes (mostly in Class A), whereas IFNβ preferentially induced non-CpG-island genes (mostly in Classes C and D).
(C) The number of genes within each of the 6 classes that were induced or were not induced by IFNβ and TNFα are depicted in a bar graph. Uninduced genes were defined as those induced to a level below 15% of the maximum induction by any of the 5 stimuli shown in panel A.
The sequences of the SWI/SNF-dependent and -independent promoters were compared to identify distinguishing features. Remarkably, 26 of the 36 SWI/SNF-independent primary response genes (72%, including only the 36 primary response genes in red in Figure 1A, column 3) contain CpG islands between −1 and −200 relative to the major start site (Figure 1A, column 4; see also Figure 1B). In contrast, CpG islands were observed in only 2 of the 16 (12.5%) SWI/SNF-dependent primary response genes. Figure 1A, columns 3 and 4 show the CpG content for the regions from −200 to −1 and from +1 to +200 (relative to the major start site reported in the DBTSS database). CpG content is indicated as the ratio of observed CpGs to the CpGs expected if this dinucleotide were randomly represented in the genome. Because CpG dinucleotides have been depleted from mammalian genomes, this ratio is generally low (0.1–0.2). CpG islands have been defined as regions containing ratios greater than or equal to 0.55, 0.60, or 0.65 (Gardiner-Garden and Frommer, 1987; Davuluri et al., 2001; Takai and Jones, 2002; Saxonov et al., 2006). For this analysis, we used the intermediate value. The overall percentage of GC bps is also shown in Figure 1A (columns 6 and 7).
Because CpG-island promoters are often found in housekeeping genes, we asked whether TLR4-induced genes containing CpG islands might be transcribed at a higher level than non-CpG-island genes in unstimulated cells. Precursor transcript levels for 30 genes were monitored by qRT-PCR in unstimulated and LPS-stimulated bone marrow-derived macrophages, using primer pairs in which one primer annealed to exonic sequences and the other to an intronic sequence. Precursor transcript levels are thought to reflect transcription rates more accurately than mRNA levels. After normalization of the RT-PCR efficiency for each gene using genomic DNA, a wide range of precursor transcript levels was observed in two independent experiments in unstimulated macrophages, with transcript levels spanning approximately four orders of magnitude (Suppl. Figure 2). The number of primary transcripts was, on average, slightly higher for CpG-island genes than non-CpG-island genes, raising the possibility that the higher basal transcription levels may contribute to the SWI/SNF-independent induction of CpG-island genes, or, alternatively, may be a consequence of their capacity for SWI/SNF-independent induction. However, no consistent trend was observed, as some SWI/SNF-dependent non-CpG-island genes exhibited basal transcript levels comparable to those observed at SWI/SNF-independent CpG-island genes. Importantly, precursor transcript levels increased two orders of magnitude or more upon LPS stimulation for most of the genes in both classes, with no consistent differences between the two classes (Suppl. Figure 2). Thus, the existence of basal transcripts and basal transcript levels cannot explain the distinction between SWI/SNF-independence and -dependence.
Assembly of CpG-Island Promoters into Constitutively Active Chromatin
To understand why TLR4 target genes containing CpG islands are almost always SWI/SNF-independent, ChIP was used to analyze chromatin at TLR4 target genes in unstimulated bone marrow-derived macrophages. To compare ChIP signals at the various promoters, primer amplification efficiencies were normalized using genomic DNA. Two housekeeping genes, Actb and Gapd, were included for the purpose of comparison. When examining total histone H3 levels, a significant but imperfect trend toward lower histone occupancy at CpG-island promoters was observed (Figure 2, top; p<0.002). Despite the reduced histone H3 levels at a large fraction of CpG-island promoters, a striking trend toward higher histone H3K9/K14 acetylation and H3K4 trimethylation levels was observed at these promoters (Figure 2). Thus, inducible CpG-island promoters appear to be assembled into chromatin containing modifications characteristic of active genes.
Figure 2. Constitutively Active Chromatin is Preferentially Found at LPS-Induced CpG-Island Promoters.
ChIP was used to monitor chromatin structure at 37 LPS-induced genes and two housekeeping genes (Gapd and Actb) in unstimulated bone marrow-derived macrophages. Genes containing CpG-island and non-CpG-island promoters are in red and black, respectively. Antibodies against unmodified histone H3, H3K9/K14ac, H3K4me3, RNA polymerase II, and TBP were examined. PCR primer pairs were normalized using genomic DNA. Normalized results are shown as a percentage of input values. Higher values were obtained with the modified histone antibodies than with the unmodified histone antibodies due to different antibody qualities. The results are averages of 3 independent experiments performed with independent chromatin preparations, with standard deviations. P-values for the differences between CpG-island and non-CpG-island promoters were: histone H3, p<0.002; H3K9/14ac, p<0.001; H3K4me3, p<0.00004; RNA polymerase II, p<0.002; and TBP, p<0.001.
