Abstract
Growth and virulence of mycobacteria requires sulfur uptake. The Mycobacterium tuberculosis genome contains, in addition to the ABC sulfate permease cysTWA, three SLC26-related SulP genes of unknown function. We report that induction of Rv1739c expression in E. coli increased bacterial uptake of sulfate, but not Cl−, formate, or oxalate. Uptake was time-dependent, maximal at pH 6.0, and exhibited a K1/2 for sulfate of 4.0 μM. Na+-independent sulfate uptake was not reduced by bicarbonate, nitrate, or phosphate, but was inhibited by sulfite, selenate, thiosulfate, N-ethylmaleimide and carbonyl cyanide 3-chloro-phenylhydrazone. Sulfate uptake was also increased by overexpression of the Rv1739c transmembrane domain, but not of the cytoplasmic C-terminal STAS domain. Mutation to serine of the three cysteine residues of Rv1739c did not affect magnitude, pH-dependence, or pharmacology of sulfate uptake. Expression of Rv1739c in a M. bovis BCG strain lacking the ABC sulfate permease subunit CysA could not complement sulfate auxotrophy. Moreover, inducible expression of Rv1739c in an E. coli strain lacking CysA did not increase sulfate uptake by intact cells. Our data show that facilitation of bacterial sulfate uptake by Rv1739c requires CysA and its associated sulfate permease activity, and suggest that Rv1739c may be a CysTWA-dependent sulfate transporter.
Introduction
Eukaryotic bicarbonate/anion transporters are encoded by the SLC4 and SLC26 gene superfamilies. Inherited human diseases are associated with mutations in multiple members of both superfamilies (Mount et al. 2004; Romero et al. 2004; Alper 2006; Kere 2006). However, the transmembrane domain structure of neither family is currently known. SLC26-related SulP genes are widely distributed among bacterial species (Felce et al. 2004; Price et al. 2004), in addition to the widespread expression of SLC26 genes among eukaryotic organisms (Mount et al. 2004). Vertebrate SLC26 polypeptides transport a wide range of anions, including Cl−, HCO3−, iodide, formate, oxalate, and sulfate, and have been implicated in teleost kidney sulfate excretion and osmoregulation (Renfro et al. 1999; Nakada et al. 2005; Katoh et al. 2006). SulP sulfate transporters of plants and yeast have been characterized functionally, and evidence of SulP-mediated HCO3− transport has been demonstrated in Synechococcus species (Price et al. 2004), but little other functional data on bacterial SulP proteins is available. Bacterial proteins have been important for catalyzing progress in the structure determination of polytopic membrane proteins of higher organisms. Functional data on prokaryotic SulP proteins is important for purification of native protein for crystallization and other structural studies.
As is true for eukaryotes, sulfur is a key element in bacterial metabolism. Rapidly growing numbers of anaerobic, sulfate-reducing chemolithoauxotrophic species have been identified in samples from deep sea hydrothermal vents, both as free-living bacteria and as thiotrophic endo- and exosymbionts of deep vent marine invertebrates (Rosenberg et al. 2006). Genes involved in sulfur metabolism have been implicated as virulence determinants in mammalian pathogens. In mycobacteria, mycothilols have been reported to contribute to anti-oxidant defense (Fan et al. 2007). Sulfolipids present in the mycobacterial outer envelope act as both positive and negative regulators of M. tuberculosis pathogenicity (Schelle et al. 2006). Overexpression screening of several bacterial SulP genes revealed inducible, stable accumulation of M. tuberculosis Rv1739c and E. coli ychM. In view of the importance of the SulP gene family in plant (Yoshimoto et al. 2007) and yeast (Shibagaki et al. 2006), and fish biology (Renfro et al. 1999; Nakada et al. 2005; Katoh et al. 2006), and the significance of the SLC26 gene superfamily in mammalian biology (Mount et al. 2004; Markovich et al. 2007), and the proposed importance of sulfate in M. tuberculosis pathogenicity (Schelle et al. 2006; Fan et al. 2007), we investigated further the function of Rv1739c (Fig. 1A) as a candidate sulfate transporter of mycobacteria.
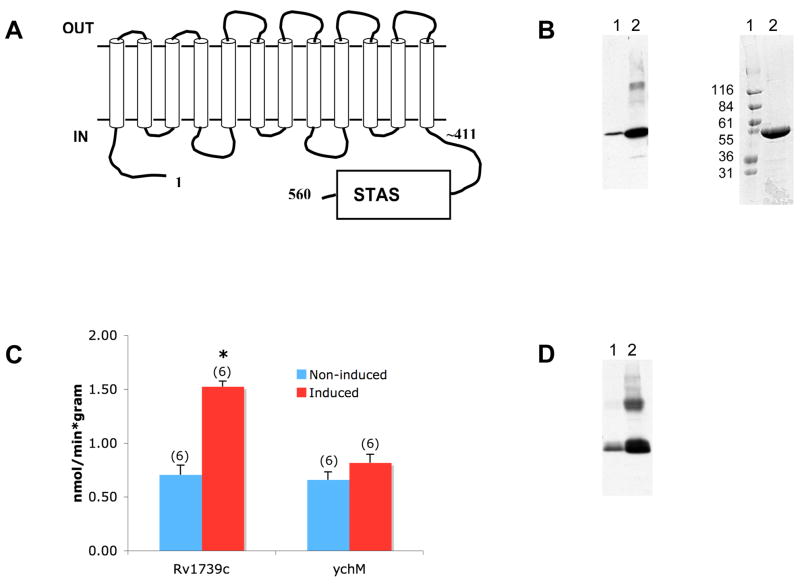
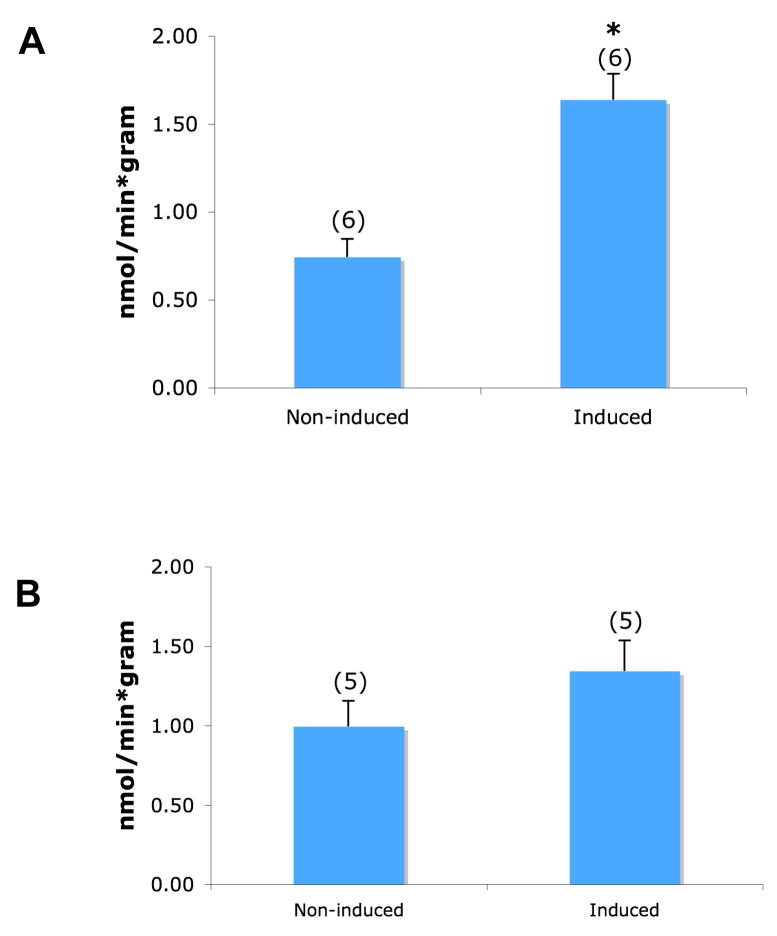
Figure 1. Overexpression of Rv1739c increases sulfate uptake.
