Abstract
PURPOSE
To perform a genome-wide linkage screen with a single-nucleotide polymorphism (SNP) linkage panel to identify regions of genetic linkage in Fuchs endothelial corneal dystrophy (FECD) and to analyze affected individuals for mutations in the COL8A2 gene.
METHODS
Ninety-two individuals from 22 families with FECD were identified from our multiplex FECD family cohort. A genome-wide linkage scan was performed using an SNP linkage panel. Parametric two-point linkage analyses were calculated and nonparametric multipoint linkage analyses were performed on chromosomes with two-point LOD scores (HLOD) > 1.0. All affected individuals were analyzed for the two previously reported FECD mutations in the COL8A2 gene (L450W and Q455K).
RESULTS
The genome-wide analysis identified five regions with linkage signals from all analyses on chromosomes 1, 7, 15, 17, and X. The highest two-point HLODs were found on the long arm of chromosome 15 with an HLOD of 3.26 for the recessive model and 2.48 for the dominant model. Multipoint linkage analysis also identified a linkage peak on the long arm of chromosome 15 with a LOD > 1. The region of linkage on chromosome 1p, driven by two multigenerational FECD families with a two-point LOD > 2, was adjacent to the previously identified COL8A2 gene; however, the two reported mutations in COL8A2 were not identified in any of the 56 affected individuals in the 92 samples tested.
CONCLUSIONS
Genome-wide linkage analysis was used to identify potential linkage regions on chromosomes 1, 7, 15, 17, and X for FECD. The previously reported mutations in the COL8A2 gene were not found in the 92 samples tested. (Invest Ophthalmol Vis Sci. 2009;50:1093–1097)
Fuchs endothelial corneal dystrophy (FECD) is a common progressive disorder of the corneal endothelium that becomes symptomatic with aging.1 It appears as guttae progressively form on Descemet’s membrane along with corneal edema and stromal clouding leading to impaired vision. FECD has been one of the main indications for corneal transplantation in many countries, including the United States,2 with the main treatment modality being a penetrating keratoplasty, until the advent of various types of endothelial keratoplasty.
FECD typically becomes symptomatic during the fifth or sixth decade of life and predominantly affects women, who comprise three of four affected individuals.3 The pathogenesis of FECD remains unknown, but the corneal endothelial cells are arrested in the G1 phase of the cell cycle and do not undergo mitosis.4 During a normal state, the cornea is kept in a state of deturgescence by endothelial cell Na+/K+ ATPases that remove fluid from the stroma and by tight junctions between endothelial cells that limit fluid entry.1 This maintains the stroma’s ordered arrangement of collagen and with it, the transparency of the cornea. In states of low endothelial cell density, the loss of the endothelium’s barrier function causes corneal edema. Damage to the endothelial cell layer therefore results in irreparable tissue damage that necessitates endothelial or penetrating keratoplasty.1
It is well established that FECD can affect multiple family members and that several genetic factors underlie the development of the disorder.5 However, the use of genetic approaches to identify the genes underlying the development of FECD is complicated by the late onset of the disease, so that parents and even siblings of the proband are often unavailable for study. In addition, younger individuals in a family may not clinically exhibit the disease because they may not be old enough to manifest the disorder.
The first progress toward identifying gene mutations associated with familial FECD was provided by a rare subtype of FECD characterized by onset in early childhood. Early-onset FECD was found to be caused by a missense mutation, Q455K, in the COL8A2 gene (MIM: 120252), which encodes for the α2 subtype of collagen VIII, a major component of Descemet’s membrane. Three other missense mutations within COL8A2—R155Q, R304Q, and R434H—were found in patients with late-onset FECD, but their role remains uncertain, as R155Q has the same frequency in normal controls.6 Another mutation in COL8A2, L450W, that is believed to be pathogenic has been detected in a family with atypical FECD.7,8 Although mutations in COL8A2 have been linked to early-onset FECD, to date, mutations in the COL8A2 gene have not been associated with the much more common late-onset FECD.9–11 Heterozygous single nucleotide polymorphisms in the SLC4A11 gene on chromosome 20 (20p13) have recently been detected in four Chinese and Indian patients with the common phenotype of FECD,12 but they contribute to only ∼5% of the genetic burden of the disease.
Family-based studies have mapped late-onset FECD susceptibility loci to 13ptel-13q12.1313 and 18q21.2-q21.32,14 in one and three large multigenerational FECD families, respectively. However, to date, there have been no FECD genetic linkage studies involving a large number of affected families. In this study, we performed the first whole-genome linkage scan for late-onset FECD in 22 families (linkage panel IV; Illumina, Inc., San Diego, CA).
METHODS
Subject Ascertainment
All subjects were identified from the Cornea Clinic at Duke University Eye Center and were examined by slit lamp biomicroscopy to determine their affection status. Grading of disease severity in affected individuals was determined using a slightly modified version of established classification systems.5 Severity was assessed on a scale of 0 to 5 (Table 1).