Most CpG-island promoters also exhibited higher levels of RNA polymerase II and TBP in unstimulated macrophages. Although association of RNA polymerase II with the inducible promoters in unstimulated cells is consistent with the existence of basal transcripts, RNA polymerase II levels did not increase or increased to only a modest extent at several of the CpG-island promoters following LPS stimulation (Suppl. Figure 3), despite increases in precursor transcript levels often exceeding 100-fold. These properties are reminiscent of those observed at Drosophila heat-shock promoters (Gilmour and Lis, 1986). It is important to emphasize, however, that the existence of significant basal transcription suggests that polymerases at the CpG-island promoters are not retained in the rigidly poised, pre-initiated state observed at Drosophila heat-shock promoters. Nevertheless, our findings suggest that LPS induction leads to greatly enhanced initiation and/or elongation by polymerase molecules that can readily associate with many of the CpG-island promoters in unstimulated cells. Further studies are needed to determine the precise mechanisms by which initiation and elongation are regulated at these genes.
The trend toward lower histone H3 levels at CpG-island promoters is interesting to consider in light of previous genome-wide studies which suggested that low nucleosome occupancy characterizes active and sometimes inactive promoters in mammalian cells (see Introduction). To determine whether similarly low histone H3 levels are found at both CpG-island and non-CpG-island promoters when they are active, ChIP experiments were performed with macrophages after LPS stimulation for 30 or 120 min. Significant decreases in ChIP signals were observed at some genes after stimulation, but histone H3 levels at several of the non-CpG-island promoters remained high (Suppl. Figure 4). This finding is consistent with our previous evidence that a positioned nucleosome at the non-CpG-island Il12b promoter becomes more accessible to nuclease cleavage, but is not evicted, upon transcriptional activation (Weinmann et al., 1999; Ramirez-Carrozzi et al., 2006). The results suggest that reduced nucleosome occupancy may primarily characterize CpG-island promoters and a limited subset of active non-CpG-island promoters from which nucleosomes have been evicted. Furthermore, the continuum of histone H3 levels observed in our analysis (Figure 2) suggests that CpG-island promoters possess nucleosome densities that are reduced to variable degrees.
Strong Constitutive DNase I Hypersensitivity at Inducible CpG-Island Promoters in Human CD4+ T Cells
To understand why CpG-island promoters often exhibit lower histone H3 levels than non-CpG-island promoters, we first hypothesized that the binding of a specific transcription factor, such as Sp1, is responsible for nucleosome loss. Indeed, constitutive Sp1 binding is detectable at many of the Class A promoters in ChIP experiments (data not shown; D.C. Hargreaves and R. Medzhitov, personal communication). However, consensus Sp1 sites are also found in some of the non-CpG-island promoters that exhibit high nucleosome occupancy (data not shown). This observation led us to consider the possibility that the full CpG-island sequence, rather than isolated transcription factor binding sites, might be responsible for the low nucleosome occupancy, analogous to the role of poly (dA:dT) tracts at yeast promoters (see Introduction). Initial support for this hypothesis was provided by previous studies which defined sequences that favor or disfavor nucleosome assembly (e.g. Drew and Travers, 1985; Satchwell et al., 1986; Segal et al., 2006; Mavrich et al., 2008b). In fact, using the computational tools of Segal et al. (2006), virtually all CpG-islands in our promoter-set are predicted to be devoid of stable nucleosomes (data not shown). Although CpG islands contain the GC-rich sequences whose minor grooves are often located at the exposed surface of stable nucleosomes, the periodic AA/TT dinucleotides that favor DNA bending and stable nucleosome assembly are usually absent.
Although the ruled defined by Segal et al. (2006) predict that CpG-island promoters are incompatible with stable nucleosome assembly, the ChIP results (Figure 2) suggest that nucleosome occupancy is variable, despite a significant trend toward low occupancy at CpG-island promoters. One possibility is that nucleosome instability does not always lead to a nucleosome deficit. As an independent strategy for comparing the physical state of nucleosomes at inducible CpG-island versus non-CpG-island promoters in vivo, we examined published data that identified DNase I hypersensitive sites at a genome-wide level in quiescent human CD4+ T cells (Boyle et al., 2008).
Strikingly, the human homologues of 18 of our 26 (69%) Class A genes exhibited high hypersensitivity scores in resting T cells, whereas only 3 of the 35 (9%) non-CpG-island genes in the other classes exhibited comparable hypersensitivity scores (Suppl. Figure 5). Furthermore, none of the 7 most strongly induced non-CpG-island genes in T cells exhibited high hypersensitivity scores, and only two of these 7 genes exhibited detectable hypersensitivity (Suppl. Figure 5). Published expression profiles from human CD4+ T cells revealed that at least 9 of the Class A genes are induced in CD4+ T cells by CD3 and CD28 antibodies (Suppl. Figure 5); this number almost certainly represents an underestimate because induction was monitored only at relatively late time points. In sum, these results, obtained with a different cell type and using a different assay, provide further evidence that nucleosomes associated with inducible CpG-island promoters are structurally different than nucleosomes associated with non-CpG-island promoters in unstimulated cells.