A. Schematic of Rv1739c polypeptide from M. tb. The hypothetical disposition of the transmembrane spans is based on hydropathy analysis described by (Felce et al. 2004). B. Left: Anti-6His immunoblot of clarified SDS lysate of uninduced (lane 1) and IPTG-induced Tuner(DE3)pLacI cells (lane 2). Right: Coomassie Blue R250-stained gel of Rv1739c eluted from Ni-NTA column by 250 mM imidazole. C. 35S-sulfate uptake in uninduced Tuner cells or cells induced to express either M. Tb. Rv1739c (left bars) or E. coli ychM (right bars). Number of measurements is indicated within parentheses. *, p < 0.05. D. Anti-6His immunoblot of clarified SDS lysate of uninduced Tuner cells (lane 1) or Tuner cells induced to express ychM polypeptide (lane 2).
Materials and Methods
Molecular cloning
M. tuberculosis H37Rv genomic cosmid MTCY4C12 containing the Rv1739c gene (NC_000962) was provided by K. Eiglmeier (Inst. Pasteur, Paris). Rv1739c DNA encoding the predicted 560 aa Rv1739c SulP polypeptide (CAB03701) was PCR-amplified from the cosmid with addition of a C-terminal 6His tag. E. coli K-12 MG1655 BamHI genomic clone DD267 in phage λCharon was provided by the G. Plunkett and F. Blattner (E. coli Genome Project, Univ. Wisconsin). ychM DNA encoding the (predicted) 550 aa (footnote 1) ychM SulP polypeptide (BAB35134) was PCR amplified from the phage DNA, also with addition of a C-terminal 6His tag. Similar approaches were used to generate DNAs encoding the isolated Rv1739c transmembrane domain (aa 1–436, in which aa 411–436 are predicted to reside in the juxtamembrane cytosol) and the isolated Rv1739c C-terminal cytoplasmic region encompassing its STAS domain (aa 437–560). PCR products were subcloned into pETBlue-1 (Perfectly Blunt Cloning Kit, Novagen) and sequenced on both strands. pETBlue-1-SulP constructs were transformed into Tuner DE3pLacI cells (Novagen) for inducible expression. Oligonucleotide primers used are listed in Supplemental Table 1.
The Cys-less Rv1739c was constructed with QuickChange Multi Site-Directed Mutagenesis Kit (Stratagene). Directed mutagenesis of single codons was performed with the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). PCR was used to introduce the Kozak consensus sequence CACTCCCGCAGG from mouse Ae2b1 (Lecanda et al. 2000) immediately 5′ to the Rv1739c initiator methionine, and the modified DNA was subcloned into the Xenopus oocyte expression vector pXT7. This Kozak sequence is active in Xenopus oocytes (Kurschat et al. 2006).
Wildtype E coli strain BW25113 (obtained from Yale E. coli Genetic Stock Center) and its cysA deletion mutant JD21556 (obtained from National Bioresource Project of the Institute of Genetics, Japan) were both grown without antibiotics [www.shigen.nig.ac.jp/ecoli/strain/nbrpStrainDetail Action.do?strainId =10554] (or =1187). Rv1739c-6His was subcloned into pBAD33 (Guzman et al. 1995) for expression in JD21556. Rv1739c-6His was also subcloned into pMV261.hyg for GroEL promoter-driven constitutive expression in the wildtype M. bovis BCG strain, grown in 7H9T liquid medium (Wooff et al. 2002). The BCG cysA deletion strain EWP44 (the kind gift of Esen Wooff, Veterinary Laboratories Agency, Addlestone, Surrey, UK), was grown in 7H9T liquid medium supplemented with kanamycin and methionine (Wooff et al. 2002). M. bovis expression plasmid pSM-101EW encoding M. tuberculosis CysA (Wooff et al. 2002) was also the gift of Esen Wooff. Bacterial strains are summarized in Supplemental Table 2.
All subclones generated by PCR were verified by DNA sequencing of both strands, and by immunoblot confirmation of IPTG-induced biosynthesis of 6His-tagged polypeptide of the appropriate Mr using anti-6His monoclonal antibody (“His-Tag”, Novagen).
Isotopic anion influx measurement
An 8 hr culture of Tuner DE3pLacI cells transformed with pET-Blue-1-Rv1739c-6His or with pET-Blue-1-ychM-6His picked from a single colony was grown in LB supplemented with 1% glucose, 50μg/ml carbenicillin, and 30 μg/ml chloramphenicol. 50 ml fresh LB medium with or without 0.5 mM isopropylthiogalactoside (IPTG) was inoculated with an aliquot of liquid culture and grown overnight at 20–22°C with shaking at 300 rpm. Cultures were harvested by centrifugation upon reaching O.D. 0.3–0.5.
35S-sulfate uptake into bacterial suspensions was measured as described (Lindblow-Kull et al. 1985), with modifications. E. coli suspensions in preweighed 50 ml Falcon tubes were pelleted and washed once in 10–15 ml “uptake medium” containing (in mM) 62 K phosphate, 47 NH4Cl, 1.7 Na citrate, 0.6 MgCl2, and 11 glucose, pH 7.0. Washed pellets were resuspended at 25 mg/ml in uptake medium supplemented with 0.5 mM Na2SO4, incubated 30 min at room temp, pelleted, and resuspended in identical volumes of uptake medium containing 0.1 mM sulfate or the indicated concentrations at indicated pH, with additional agents as indicated. Uptake was initiated by addition of 1–2 μCi/ml 35S-sulfate (ICN) with vortexing. 0.1 ml samples were removed at 0 and 5 min, or at the times indicated, rapidly filtered through Millipore GSTF02500 filters mounted on a manifold (Hoeffer FH224V). Filters were immediately washed twice with 2.5 ml sulfate-free uptake medium and, after drying, 35S was measured by scintillation counting. All uptake experiments were performed at room temperature. All uptake data presented in individual figure panels represents experiments in which uninduced and induced bacteria were prepared in parallel and assayed the same day.
Assay for complementation of M. bovis BCG auxotrophy
Competent cells of the cysA deletion strain EWP44 (Wooff et al. 2002) were electroporated with pMV261.hyg Rv1739c or with the empty plasmid, and plated on 7H10 agar (Wooff et al. 2002) with kanamycin and hygromycin in the presence or absence of methionine. EWP44 was also electroporated with the CysA-expressing plasmid pSM101EW and plated on 7H10 agar with kanamycin, in the presence or absence of methionine. Growth was evaluated ~3 weeks later.
Results
Induced overexpression of M.tb. Rv1739c, but not of E. coli ychM, increases sulfate uptake by E. coli
IPTG-induction of the M. tb. sulP gene Rv1739c in E. coli increased abundance of the 560 aa Rv1739c polypeptide (Fig. 1A) as detected by 6His-immunoblot of lysate and by Coomassie stain of Ni-NTA-enriched protein (Fig. 1B). The Rv1739c polypeptide migrated on SDS-PAGE as expected for a monomer. Increased expression of Rv1739c was accompanied by increased sulfate uptake into intact bacteria (Fig. 1C). Increased sulfate uptake did not accompany increased expression of ychM at similar abundance (Fig. 1C,D). As evident in the immunoblots, the uninduced T7 promoter exhibited leaky expression of both Rv1739c and ychM.