TABLE 1.
Fuchs Corneal Dystrophy Grading System
| Grade | Findings |
|---|---|
| 0 | No central cornea guttae |
| 1 | Scattered central cornea guttae |
| 2 | 1 or 2 mm of central corneal guttae |
| 3 | >2 to 5 mm of grouped cornea guttae |
| 4 | >5 mm of grouped central cornea guttae |
| 5 | Cornea guttae with corneal edema |
A subject was classified as affected if one or more of the following criteria were met: (1) The individual had a corneal transplant with a histopathologic diagnosis of FECD; and (2) the individual had corneal guttae grades 2 to 5 and at least one family member had undergone a corneal transplant with a histopathologic diagnosis of FECD.
If the corneal guttae were grades 0 to 1, the disease status was considered to be uncertain. Individuals were classified as unaffected if a corneal examination was normal.
Any subject with a history of intraocular surgery, previous keratitis or anterior segment inflammation, or previous corneal trauma with resulting corneal scar was excluded from the study. The protocol was in accordance with Duke University Institutional Review Board guidelines, and informed consent was obtained from all study participants, in compliance with the tenets of the Declaration of Helsinki.
Genotyping for Single Nucleotide Polymorphisms (SNPs)
Standard protocols were used to collect blood from affected individuals and their relatives. Briefly, approximately 10 mL of blood was collected and frozen at −20°C. DNA was extracted from whole blood after isolation of blood lymphocytes (Puregene system; Gentra Systems, Minneapolis, MN). DNA samples were genotyped according to the manufacturer’s instruction on a bead array (Illumina), SNP linkage panel IV, which includes 5858 markers.
Each sample included in the analysis was genotyped in four multiplexed assays containing up to 1536 markers (SNPs) per tube. The assay products from these were hybridized to high-density, bead-based microarrays, and were imaged on a submicron-resolution scanner. The Illumina genotyping system is fluorescence based, with one color specifying the first allele (A) and a second color specifying the second allele (B). Homozygous samples (AA or BB) therefore generate a signal exclusively/ predominantly in the corresponding single color only, whereas heterozygous samples (AB) generate a signal in both colors. Since samples vary in their performance, each marker produces three clusters of results within color-signal space, one for each genotype. As part of the Illumina genotyping process, a quality measure known as a “GenCall score” is determined for each genotype. This metric measures how close a genotype is to the center of the cluster of other samples assigned the same genotype, compared with the centers of the clusters for the two other genotypes. The metric varies from 0 to 1, such that the higher the value, the more reliable the genotype. A sample giving a signal close to the center of a cluster that is tight and distinct from the other two clusters will have a high GenCall score.
Linkage Analysis
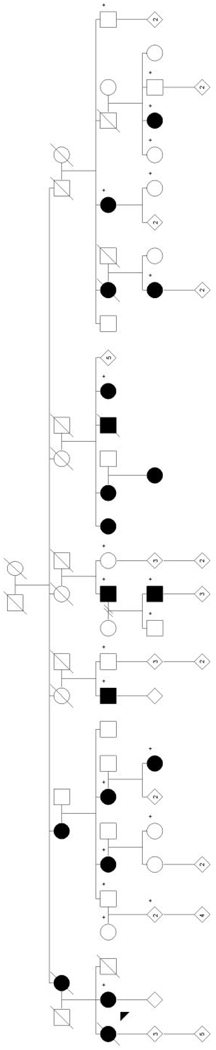
Ninety-two individuals (56 affected and 36 unaffected) from 22 families with FECD were used as the basis for our linkage screen. The cohort included one large multigenerational family (family 1), which included 18 family members (12 affected) (Fig. 1) and 21 small multiplex families that contained two to eight family members.
FIGURE 1.
Pedigree of the large multigenerational family in this study (family 1). +, individuals from whom DNA was obtained; filled symbols: affected individuals; numbers in diamond symbols: unaffected individuals; arrowhead: the proband.
The FASTLINK and HOMOG programs (http://linkage.rockefeller.edu/providedinthepublicdomainbyRockefellerUniversity, New York, NY) were used for parametric two-point linkage analysis. Because the mode of inheritance for FECD is unknown, parametric analyses were performed using both autosomal dominant and autosomal recessive penetrance models with disease allele frequencies of 0.001 and 0.01, respectively. This analysis generates a heterogeneity LOD (HLOD) score, the maximum θ, for each marker. For multipoint linkage analysis, in which multiple markers are used to infer linkage information Allegro15 was used to perform nonparametric multipoint analysis on the chromosomes with significant two-point HLOD scores greater than 1.5. All four Allegro analyses models were used: exponential all, exponential pairs, linear all, and linear pair. All linkage analyses were performed for the overall dataset (22 families [22Fam]), the single large family identified (family 1 [Fam1]), and all families excluding family 1 (21 families [21Fam]). This analysis allowed us to address the hypothesis that different genetic mechanisms underlie the development of FECD in multigenerational families compared with sporadic occurrence within singleton families.