Reduced Assembly of CpG-Island Promoters into Nucleosomes In Vitro
Although the above results suggest that nucleosomes at CpG-island promoters may be unstable, perhaps contributing to their SWI/SNF-independent activation, in vivo studies cannot distinguish between intrinsic instability due to nucleotide content and reduced nucleosome occupancy due to the activities of constitutively associated transcription factors. Therefore, we compared intrinsic nucleosome stabilities at CpG-island and non-CpG-island promoters using an in vitro nucleosome assembly/solution competition assay that makes use of purified recombinant histone octamers from Xenopus laevis (Figure 3A). Pools of 300-bp DNA fragments spanning 27 CpG-island and non-CpG-island promoters were mixed and assembled into nucleosomes using limiting concentrations of recombinant histone octamers. High-affinity promoters were isolated from the nucleosomal band obtained with reactions in which 10% of the promoter fragments were assembled; low-affinity fragments were isolated from the “free” band obtained in reactions in which 80–90% of the fragments were assembled (Figure 3A). The fragments were PCR amplified using common primers, and were again subjected to nucleosome assembly and EMSA. After each round of assembly, EMSA, and fragment elution, the fraction of each DNA fragment present in the assembled and free DNA pools was quantified by qPCR.
Figure 3. CpG-Island Promoters Compete Less Effectively than Non-CpG-Island Promoters for Nucleosome Assembly In Vitro.
(A) A sequential assembly and amplification assay was used to compare the stabilities of nucleosomes assembled on CpG-island and non-CpG-island promoters. 300-bp DNA fragments were pooled from 23 LPS-induced promoters, 3 housekeeping promoters (Gapd, Actb, and Dhfr), and a synthetic DNA fragment previously shown to assemble into unusually stable nucleosomes (601; Lowary and Widom, 1998). After assembly into nucleosomes with recombinant histones and separation of nucleosomal fragments from free fragments by gel shift, the nucleosomal and free fragments were isolated. A portion of each resulting pool was re-assembled, with another portion used for qPCR to determine the relative amount of each DNA fragment in each pool. Four rounds of assembly, elution, and amplification were performed.
(B) The ratio of each promoter fragment found in the nucleosomal (bound) band to the free band in the gel shift experiments after each assembly and elution cycle is shown. CpG-island promoters are in red and non-CpG-island promoters in black. The Cxcl10 fragment used for this analysis is depicted as a CpG-island, although the Cxcl10 promoter from −1 to −200 contains an observed:expected CpG ratio of only 0.4 (Figure 1). The reason for this difference is that the 300-bp fragment used for in vitro assembly extends into the CpG-rich transcribed region (−161/+139) and, with the adaptor, possesses a CpG ratio of 0.7. The P-value for the difference between CpG-island and non-CpG-island promoters is p<0.01.
After four rounds of selection, a clear difference in the competition for nucleosome assembly was observed, with non-CpG-island sequences competing much more successfully than CpG-island sequences (Figure 3B). It is important to note that a DNA sequence referred to as 601 was used as a control in this experiment. This sequence was previously selected on the basis of its ability to assemble into unusually stable nucleosomes (Lowary and Widom, 1997). Consistent with the previous data, the 601 sequence exhibited greater enrichment in the nucleosomal fraction than any of the native promoters. Interestingly, the 601 sequence conforms to the definition of a CpG island. However, unlike the native CpG-island promoters, it contains properly phased AT-bps to promote the assembly of stable nucleosomes (Lowary and Widom, 1997).
These results provide strong support for a model in which the reduced nucleosome occupancy and enhanced accessibility observed at CpG-island promoters in vivo are largely due to the reduced stability of nucleosomes at these promoters, as a direct result of their nucleotide content. We hypothesize that the reduced nucleosome stability is responsible, at least in part, for the SWI/SNF-independent activation of these genes. Importantly, this hypothesis is consistent with well-established evidence that nucleosome destabilization in S. cerevisiae Sin mutants can result in SWI/SNF-independent activation of genes that normally are SWI/SNF dependent (Muthurajan et al., 2004 and references therein).
It is important to note that, although assembly into unstable nucleosomes may play a major role in the reduced nucleosome occupancy, constitutive DNase I hypersensitivity, and SWI/SNF-independence of inducible CpG-island promoters, instrinsic nucleosome instability is unlikely to be sufficient for constitutive histone acetylation and H3K4 trimethylation at these promoters. Most likely, the active chromatin state that characterizes CpG-island promoters benefits from both intrinsic nucleosome instability and the pre-association of transcription factors like Sp1.