Tuner DE3pLacI cells exhibit strong density-dependent induction of native sulfate uptake
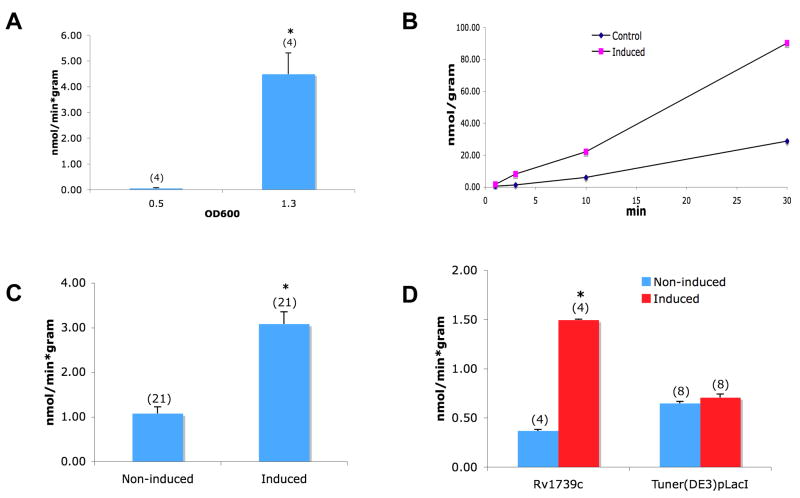
While optimizing the sulfate uptake assay, we noted that Tuner DE3pLacI cells exhibit strong upregulation of sulfate uptake in high density liquid culture (Fig. 2A). Rv1739c-expressing bacteria harvested from saturation culture did not exhibit an increase of sulfate uptake over this high background of saturation-cultured cells. However, cultures at early-mid log phase (O.D. 0.3–0.5) reliably exhibited stimulation of sulfate uptake associated with induction of Rv1739c expression. This Rv1739c-associated sulfate uptake was time-dependent, roughly linear over 30 min (Fig. 2B), highly reproducible (Fig. 2C), and was not evident in untransformed Tuner cells exposed to IPTG (Fig. 2D). The magnitude of sulfate uptake was not altered by omission of the 30 min loading in 0.5 mM Na sulfate prior to the influx assay or by a 30 min pre-incubation period in the absence of sulfate (n=4, not shown).
Figure 2.
A. Growth phase-dependence of 35S-sulfate uptake by native Tuner cells untreated with IPTG. B. Time-dependence of 35S-sulfate uptake by uninduced Tuner cells (diamonds) and by cells induced to express Rv1739c (squares). C. 35S-sulfate uptake into uninduced cells and cells induced to express Rv1739c. D. Sulfate uptake into cells harboring (Rv1739c) or lacking the Rv1739c expression plasmid (Tuner), after treatment without (uninduced) or with IPTG (induced). Number of measurements is indicated within parentheses. *, p < 0.05.
Rv1739c-associated sulfate uptake is maximal at pH 6 and is of high apparent affinity
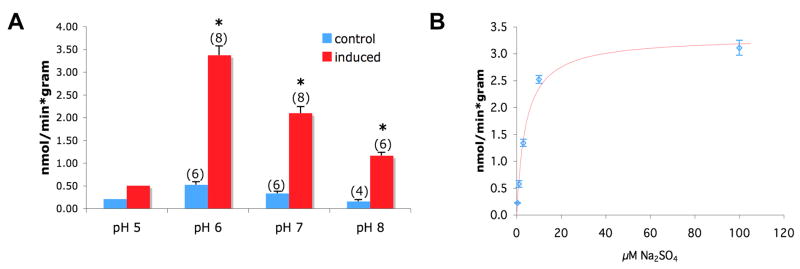
Rv1739c-associated sulfate uptake was inhibited at pH 8.0 and maximal at pH 6.0 (Fig. 3A), but nearly abolished by further acidification to pH 5.0. Sulfate uptake in uninduced Tuner cells transformed showed qualitatively similar changes at low background levels. Rv1739c-associated sulfate influx exhibited a K1/2 for extracellular sulfate of 4.04 μM (Fig. 3B). Tuner cells induced to express Rv1739c did not exhibit increased uptake of 36Cl−, 14C-formate, 14C-oxalate at pH 6.0 or pH 7.0 (not shown). Thus, Rv1739c-associated sulfate uptake was of high affinity, maximal at pH 6, and sulfate-specific.
Figure 3.
A. pH-dependence of 35S-sulfate uptake into cells induced to express Rv1739c. Number of measurements is indicated within parentheses. *, p < 0.05. B. Sulfate concentration dependence of 35S-sulfate uptake into cells expressing Rv1739c (n=6), fit with a hyperbolic equation (SigmaPlot).
As rare examples of bacterial transporters have been reported to function when expressed in Xenopus oocytes (Quick et al. 2002), Rv1739c was subcloned into the oocyte transcription vector pXT7 behind the Kozak sequence from Kcc1/Slc12a1, and functionally tested in Xenopus oocytes for 35S-sulfate influx and efflux. The 30 min sulfate uptake into individual oocytes from ND-96 medium at pH 6.0 was slightly increased over background (0.23±0.01 pmol min−1, n=30) by Rv1739c expression in oocytes injected with 10 ng RNA (0.31±0.02 pmol min−1, n=48, p<0.005), but not at pH 7.4 (n=30). However, interpretation of this cRNA-associated stimulation was confounded by lack of increased sulfate uptake into oocytes injected with 25 ng Rv1739c cRNA (n=30). Moreover, oocytes injected with 10 ng cRNA exhibited no sulfate-induced current at extracellular pH 6.0 (n=4). At pH 7.4, the small increase in inward current (measured at −100 mV) elicited by bath sulfate addition (from −122±16 nA to −183±9 nA, n=4, p<0.05) was unaccompanied by significant change in reversal potential. Taken together, the data suggest that Rv1739c function is expressed at very low level, if at all, in Xenopus oocytes.