Class B Promoters Exhibit SWI/SNF Independence without a CpG Island
Although 26 of 36 LPS-induced, SWI/SNF-independent primary response genes contain CpG-island promoters (Figure 1A, Class A), the remaining 10 do not have a high CpG-content between −1 and −200. These SWI/SNF-independent, non-CpG-island genes were placed in Class B, along with a gene with an ambiguous SWI/SNF-dependence (Figure 1A). ChIP data for four Class B genes (Traf1, Csf2, Il23a, and Il1b) are included in Figure 2, revealing an absence of constitutively active chromatin. Furthermore, stable nucleosomes readily assembled in vitro at the two Class B promoters examined (Figure 3; Il1b and Traf1). This finding is consistent with the prediction that stable nucleosomes can readily assemble on all Class B promoters using the computation tools of Segal et al. (2006). Thus, the reason Class B genes are activated in a SWI/SNF-independent manner will require further investigation (see Discussion).
Most Primary Response Genes that Require IRF3 for Activation are SWI/SNF-Dependent
Although most LPS-induced primary response genes were SWI/SNF-independent, 29% (16 of 55) exhibited substantial SWI/SNF-dependence, with all but two of these genes lacking CpG-island promoters. Notably, several of these genes are known to require interferon regulatory factor 3 (IRF3) for activation in LPS-stimulated macrophages (Doyle et al., 2002). IRF3 activity is induced by a select subset of TLRs, including TLR4, in contrast to NF-κB and AP-1, whose activities are induced by all TLRs (Kawai and Akira, 2007). An analysis of mRNA levels of all 67 genes in LPS-stimulated macrophages from IRF3−/− mice (in the presence of CHX to eliminate redundancy due to factors like IRF7 that are newly synthesized in response to LPS) revealed strong IRF3-dependent expression of 50% (8 of 16) of the SWI/SNF-dependent primary response genes (Figure 1A, column 8). These genes were placed in Class D, along with two additional genes (Ifit2 and Cxcl10) that exhibited intermediate SWI/SNF dependence (Figure 1A). Importantly, mRNA levels for only 4 of the 36 SWI/SNF-independent primary response genes were reduced by more than 3-fold in IRF3−/− macrophages, and three of these genes remained strongly induced (Figure 1A and data not shown). Thus, genes that are dependent on IRF3 activity for expression in LPS-stimulated macrophages are generally SWI/SNF-dependent.
The strong IRF3 dependence in the presence of CHX suggests that the Class D genes are direct targets of IRF3. Consistent with this hypothesis, consensus IRF3 binding sites were readily observed in 6 of the 10 Class D promoters, but in only 6 of the 57 promoters in the remaining classes (Suppl. Figure 6). In addition, ChIP experiments confirmed that IRF3 can directly associate with the promoters of representative Class D genes (Suppl. Figure 6).
Biological Classification of SWI/SNF-Dependent and -Independent Genes
The finding that IRF3-dependent primary response genes generally contain non-CpG-island promoters and are SWI/SNF-dependent suggests that these promoter properties are primarily used to restrict transcriptional activation of genes that require tight regulation. In contrast, genes that are induced by a wide range of stimuli may be more compatible with CpG-island promoters and SWI/SNF independence.
An examination of the biological functions of our set of LPS-induced genes provides additional support for this model. All 10 genes that encode transcriptional regulators among the 55 primary response genes are found within Class A (Figure 1A, column 9; Figure 1C). Most of these transcription-factor genes, including Egr1, Egr2, Junb, Fos, Fosb, and Bcl3, are known to be induced by diverse stimuli (Herschman, 1991). In contrast, only 3 of the 15 genes encoding cytokines, which are induced more selectively, are found in Class A (Figures 1A and 1C). These findings suggest that CpG-island SWI/SNF-independent promoters are often associated with promiscuous activation, and that non-CpG-island SWI/SNF-dependent promoters correlate with selective activation. It is noteworth that Class B consists primarily of cytokine genes that require selective regulation, despite the SWI/SNF independence of this class.
IRF3 is Required for Nucleosome Remodeling at IRF3-Dependent Genes
To explore the relationship between SWI/SNF dependence and IRF3, a restriction enzyme accessibility/Southern blot assay was used to monitor nucleosome remodeling at two IRF3-dependent genes, Ccl5 and Ifit1. Like the mRNA analysis, this analysis was performed in cells stimulated with LPS in the presence of CHX, which eliminates the secondary activation of the interferon pathway that partially compensates for the loss of IRF3. In wild-type macrophages, a strong increase in restriction enzyme cleavage was observed in stimulated cells at both the Ccl5 and Ifit1 promoters (Figure 4B and 4C, lanes 1 and 2). This inducible cleavage was greatly reduced in IRF3−/− macrophages (Figure 4B and 4C, lanes 3 and 4). The strong dependence of nuclease accessibility on IRF3 supports the notion that the assembly of these promoters into stable nucleosomes confers a requirement for remodeling by SWI/SNF complexes, with remodeling dependent on a specialized TLR4-activated factor, IRF3.
Figure 4. IRF3 is Required for Nucleosome Remodeling at Class D Promoters.
(A) Macrophages from C57BL/6 mice and IRF3−/− mice were stimulated with LPS in the presence of CHX. mRNA levels for the Ccl5 and Ifit1 genes were strongly reduced in the IRF3−/− cells.
(B) Restriction enzyme accessibility at the Ccl5 promoter was monitored using a Southern blot assay. Results are shown from three independent experiments, with the average percentage of alleles cleaved in the nuclei shown in the bar graph. The larger DNA fragment (*) results from cleavage of the purified genomic DNA by EcoRI and HindIII, which cleave sites flanking the Ccl5 promoter. The smaller fragment (arrow) was generated when EcoNI, which was added to the isolated nuclei, cleaved within the Ccl5 promoter.
(C) Restriction enzyme accessibility was monitored at the Ifit1 promoter, as described above for the Ccl5 promoter. Results from two independent experiments are shown. DraIII was used for digestion of purified DNA at sites flanking the Ifit1 promoter, with DraI used for digestion of nuclear DNA within the Ifit1 promoter.
A fourth class of primary response genes, Class C, includes SWI/SNF-dependent genes that do not require IRF3 for expression (Figure 1A). We hypothesize that one or more specialized LPS-induced transcription factors other than IRF3 promote nucleosome remodeling at promoters within this class, contributing to their selective activation.
Preferential Activation of SWI/SNF-Dependent Versus SWI/SNF-Independent Genes by Other Stimuli
To examine the broader significance of the distinction between SWI/SNF-independent CpG-island and SWI/SNF-dependent non-CpG-island primary response genes, we analyzed the 67 genes after stimulating bone marrow-derived macrophages with other inducers, including peptidoglycan (TLR2), poly I.C (TLR3), IFNβ, and TNFα. The mRNA levels for each gene at three different time points in response to each stimulus are presented as a percentage of the maximum level of induction by any of the stimuli (100%) (Figure 5; see also Suppl. Figure 7).
Striking differences were found in the preferences of some stimuli for SWI/SNF-independent versus SWI/SNF-dependent genes. Of particular relevance, TNFα induction was strongly biased toward Class A genes. TNFα stimulated 23 of the 24 Class A genes to a level that was at least 15% of the maximum induction (Figure 5A). However, only 9 of the remaining 37 genes were activated to this level, with these 9 genes scattered among the other classes (Figure 5A–C). This finding is consistent with the fact that TNFα signaling does not induce IRF3 and suggests that TNFα may not directly induce any other transcription factors that can promote efficient nucleosome remodeling in macrophages, thereby restricting strong activation to SWI/SNF-independent primary response genes. We cannot exclude the possibility that TNFα activates a distinct set of SWI/SNF-dependent non-CpG-island primary response genes via transcription factors that differ from those induced by LPS. However, independent microarray studies of fetal-liver derived macrophages activated with TNFα failed to reveal a compelling set of non-CpG-island primary response genes (C.C. and A.H., unpublished results).
In striking contrast to the preferential induction of Class A genes by TNFα, IFNβ exhibited a strong preference for SWI/SNF-dependent genes in Classes C and D (Figure 5A–C). This finding is consistent with the view that IFNβ induces transcription via IRF proteins and STAT proteins; both of these protein families have been suggested to promote nucleosome remodeling by SWI/SNF complexes (see Figure 4 and Liu et al., 2002; Huang et al., 2002; Cui et al., 2004). Therefore, IFN-induced factors appear to be well-suited for the selective activation of SWI/SNF-dependent genes assembled into stable nucleosomes, with no need for constitutively active chromatin or a CpG island.
Although TNFα and IFNβ exhibited strong preferences, TLR2 and TLR3 signaling resulted in the induction of nearly all genes induced by TLR4. The only clear difference was that TLR2 signaling failed to induce the IRF3-dependent genes in Class D, as well as some secondary response genes dependent on IFN signaling, consistent with knowledge that TLR2 signaling does not activate IRF3 (Kawai and Akira, 2007).
Further support for the hypothesis that some stimuli preferentially induce SWI/SNF-independent CpG-island genes during a primary response, perhaps due to the inability of these stimuli to activate transcription factors capable of promoting nucleosome remodeling, was provided from a literature analysis of well-documented primary response genes induced by serum and the tumor promoter TPA. Collections of bona fide primary response genes induced by these stimuli were compiled by Herschman (1991) before promoter sequences for most genes were available. Remarkably, every serum- and TPA-induced gene compiled by Herschman (1991) contains a CpG-island promoter (Figures 6A and 6B). Independent microarray experiments failed to uncover any non-CpG-island genes that are potently induced during the primary response to serum in serum-starved NIH 3T3 cells (data not shown). In contrast, 74% of primary response genes induced by IFNβ by at least 5-fold in real-time RT-PCR experiments lacked CpG islands between −200 and −1 (Figure 6C).