Pharmacological properties of Rv1739c-associated sulfate uptake
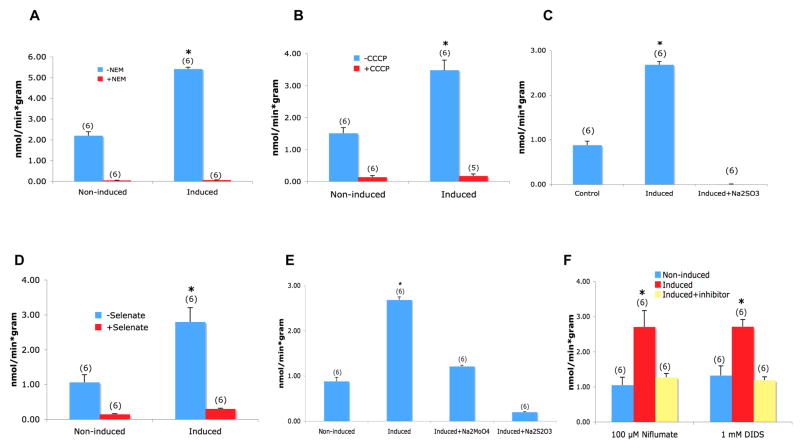
As shown in Figs. 4A and 4B, sulfate uptake associated with induction of Rv1739c expression was abolished by the inhibitors of the F1/F0 H+-ATPase, N-ethylmaleimide (NEM, 1 mM) and the oxidative phosphorylation uncoupler, carbonyl cyanide 3-chloro-phenylhydrazone (cccp, 50 μM). Rv1739c-associated sulfate uptake was also abolished or nearly so by 5 mM of the sulfate analogs sulfite (Fig. 4C), selenate (Fig. 4D), and thiosulfate (Fig. 4E). But none of these inhibitors discriminated between native sulfate uptake and sulfate uptake attributable to Rv1739c expression. 5 mM of the sulfate analog molybdate, and the general anion transport inhibitors niflumate (100 μM, Fig. 4E) and di-isothiocyanato-4′,4″-disulfono-stilbene disulfonate (DIDS, 1 mM, Fig. 4F) lowered induced sulfate uptake to uninduced levels. However, as 1 mM DIDS also nearly completely inhibited sulfate uptake by uninduced bacteria (n=14, not shown), it provided at this concentration little discrimination between native and Rv1739c-associated sulfate uptake. The anion transport inhibitor NS3623 (100 μM) did not inhibit uninduced or induced sulfate uptake (n=2, not shown).
Figure 4.
Tests of inhibition of 35S-sulfate uptake into cells expressing Rv1739c by (A) 1 mM NEM, (B) 50 μM CCCP, (C) 5 mM Na sulfite, (D) 5 mM Na selenate, (E) 5 mM Na molybdate or Na thiosulfate, (F) 1 mM DIDS or 100 μM niflumate. 35S-sulfate concentration was 100 μM in the presence of sulfite, molybdate, or thiosulfate, and 50 μM in the presence of the other antagonists. Number of measurements is within parentheses.
The transmembrane domain of Rv1739c is required for the increased sulfate uptake associated with induced overexpression
Rv1739c consists of two principal structural domains, the N-terminal transmembrane domain with a short predicted juxtamembrane cytoplasmic stretch, encompassed by aa 1–436, and the C-terminal cytoplasmic domain (aa 437–560) encompassing the “sulfate transporter and anti-sigma factor antagonist” (STAS) domain. Overexpression of 6His-tagged aa 1–436 transmembrane domain was associated with increased sulfate transport (Fig. 5A), whereas overexpression of the C-terminal aa 437–560 cytoplasmic STAS domain did not enhance sulfate uptake. This result is consistent with the membrane domain either mediating or being necessary for transport mediated by an associated native polypeptide. However, expression of the just the STAS domain itself, presented as aa 437–560, was not sufficient to support increased sulfate uptake by native transport proteins.
Figure 5.
A. 35S-sulfate uptake is increased by induction of expression of the Rv1739c transmembrane domain (aa 1–436). B. 35S-sulfate uptake is not increased by induction of expression of the C-terminal cytoplasmic domain of Rv1739c (aa 437–560) including its STAS domain. Number of measurements is within parentheses. *, p < 0.05.
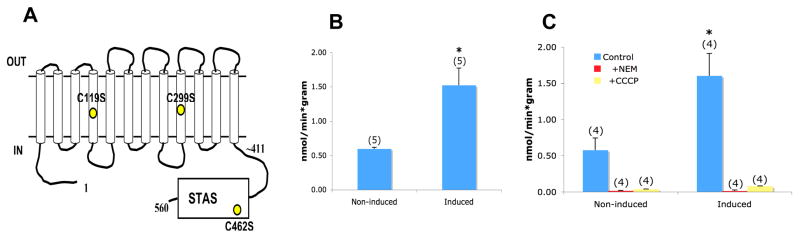
Rv1739c cysteine residues are not required for Rv1739c-associated increased sulfate uptake
Complete inhibition of sulfate uptake by NEM suggested that one or more of the three cysteine residues of Rv1739c might be essential for Rv1739c-associated sulfate uptake. Creation of Cys-less Rv1739c would provide not only a test of this hypothesis, but would also represent a valuable substrate for scanning cysteine accessibility mutagenesis. Therefore, the two cysteine residues of the transmembrane domain and the single cysteine residue of the STAS domain were mutagenized to serine (Fig. 6A). However mutagenesis of all three cysteine residues neither reduced the IPTG-induced increase in sulfate uptake (Fig. 6B) nor altered the complete inhibition of either uninduced or induced levels of sulfate uptake by NEM (Fig. 6C). Moreover, the pH-dependence of Cys-less Rv1739c was unchanged (pH 6.0 > pH 7.0 > pH 8.0 > pH 5.0, n=2, not shown).
Figure 6.
A. Schematic of the Cys-less Rv1739c polypeptide with its three cysteine-to-serine substitutions indicated. B. Induction of 35S-sulfate uptake in cells expressing Cys-less Rv1739c. C. Inhibition by NEM (1 mM) and by CCCP (50 μM) of 35S-sulfate uptake into cells induced to express Cys-less Rv1739c. Number of measurements is within parentheses. *, p < 0.05.
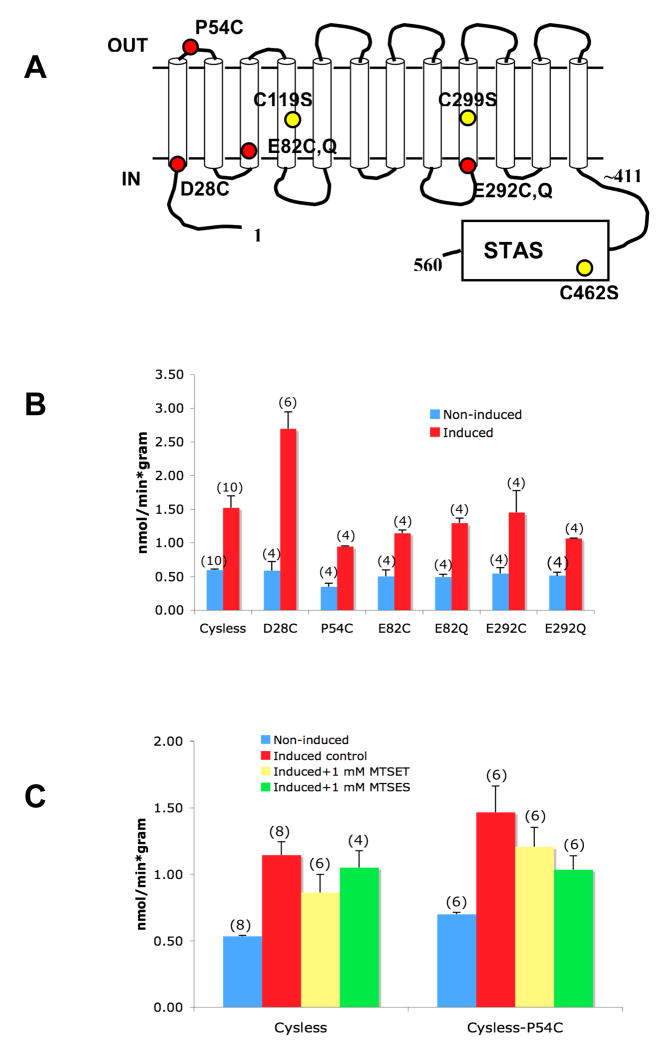
The Cys-less Rv1739c provided an opportunity to reintroduce single cysteine residues into the polypeptide (Fig. 7A) for subsequent tests of targeted sulfhydryl modification with methanethiosulfonate (MTS) reagents. Similar cysteine scanning mutagenesis has defined residues important for the function or folding of the phylogenetically related sulfate transporters Sultr1;2 of A. thaliana (Shibagaki et al. 2004; Shibagaki et al. 2006) and SHST1 of the tropical legume Stylosanthes hamata (Howitt 2005). The IPTG-induced increment in Rv1739c-associated sulfate uptake was preserved in the protein products of all mutations introduced into the Cys-less polypeptide (Fig. 7B). These mutants could therefore be used, on the hypothesis that Rv1739c was mediating sulfate influx, to test the topographical disposition and the requirement for sulfate uptake of individual residues. Fig. 7C shows that sulfate uptake associated with expression of Cys-less Rv1739c mutant P54C, but not by the Cys-less parent protein, was modestly inhibited by 1 mM MTSES. 1 mM concentrations of MTSET inhibited neither Cys-less Rv1739c nor its P54C mutant. However, 10 mM MTSET and MTSES each inhibited Cys-less Rv1739c-associated sulfate uptake by 60% (n=6, not shown). Cys-less Rv1739c was also inhibited 50% by 100mM MTSEA, and completely by 1 mM, and the P54C mutant showed roughly similar sensitivity (n=4, not shown). Thus, the effects of MTS reagents on sulfate uptake were not mediated directly through Rv1739c.