Figure 6. Differential Induction of CpG-Island Versus Non-CpG-Island Genes.
(A) A collection of well-characterized primary response genes induced by serum is shown, along with the CpG-content and GC-content of their promoters. The list includes every serum-induced gene described in Herschman (1991).
(B) A collection of well-characterized primary response genes induced by TPA is shown, along with the CpG-content and GC-content of their promoters. Every TPA-induced gene described in Herschman (1991) is included.
(C) A set of primary response genes induced by IFNβ in mouse bone marrow-derived macrophages is shown. The list includes all genes from the set of 67 LPS-induced genes that were induced by IFNβ by at least 5-fold in qRT-PCR experiments.
Cell-Type-Specific Classification of an LPS-Induced Gene
Finally, an analysis of gene induction in primary mouse embryonic fibroblasts (MEFs) demonstrated that genes induced by a given stimulus can be assigned to different classes in different cell types. This fundamental property was revealed through an analysis of the Il6 gene. In LPS-stimulated macrophages, Il6 is a SWI/SNF-dependent secondary response gene (see Figure 7A, Figure 1, and Ramirez-Carrozzi et al. 2006). In contrast, Il6 was induced in a protein synthesis-independent, SWI/SNF-independent manner in primary MEFs (Figure 7A and 7C). Interestingly, a restriction enzyme accessibility analysis revealed that the Il6 promoter is highly accessible in unstimulated MEFs, with little change following stimulation, in contrast to its inducible accessibility in macrophages (Figure 7B). Thus, despite the assignment of Il6 to secondary response Class F in macrophages, its properties are more appropriate for primary response Class B in MEFs. This dramatic change appears to be unusual, as none of the other Class F secondary response genes exhibited properties of a primary response gene in MEFs (data not shown; see Discussion).
Figure 7. Il6 is SWI/SNF-Independent in LPS-Stimulated MEFs.
(A) Il6 mRNA levels were monitored by qRT-PCR in J774 macrophages or primary MEFs following stimulation with LPS in the presence of CHX or in the presence of the DMSO solvent. Results shown are averages of three independent experiments, with standard deviations. The CHX-sensitivity observed in the J774 line was also observed in primary bone marrow-derived macrophages (Ramirez-Carrozzi et al., 2006).
(B) Restriction enzyme accessibility at the Il6 promoter was examined in J774 macrophages and primary MEFs as described (Ramirez-Carrozzi et al., 2006). Cells were left unstimulated or were stimulated for different time periods. Cells were also stimulated for 120 min in the presence of CHX.
(C) An shRNA that simultaneously targets the Brg1 and Brm mRNAs for degradation was introduced into primary MEFs using a retroviral vector (Ramirez-Carrozzi et al., 2006). Efficient knockdown of Brg1 and Brm was monitored by Western blot (data not shown). Cells were stimulated with LPS and Il6 mRNA levels were monitored by qRT-PCR. Results represent averages of three independent experiments.
DISCUSSION
We have provided a framework for understanding the relationship between CpG islands, nucleosome remodeling, and nucleosome stability during inducible gene transcription. CpG-island promoters were generally associated with primary response genes induced by a broad range of stimuli in a SWI/SNF-independent manner. The high CpG-content appeared to be responsible for promoter assembly into unstable nucleosomes, which may directly contribute to the SWI/SNF independence, analogous to the relationship between nucleosome instability and SWI/SNF independence in S. cerevisiae Sin mutants (Muthurajan et al., 2004). In striking contrast, SWI/SNF-dependent genes lacked CpG-island promoters and assembled into stable nucleosomes. Assembly into stable nucleosomes conferred the capacity for tight regulation, with activation dependent on specialized transcription factors that promote nucleosome remodeling.
We hypothesize that, during the evolution of some genomes, CpG islands provided an attractive platform for promoters of constitutive and broadly induced genes for two reasons. First, the instability of nucleosomes assembled on CpG islands facilitated constitutive expression and rapid induction without an energy requirement for nucleosome remodeling or a requirement for factors that can promote remodeling. Second, CpG-island promoters contained binding sites for ubiquitous factors like Sp1, which are likely to facilitate the establishment of constitutively active chromatin. This dual benefit may have provided selective pressure that contributed to the maintenance of CpG-island promoters through evolution.
The striking differences in the properties of promoters induced by different stimuli have broad biological relevance. Many CpG-island SWI/SNF-independent genes are activated by “generic” signaling pathways, such as NF-κB and MAP kinase pathways, which are targeted by a large number of growth factors, cytokines, and microbial stimuli. The transcription factors induced by these pathways may not readily promote nucleosome remodeling and may be well-suited for the activation of promiscuously induced genes. In contrast, IFNβ, which is known to activate genes with highly specialized functions, preferentially targets non-CpG-island SWI/SNF-dependent genes. The activation of these genes is restricted by the assembly of their promoters into stable nucleosomes.