Figure 7.
A. Schematic of single cysteine substitutions introduced into the Cys-less Rv1739c polypeptide. B. Increased 35S-sulfate uptake into cells induced to express Cys-less Rv1739c polypeptide containing the indicated single reintroduced cysteine residues. C. Effects of MTSET and MTSES on sulfate uptake induced in Cys-less Rv1739 and its P54C variant. Number of measurements is indicated within parentheses.
Constitutive expression fails to rescue sulfate auxotrophy in the BCG cysA auxotroph strain EW44
The M. bovis cysA gene product has been previously defined by a global genetic complementation screen in rich medium as the unique sulfate uptake pathway of M. bovis (Wooff et al. 2002). We tested the hypothesis that constitutive overexpression of candidate sulfate transporter Rv1739c might complement the sulfate auxotrophy of the EWP44 strain of M. bovis lacking functional cysA. However, heterologous expression of Rv1739c-6His did not rescue EWP44 sulfate auxotrophy, whereas complementation with the cysA gene did so (not shown). Moreover, constitutive Rv1739c expression inhibited growth of EWP44 even in the presence of methionine. Thus, expression of Rv1739c in this setting appeared toxic.
Density-dependent induction of native sulfate uptake and Rv1739c-associated increased sulfate uptake are both abolished in the absence of the CysA component of the ABC sulfate uptake pathway
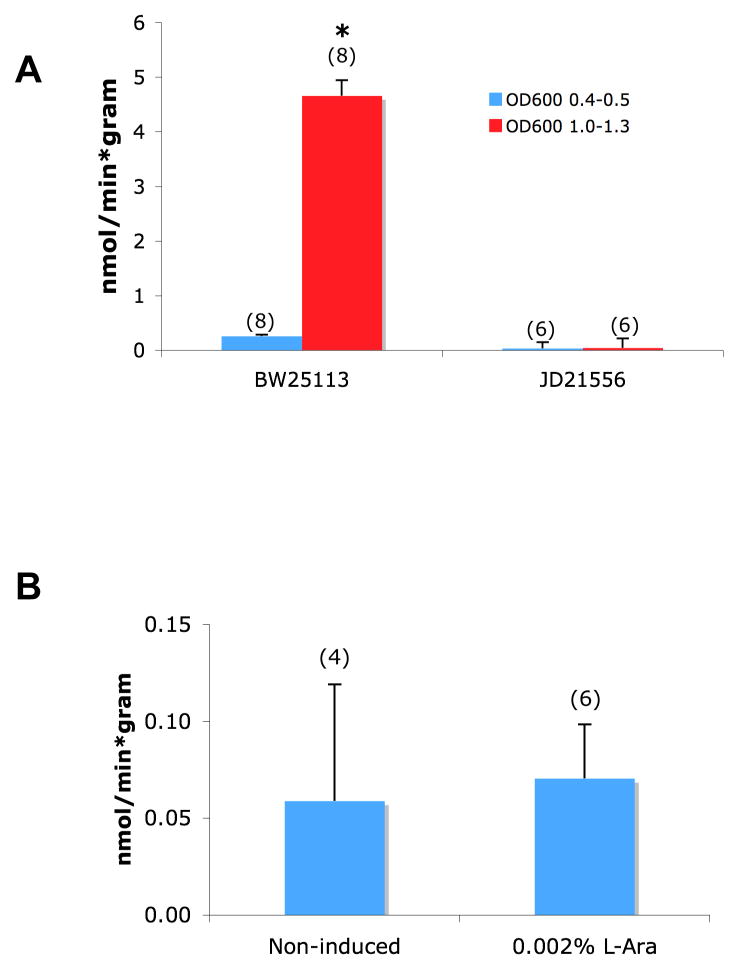
We tested the hypothesis that expression of CysA in E. coli might be required for enhancement of sulfate uptake by overexpression of Rv1739c. This experiment was of additional interest in testing the role of CysA in the density-dependent endogenous sulfate uptake system of E. coli. The E. coli strain BW25113 (the parent strain for the global library of insertional single gene knockouts (Baba et al. 2006)) displays growth density-dependent increase in sulfate uptake (Fig. 8A) similar to that observed in Tuner cells. This density-stimulated sulfate uptake was completely abrogated in the JD21556 strain derived from BW25113 and lacking a functional cysA gene (Fig. 8A). This result suggested the possibility that not only the basal sulfate uptake but also the Rv1739c-stimulated sulfate uptake might be mediated by the CysA pathway.
Figure 8.
A. Cell density-dependent induction of 35S-sulfate transport in E. coli strain BW25113, but not in the cysA deletion strain JD21556. B. Arabinose treatment does not increase 35S-sulfate uptake into JD21556 bacteria expressing Rv1739c in plasmid pBAD33. Number of measurements is indicated within parentheses. *, p < 0.05.
Since the JD21556 strain has very low background sulfate flux, we tested whether improved signal-to-noise might benefit the stimulation of sulfate uptake by expression of Rv1739c. Rv1739c-6His was subcloned into pBAD33, transformed into JD21556, and incubated in the absence or presence of 0.002% L-arabinose. As shown in Fig. 8B, arabinose did not induce elevated expression of sulfate uptake in JD21556 cells harboring Rv1739c. Despite immunoblot evidence of Rv1739c induction (not shown), arabinose induction at 0.002% or (not shown) at concentrations from 100-fold lower to 100-fold higher) failed to increase Rv1739c-associated sulfate uptake in JD21556 cells. Thus, Rv1739c expression did not increase sulfate uptake in the absence of CysA expression.
Discussion
The large SLC26-related SulP family includes sulfate transporters in yeast and plants, but has been minimally characterized in bacteria. The only functional analysis of a bacterial sulP protein reported to date is that for the Na+-dependent HCO3− transporter BicA of euryhaline cyanobacterial Synechococcus species (Price et al. 2004), a representative of the small class of SulP/CynT rosetta protein fusions of an SLC26-like anion transporter sequence with a type β-carbonic anhydrase sequence (Felce et al. 2004). The transport function of polypeptides of the more abundant SulP class characterized by presence of a C-terminal cytoplasmic STAS domain has not been reported. In addition, no bacterial or yeast sulP protein has been studied in isolated membranes or in reconstituted proteoliposomes.
Mycobacterial SulP transporters offer potential insight into the sulfur assimilation pathways leading to biosynthesis of sulfolipid pathogenicity determinants. The bacterial SulP polypeptides also provide attractive targets for structural analysis of protein folds homologous to those of the mammalian SLC26 anion transporter disease genes. Therefore we investigated the function of Rv1739c that, unique among the three examined recombinant SulP genes of the M. tb. genome, accumulated to substantial levels after IPTG induction in Tuner DE3pLacI cells.
Although endogenous sulfate uptake in Tuner cells increases dramatically in late-log phase, Rv1739c overexpression in Tuner cells led to 2–5 -fold increased sulfate uptake in early-to-mid log phase cells. This incremental sulfate uptake was maximal at pH 6 but abolished at pH 5, and was of high affinity with K1/2 of 4 μM. Rv1739c expression did not increase uptake of Cl−, formate, or oxalate, and neither HCO3− nor nitrate inhibited sulfate uptake. Sulfate uptake was nearly abolished by the metabolic inhibitors NEM and CCCP, and the sulfate analogs sulfite, selenate, and thiosulfate. Sulfate uptake was inhibited to lesser extent by molybdate and the general anion transport inhibitors, niflumate and DIDS, but not by NS3623. Expression of the Rv1739c transmembrane domain was sufficient to increase sulfate uptake, but expression of the isolated C-terminal cytoplasmic domain did not affect sulfate uptake. Rv1739c-associated sulfate uptake was specific, insofar as no increase in sulfate uptake was associated with equivalent expression levels of E. coli SulP polypeptide ychM.
Role of the STAS domain in Rv1739c function
The function of the Rv1739c STAS domain remains unknown. A clue resides in the preliminary report that the A. thaliana Sultr1;2 STAS domain binds to the O-acetyl serine (thiol) lyase/cysteine synthase (OASTL) cytosolic isoform expressed in cells of the root cortex. This complex may constitute a sulfate assimilation metabolon (Shibagaki 2007), deletion or mutation of which increases resistance to selenate toxicity (El Kassis et al. 2007). Interestingly, E. coli CysA also can bind to OASTLa/CysK (Arifuzzaman et al. 2006). STAS domains in plant SulP and mammalian SLC26 transporters are essential for plasma membrane targeting and for transport function. In SLC26A3, the STAS domain binds the R domain of the cystic fibrosis transmembrane regulator (Ko et al. 2004). Carbonic anhydrase II binding by SLC26A6 STAS domain is associated with increased SLC26A6-associated Cl−/HCO3− exchange activity (Alvarez et al. 2005).
The three cysteine residues of Rv1739c were non-essential for increased sulfate uptake. This finding resembles results reported for the SHST1 SulP sulfate transporter of a tropical legume (Howitt 2005). However, unlike the STAS domain requirement for sulfate transport by A. thaliana Sultr1;2 (Shibagaki et al. 2004), the STAS domain was dispensable for RV1739c-associated enhancement of sulfate uptake. Moreover, modification of selected conserved residues homologous to those found important for sulfate transport by the plant SulP polypeptides SHST1 (Khurana et al. 2000; Shelden et al. 2003) and Sultr1;2 polypeptides (Shibagaki et al. 2006) also appeared unimportant for Rv1739c-associated sulfate uptake. Expression of the Rv1739c STAS domain in the absence of its transmembrane domain did not enhance sulfate uptake.
Relation of the Rv1739c polypeptide to other SulP polypeptides
Rv1739c shares only 25% amino acid identity with the STAS domain-containing M. tb. SulP polypeptide Rv1707, and only 23% identity with the cynT domain-containing M. tb. SulP polypeptide Rv3273, a putative HCO3− transporter. Rv3273 polypeptide has been detected on Coomassie Blue G250-stained SDS-PAGE gels of alkali-extracted inner membrane fraction from M. tb. (Mattow et al. 2007), and is present in M. bovis BCG membranes at 10-fold greater abundance (Schmidt et al. 2004). In contrast, the two STAS-domain-containing SulP proteins Rv1739c and Rv1707 have not been detected in proteomic screens of M. tb.
Rv1739c has a predicted pI 10.39, with basic residues distributed throughout both transmembrane and cytoplasmic domains. The distantly related SulP homolog Rv1707 also has a pI of 10.24. These alkaline pI values are rare among the M. Tb. proteome, in which the majority of polypeptides is highly acidic (Mattow et al. 2003). Rv1739c is marked by a 6 residue internal direct repeat, AVLAAT at aa 105–110 and again at 213–218, with AAVLA also at aa 90–94. The significance of these sequences beyond their hydrophobicity is unknown. Rv1739c from M. tb. Strain H37Rv differs in the single residue R134 from the L134 present in strains CDC1551, F11, and c, as well as in the M. bovis ortholog. The smaller genome of M. leprae lacks all three SulP genes. The closest non-mycobacterial homolog of Rv1739c detected by BLAST is SulP polypeptide NP_600277 from Corynebacterium glutamicum, with 51% amino acid sequence identity.
The P. aeruginosa SulP polypeptide PA1647 shares 41% amino acid identity with M. Tb. Rv1739c. PA1647 mRNA abundance increased 3-fold upon sulfate deprivation, the same degree of increase as exhibited by the P. aeruginosa sulfate permease gene products CysT, CysW, and CysA. Moreover, PA1647 mRNA was increased 6-fold by growth of the cystic fibrosis P. aeruginosa isolate E601 on (sulfate-rich) mucin in sulfate-free conditions (Tralau et al. 2007). Sulfate starvation experiments revealed no upregulation of the ychM gene in E. coli (Gyaneshwar et al. 2005 ) or of any SulP gene product in B. subtilis (Auger et al. 2002), but have not been published for M. tb. Nonetheless, the P. aeruginosa data provide evidence for a role of a Rv1739c-related polypeptide in the sulfur assimilation response, and suggest a direct or supporting role in sulfate transport function.
M. Tb. Rv1739c shares 23% amino acid sequence identity and 44% homology (NCBI BLAST) with trout Onchorhynchus mykiss Slc26a1 polypeptide (Katoh et al. 2006) across their aligned putative transmembrane domains and juxtamembrane regions, and 24% identity (45% homology) with eel Anguilla japonica kidney Slc26a1 (Nakada et al. 2005).
Regulation of Rv1739c gene expression
Several reports have demonstrated control of M. tb. Rv1739c expression by environmental stressors believed to mimic the stresses of the host environment (Murphy et al. 2007). Thus, 24 hrs after M. tb. infection of interferon-γ-activated macrophages, Rv1739c and Rv1707 mRNAs increased 2.5 and 4-fold, respectively. Whereas the increased expression of Rv1739c was independent of macrophage inducible nitric oxide synthase (iNOS) activity, induction of Rv1707 required iNOS activity (Schnappinger et al. 2003). An in vitro study noted 4-fold upregulation of Rv1739c mRNA in response to imposition of hypoxia in early log-phase liquid culture (Sherman et al. 2001). If in hypoxic conditions the ATP required for the ABC sulfate permease activity is limiting, an ATP-independent sulfate transporter might confer advantage. Although exposure in liquid culture to nitric oxide donors at high concentration led to 20-fold upregulation of Rv1739c mRNA levels (Ohno et al. 2003), lower concentrations believed better to model a “dormancy response” did not increase Rv1739c mRNA (Voskuil et al. 2003). E. coli SulP ychM mRNA levels have been shown to be upregulated 2–3 fold in glass wool biofilms (Ren et al. 2004).