In addition to facilitating highly selective activation, a second potential benefit of promoter assembly into stable nucleosomes may be to help minimize basal transcription, thereby preventing synthesis of gene products that may be detrimental to the cell when constitutively present at low levels. The higher basal transcription levels observed with some CpG island genes may be less detrimental and perhaps of some benefit. However, some of these genes are likely to be regulated at the level of mRNA stability (data not shown), allowing little expression of their gene products in quiescent macrophages, despite substantial precursor transcript levels.
It is noteworthy that the SWI/SNF-independent activation of many genes suggests that these genes do not contain distant enhancers that require SWI/SNF-dependent remodeling. Perhaps, SWI/SNF-independent primary response genes do not require distant enhancers at all for their activation. Alternatively, the enhancers for these genes may be constitutively active. It is also important to consider the possibility that other ATP-dependent nucleosome remodeling complexes may contribute to remodeling at enhancers for these genes.
Previous studies have suggested that reduced nucleosome occupancy may be a general property of mammalian promoters (Heintzman et al., 2007; Ozsolak et al., 2007; Schones et al., 2008). We propose that nucleosome occupancy is reduced to variable degrees at CpG-island promoters as a result of the destabilizing effect of the CpG-island sequence, with nucleosomes evicted from a subset of non-CpG-island promoters during transcriptional activation. The role of CpG islands in generating a nucleosome deficit appears analogous to the role of poly (dA:dT) tracts at S. cerevisiae promoters (Iyer and Struhl, 1995; Mavrich et al., 2008b). However, the precise role of CpG island-induced nucleosome instability in conferring SWI/SNF-independence awaits studies to determine whether a SWI/SNF-dependent promoter can be converted to a SWI/SNF-independent promoter by destabilizing nucleosomes through changes in the DNA sequence. Thus far, our efforts to achieve this goal have been unsuccessful, due to the challenge of altering promoter sequences to a sufficient extent to destabilize nucleosomes without disrupting or introducing binding sites for specific transcription factors.
Although the assembly of CpG-island promoters into unstable nucleosomes may contribute to their SWI/SNF-independence, these promoters possess other features of transcriptionally active chromatin in unstimulated cells. Unstable nucleosomes may be intrinsically susceptible to acetylation and methylation in the absence of transcription factor targeting. However, a more likely scenario is that constitutively expressed transcription factors play a role in targeting histone modifications. Although CpG-island promoters do not exhibit a functional requirement for SWI/SNF complexes during their activation, we previously found that these promoters are constitutively associated with Brg1 (Ramirez-Carrozzi et al., 2006). We favor the view that constitutive association results from non-specific binding of SWI/SNF complexes to genomic regions assembled into relatively open chromatin structures. However, we cannot exclude the possibility that SWI/SNF complexes play a role in establishing a constitutively open chromatin structure at CpG-island promoters that is sufficiently stable to permit activation following Brg1/Brm knockdown. We also must consider the possibility that non-catalytic subunits of the SWI/SNF complexes play roles that have not yet been revealed.
Although our current characterization provides considerable insight into the regulation of Class A and Class D promoters, promoters in Classes B and C remain poorly understood. A different nucleosome remodeling complex may be responsible for the SWI/SNF-independent activation of Class B promoters. Alternatively, the binding of specific transcription factors to Class B promoters in unstimulated cells may facilitate their assembly into constitutively open chromatin, allowing transcriptional activation in the absence of inducible nucleosome remodeling.
The evidence that the Il6 gene can switch from Class F to Class B reveals that genes are not fixed in their classification. Il6 was the only Class F gene in macrophages converted to a Class B gene in MEFs, which may be related to the need for unusually versatile regulation of Il6 expression because of its diverse biological functions (Kishimoto 2006). We hypothesize that the constitutive expression of a factor in MEFs that is inducibly expressed in macrophages is responsible for this switch. Although this hypothetical factor remains to be identified, the classification scheme and mechanistic insights provided by this analysis provide a consistent framework toward a global understanding of the diverse mechanisms responsible for inducible gene transcription, and of the biological necessity for this diversity.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Bone marrow-derived macrophages were prepared from C57BL/6 and IRF3−/− mice. MEFs were from D13.5–14.5 C57BL/6 embryos and were maintained in DMEM with 10% FBS and 0.05mM β-mecaptoethanol. Macrophages were activated on day 6 with S. aureus peptidoglycan (Sigma-Aldrich) (PGN) (20 µg/ml), poly I:C (1 µg/ml), S. typhosa LPS (Sigma-Aldrich) (10 µg/ml), IFNβ (PBL Biomedical Laboratories) (250 U/ml), or TNFα (BD Pharmingen) (10 ng/ml). MEFs were activated at passage 4. When indicated, cells were pre-incubated for 15 min with CHX (10 µg/ml).