Is the M. tb. SulP polypeptide Rv1739c a sulfate transporter?
Both Tuner cells and the E. coli strain BW25113 exhibited strong induction of sulfate uptake in late log and saturation phase of growth in liquid culture (Figs. 2 and 8). In the JD21556 derivative of BW25113 lacking CysA, both basal and density-induced sulfate uptake was almost completely abrogated (Fig 8). Thus, CysA-mediated sulfate uptake in E. coli is strongly regulated by cell density in rich medium. This density-dependence may in part reflect sulfate depletion from the medium, since sulfate starvation of E. coli altered expression of the genes encoding the ABC sulfate permease subunits (Gyaneshwar et al. 2005).
Our experiments in rich medium showed that Rv1739c does not complement sulfate auxotrophy in the EWP44 BCG mutant deficient in CysA. However, interpretation of these experiments is confounded by the toxicity of constitutive Rv1739c expression in BCG strain EWP44. Similar toxicity was also reported for expression of the GFP-fusion protein of E. coli SulP polypeptide ychM (http://ecoli.aist.nara.ac.jp/GB6/info.jsp?id=JW5189). The presence of CysA also proved necessary for Rv1739-associated increased sulfate uptake in E. coli. Initial attempts to demonstrate intrinsic sulfate transport function of Rv1739c through sulfate flux studies in rightside-out vesicles prepared from IPTG-induced Tuner cells were unsuccessful. Thus, we conclude from our data that overexpression of Rv1739c in E. coli increases cellular sulfate uptake in a manner dependent upon expression (and presumed function) of CysA, the cytoplasmic ATP-binding subunit of the ABC sulfate permease. Our results suggest that Rv1739c may be a sulfate transporter that acts in conjunction with the CysTWA ABC sulfate permease.
If Rv1739c is indeed a sulfate transporter, its activity optimum at pH 6.0 is potentially consistent with the H+/sulfate cotransport mechanism proposed for plant (Buchner et al. 2004) and yeast SulP transporters, but the complete inhibition of Rv1739c-associated sulfate uptake at pH 5 remains unexplained. The lack of inhibition by extracellular HCO3− of Rv1739c overexpression-associated sulfate uptake into E. coli does not support a sulfate/bicarbonate exchange mechanism such as proposed for vertebrate Slc26a1 (Xie et al. 2002; Markovich et al. 2007) and consistent with carbonic anhydrase-dependence of sulfate excretion by teleost kidney (Renfro et al. 1999; Nakada et al. 2005; Pelis et al. 2005; Katoh et al. 2006). In addition, the lack of upregulated oxalate transport by Rv1739c overexpression and lack of inhibition by oxalate of associated sulfate uptake both suggest a substrate specificity distinct from that of oxalate-transporting mouse Slc26a1 (Xie et al. 2002).
Wooff et al. (Wooff et al. 2002) showed that the CysTWA sulfate permease system was the only essential sulfate transporter of BCG in rich medium and in amino acid-deprived medium. However, deletion of the cysA gene did not impair BCG survival and proliferation in normal mice. Thus, it remains possible that Rv1739c and/or other SulP proteins of mycobacteria play important roles mediating or regulating sulfate transport during host infection, a condition not optimally modeled by standard in vitro culture. Sulfate uptake mediated by cysTWA and Rv1739c polypeptides may direct sulfate into different metabolic pathways built around distinct enzymatic assemblies that have been described as “enzoskeletons” (Norris et al. 1996).
Conclusion
The SLC26-related SulP polypeptide Rv1739c of M. tb. has been overexpressed in E. coli and shown to increase sulfate uptake into intact bacteria. The increased sulfate uptake occurs by a mechanism requiring the cytoplasmic CysA subunit of the ABC sulfate permease. The transmembrane domain of Rv1739c suffices for this activity. Further experiments will be needed to test intrinsic sulfate transport activity of isolated Rv1739c polypeptide.
Supplementary Material
Supplemental Table 1. Oligonucleotide primers.
Supplemental Table 2. Bacterial strains.
Acknowledgments
This work was supported by NIH grants R01 DK43495 (SLA), DK34854 (Harvard Digestive Diseases Center funds to SLA) and T32 DK07199 (Beth Israel Deaconess Renal Training Grant to JSC). We thank Drs. E. Wooff and P. Wheeler (Veterinary Laboratories Agency, Surrey, UK) for the kind gifts of M. bovis strain EWP44 and M. bovis expression plasmid pSM-101EW.
Footnotes
The ychM open reading frame of 550 aa has been annotated as initiating from an AUG codon in E. coli O157:H7 (BAB35134) and in E. coli APEC 01 (YP_852337). The ychM open reading frame has also been annotated as 559 aa, initiating from an inframe upstream GUG codon in E. coli W3110 (AP_001831) and K12 (NP_415724). The overlapping portions of the two amino acid sequences are identical. Yet another possible initiation site is the GUG initiation codon giving rise to a predicted ychM ORF of 551 aa, and the corresponding initiation site has been annotated in the closely related S. typhimurium ychM (AAL20696; 553 aa, 93% aa identity with E. coli ychM). None of the possible translational initiation sites has been validated experimentally. We amplified the 550 aa ORF of ychM, since the overexpression of the 559 aa ORF (JW5189) was reported by the ASKA collection curators to be toxic (http://ecoli.naist.jp/GB6/info.jsp?id=JW5189), and was among the polypeptides that could not be purified as a bait protein in a global protein-protein interaction screen (Arifuzzaman et al. 2006).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alper SL. Molecular physiology of SLC4 anion exchangers. Exp Physiol. 2006;91:153–161. doi: 10.1113/expphysiol.2005.031765. [DOI] [PubMed] [Google Scholar]
- Alvarez BV, Vilas GL, Casey JR. Metabolon disruption: a mechanism that regulates bicarbonate transport. Embo J. 2005;24:2499–2511. doi: 10.1038/sj.emboj.7600736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger S, Danchin A, Martin-Verstraete I. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J Bacteriol. 2002;184:5179–5186. doi: 10.1128/JB.184.18.5179-5186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot. 2004;55:1765–1773. doi: 10.1093/jxb/erh206. [DOI] [PubMed] [Google Scholar]
- El Kassis E, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC. Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiol. 2007;143:1231–1241. doi: 10.1104/pp.106.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Luxenburger A, Painter GF, Blanchard JS. Steady-State and Pre-steady-State Kinetic Analysis of Mycobacterium smegmatis Cysteine Ligase (MshC) Biochemistry. 2007;46:11421–11429. doi: 10.1021/bi7011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felce J, Saier MH., Jr Carbonic anhydrases fused to anion transporters of the SulP family: evidence for a novel type of bicarbonate transporter. J Mol Microbiol Biotechnol. 2004;8:169–176. doi: 10.1159/000085789. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Paliy O, McAuliffe J, Popham DL, Jordan MI, Kustu S. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J Bacteriol. 2005;187:1074–1090. doi: 10.1128/JB.187.3.1074-1090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt SM. The role of cysteine residues in the sulphate transporter, SHST1: construction of a functional cysteine-less transporter. Biochim Biophys Acta. 2005;1669:95–100. doi: 10.1016/j.bbamem.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Katoh F, Tresguerres M, Lee KM, Kaneko T, Aida K, Goss GG. Cloning of rainbow trout SLC26A1: involvement in renal sulfate secretion. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1468–1478. doi: 10.1152/ajpregu.00482.2005. [DOI] [PubMed] [Google Scholar]
- Kere J. Overview of the SLC26 family and associated diseases. Novartis Found Symp. 2006;273:2–11. discussion 11–18, 261–264. [PubMed] [Google Scholar]
- Khurana OK, Coupland LA, Shelden MC, Howitt SM. Homologous mutations in two diverse sulphate transporters have similar effects. FEBS Lett. 2000;477:118–122. doi: 10.1016/s0014-5793(00)01783-x. [DOI] [PubMed] [Google Scholar]
- Ko SB, Zeng W, Dorwart MR, Luo X, Kim KH, Millen L, Goto H, Naruse S, Soyombo A, Thomas PJ, Muallem S. Gating of CFTR by the STAS domain of SLC26 transporters. Nat Cell Biol. 2004;6:343–350. doi: 10.1038/ncb1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurschat CE, Shmukler BE, Jiang L, Wilhelm S, Kim EH, Chernova MN, Kinne RK, Stewart AK, Alper SL. Alkaline-shifted pHo sensitivity of AE2c1-mediated anion exchange reveals novel regulatory determinants in the AE2 N-terminal cytoplasmic domain. J Biol Chem. 2006;281:1885–1896. doi: 10.1074/jbc.M509734200. [DOI] [PubMed] [Google Scholar]
- Lecanda J, Urtasun R, Medina JF. Molecular cloning and genomic organization of the mouse AE2 anion exchanger gene. Biochem Biophys Res Commun. 2000;276:117–124. doi: 10.1006/bbrc.2000.3439. [DOI] [PubMed] [Google Scholar]
- Lindblow-Kull C, Kull FJ, Shrift A. Single transporter for sulfate, selenate, and selenite in Escherichia coli K-12. J Bacteriol. 1985;163:1267–1269. doi: 10.1128/jb.163.3.1267-1269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovich D, Aronson PS. Specificity and regulation of renal sulfate transporters. Annu Rev Physiol. 2007;69:361–375. doi: 10.1146/annurev.physiol.69.040705.141319. [DOI] [PubMed] [Google Scholar]
- Mattow J, Schaible UE, Schmidt F, Hagens K, Siejak F, Brestrich G, Haeselbarth G, Muller EC, Jungblut PR, Kaufmann SH. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis. 2003;24:3405–3420. doi: 10.1002/elps.200305601. [DOI] [PubMed] [Google Scholar]
- Mattow J, Siejak F, Hagens K, Schmidt F, Koehler C, Treumann A, Schaible UE, Kaufmann SH. An improved strategy for selective and efficient enrichment of integral plasma membrane proteins of mycobacteria. Proteomics. 2007;7:1687–1701. doi: 10.1002/pmic.200600928. [DOI] [PubMed] [Google Scholar]
- Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Brown JR. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis. 2007;7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Zandi-Nejad K, Kurita Y, Kudo H, Broumand V, Kwon CY, Mercado A, Mount DB, Hirose S. Roles of Slc13a1 and Slc26a1 sulfate transporters of eel kidney in sulfate homeostasis and osmoregulation in freshwater. Am J Physiol Regul Integr Comp Physiol. 2005;289:R575–R585. doi: 10.1152/ajpregu.00725.2004. [DOI] [PubMed] [Google Scholar]
- Norris V, Turnock G, Sigee D. The Escherichia coli enzoskeleton. Mol Microbiol. 1996;19:197–204. doi: 10.1046/j.1365-2958.1996.373899.x. [DOI] [PubMed] [Google Scholar]
- Ohno H, Zhu G, Mohan VP, Chu D, Kohno S, Jacobs WR, Jr, Chan J. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 2003;5:637–648. doi: 10.1046/j.1462-5822.2003.00307.x. [DOI] [PubMed] [Google Scholar]
- Pelis RM, Edwards SL, Kunigelis SC, Claiborne JB, Renfro JL. Stimulation of renal sulfate secretion by metabolic acidosis requires Na+/H+ exchange induction and carbonic anhydrase. Am J Physiol Renal Physiol. 2005;289:F208–216. doi: 10.1152/ajprenal.00468.2004. [DOI] [PubMed] [Google Scholar]
- Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci U S A. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Wright EM. Employing Escherichia coli to functionally express, purify, and characterize a human transporter. Proc Natl Acad Sci U S A. 2002;99:8597–8601. doi: 10.1073/pnas.132266599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- Renfro JL, Maren TH, Zeien C, Swenson ER. Renal sulfate secretion is carbonic anhydrase dependent in a marine teleost, Pleuronectes americanus. Am J Physiol. 1999;276:F288–294. doi: 10.1152/ajprenal.1999.276.2.F288. [DOI] [PubMed] [Google Scholar]
- Romero MF, Fulton CM, Boron WF. The SLC4 family of HCO 3 - transporters. Pflugers Arch. 2004;447:495–509. doi: 10.1007/s00424-003-1180-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg NK, Lee RW, Yancey PH. High contents of hypotaurine and thiotaurine in hydrothermal-vent gastropods without thiotrophic endosymbionts. J Exp Zoolog A Comp Exp Biol. 2006;305:655–662. doi: 10.1002/jez.a.316. [DOI] [PubMed] [Google Scholar]
- Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. Chembiochem. 2006;7:1516–1524. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- Schmidt F, Donahoe S, Hagens K, Mattow J, Schaible UE, Kaufmann SH, Aebersold R, Jungblut PR. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol Cell Proteomics. 2004;3:24–42. doi: 10.1074/mcp.M300074-MCP200. [DOI] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden MC, Loughlin P, Tierney ML, Howitt SM. Interactions between charged amino acid residues within transmembrane helices in the sulfate transporter SHST1. Biochemistry. 2003;42:12941–12949. doi: 10.1021/bi034827s. [DOI] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N, Grossman AR. Probing the function of STAS domains of the Arabidopsis sulfate transporters. J Biol Chem. 2004;279:30791–30799. doi: 10.1074/jbc.M403248200. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Grossman AR. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem. 2006;281:22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Grossman AR. Functions of the STAS domain in the Arabidopsis sulfate transporter, Sultr1;2. Botany and Plant Biology; 2007; Joint Congress; 2007. Abst. 1462 poster P11018. [Google Scholar]
- Tralau T, Vuilleumier S, Thibault C, Campbell BJ, Hart CA, Kertesz MA. Transcriptomic Analysis of the Sulfate Starvation Response of Pseudomonas aeruginosa. J Bacteriol. 2007;189:6743–6750. doi: 10.1128/JB.00889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooff E, Michell SL, Gordon SV, Chambers MA, Bardarov S, Jacobs WR, Jr, Hewinson RG, Wheeler PR. Functional genomics reveals the sole sulphate transporter of the Mycobacterium tuberculosis complex and its relevance to the acquisition of sulphur in vivo. Mol Microbiol. 2002;43:653–663. doi: 10.1046/j.1365-2958.2002.02771.x. [DOI] [PubMed] [Google Scholar]
- Xie Q, Welch R, Mercado A, Romero MF, Mount DB. Molecular characterization of the murine Slc26a6 anion exchanger: functional comparison with Slc26a1. Am J Physiol Renal Physiol. 2002;283:F826–838. doi: 10.1152/ajprenal.00079.2002. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Watanabe-Takahashi A, Saito K, Takahashi H. Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 2007;145:378–388. doi: 10.1104/pp.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Oligonucleotide primers.
Supplemental Table 2. Bacterial strains.