RT-PCR, Real-Time PCR, and RNAi
RNA was extracted using TRI-reagent (Molecular Research Center), treated with RNase-free DNaseI, and purified using an RNeasy kit (Qiagen). Quantified RNA (2 µg) was reverse-transcribed using Omniscript RT Kit (Qiagen) and random hexamer primers. cDNA fragments were analyzed by qPCR using SensiMix Plus (Quantace) and the iCycler System (Bio-Rad) or a 7900HT (Applied Biosystems). PCR amplification conditions were 95°C (3 min) and 45 cycles of 95°C (15 sec), 60°C (30 sec), and 72°C (30 sec). Primer pairs (see Suppl. Table 1A) were designed to amplify 80–150 bp mRNA-specific fragments, and unique products were tested by melt-curve analysis.
The Brg1/Brm shRNA was expressed from a retroviral vector as described (Ramirez-Carrozzi et al., 2006). The efficiency of Brg1 and Brm knockdown was monitored by Western blot as described (Ramirez-Carrozzi et al., 2006). Transduced J774 cells and MEFs were stimulated 5 and 3 days after infection, respectively.
Restriction Enzyme Accessibility and ChIP
Restriction enzyme accessibility was performed as described (Ramirez-Carrozzi et al., 2006). Cell nuclei were incubated with restriction enzyme (100 U) (EcoNI for Ccl5 and DraI for Ifit1) for 15 min at 37°C. Purified DNA (10–15 µg) was then digested to completion to generate reference cleavage products using EcoRI and HindIII for Ccl5 and DraIII for Ifit1. Samples were analyzed by Southern blot with 32P-labeled probes corresponding to the following regions: Ccl5 promoter (−297 to −667) and Ifit1 promoter (−822 to −471).
ChIP experiments were performed as described (Ramirez-Carrozzi et al., 2006) with anti-H3 (Abcam ab1791), anti-trimethyl H3K4 (Abcam ab8580), anti-Acetyl H3 (Milipore 06–599), anti-RNA Pol II (Santa Cruz sc-899), and anti-TBP (Santa Cruz sc-204). Primer sequences are shown in Suppl. Table 1B. P-values were calculated by two-tailed Student’s t-test, using average values for each gene within each group.
Nucleosome Affinity Measurements
300-bp promoter fragments were cloned into pUC19. DNA fragments for nucleosome assembly were generated from these plasmids by PCR using vector-specific primers. PCR products were gel purified using Gel Extraction Kit (Qiagen). Equivalent amounts of each promoter fragment were pooled and 100 ng of the pool was assembled into nucleosomes by incubating with recombinant Xenopus laevis histones (Luger et al., 1997; Thåström et al., 2004) at 37 °C for 30 min in 10 µl of a 1M NaCl reaction containing 100 ng BSA. Low salt buffer (20 mM Tris, pH 7.6, 0.1%Triton X-100, 100 µg/ml BSA, 1mM EDTA, 0.5 mM PMSF, 5 mM DTT) was slowly added in volumes of 5, 10, 15, 30, and 30 µl, with 10 min incubations at room temperature after each addition. Samples were then run on a 6% 0.5X TBE native polyacrylamide gel and subsequently stained with 5X SYBR Green (Invitrogen). Free DNA and nucleosomal DNA bands were excised and electroeluted into 1X TE. Recovered DNA fragments were PCR amplified for 18–20 cycles. After determining the DNA concentration by OD analysis, the fragments were either re-assembled into nucleosomes or analyzed by qPCR using promoter-specific primers. P-values were calculated by two-tailed Student’s t-test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Genhong Cheng, Diana Hargreaves, Harvey Herschman, Siavash Kurdistani, Ruslan Medzhitov, Amy Weinmann, Jonathan Widom, and Michael Zhang for helpful discussions. V.R.R.C. was supported by a Giannini Family Foundation Fellowship, D.B. by the German Academic Exchange Service (Deutscher Akademischer Austausch Dienst, DAAD) and a Jonsson Cancer Center Foundation Fellowship, D.M.B. by NIH T32AI007126, K.R.D. by NIH T32GM07185, and S.T.S. by NIH R01GM086372, R01CA127279 and P50CA092131.
REFERENCES
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell. Biol. 2004;24:4476–4486. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2002;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell. Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Semin. Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res. Ther. 2006;8 Suppl 2:S2. doi: 10.1186/ar1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 2002;22:6471–6479. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008a;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008b;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thåström A, Gottesfeld JM, Luger K, Widom J. Histone-DNA binding free energy cannot be measured in dilution-driven dissociation experiments. Biochemistry. 2004;43:736–741. doi: 10.1021/bi0302043. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.