Sequence Analysis of COL8A2
We generated de novo sequence from exon 2 of COL8A2 containing the previously identified mutations. Oligonucleotide primers were designed to completely incorporate exons 2 and intron boundaries using Primer 3,16 and the PCR amplicons generated from all 92 individuals in our dataset were sequenced (model 3730 sequencer; Applied Biosystems, Inc., Foster City, CA) using the manufacturer’s recommended sequencing protocols. Sequence data were assembled and analyzed (Sequencher; Gene Codes, Ann Arbor, MI).
RESULTS
Analysis of Linkage Data
In our samples, 5800 (98%) of the markers generated potentially usable data that satisfied minimum Illumina quality standards, which include one marker on the Y and 26 markers on the XY region. A total of 5754 markers (5455 autosomal markers and 299 X-linked markers) remaining in the linkage analysis after several quality control check points including GenCall scores (>0.7), sex errors, Mendelian errors, and Hardy-Wein-berg equilibrium. All analyses were based on 5755 SNPs.
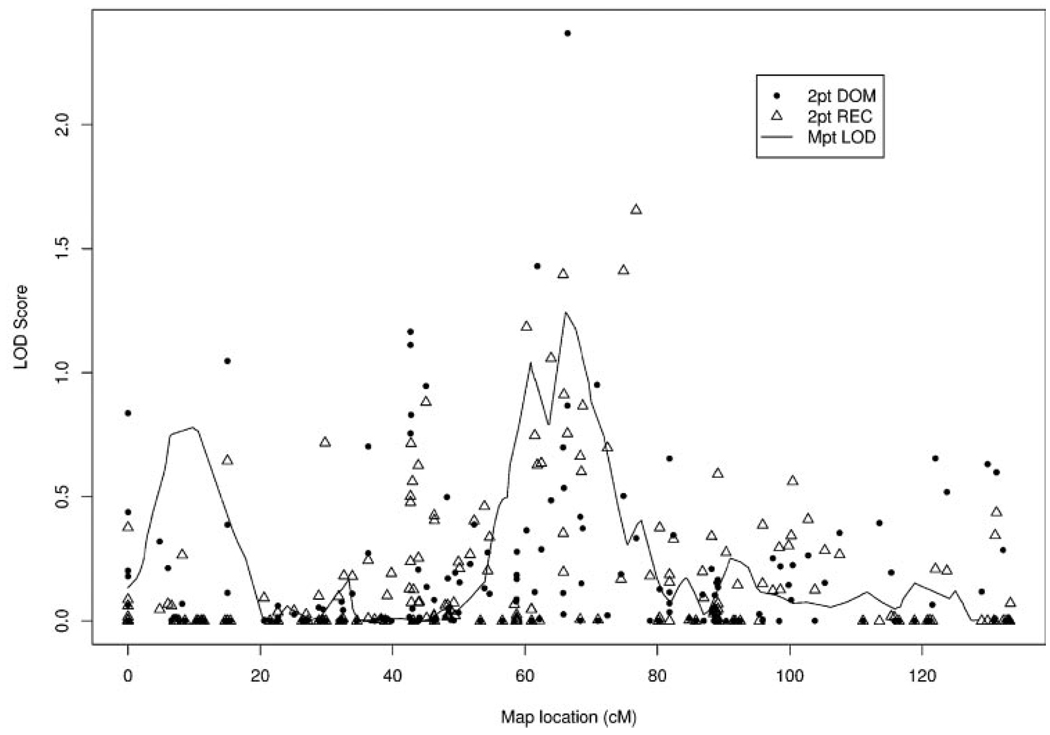
Two-point analysis from FAST LINK and HOMOG program with HLOD scores > 1.5 were found with markers in chromosomes 1, 7, 15, 17, and X (Table 2). Among all families studied, 15q showed the highest two-point HLOD score (3.26) for the recessive model and 2.48 for the dominant models (Fig. 2).
TABLE 2.
Summary of Two-Point Analysis
| Two-Point HLOD Results Dominant and (Recessive) Models† |
|||||
|---|---|---|---|---|---|
| Chromosome | SNP Marker | Location (cM)* | All 22Fam LOD Score | 21Fam LOD Score | Faml LOD Score |
| 1 | rs760594 | 52.94 | 2.02 (2.21) | 1.73 (1.82) | −0.01 (0.28) |
| 7 | rsl047035 | 108.67 | 1.22 (2.69) | 0.70 (2.13) | 0.3 (0.25) |
| rs257376 | 116.75 | 1.92 (3.22) | 139 (3.01) | 0.64 (0.04) | |
| 15 | rs352476 | 66.40 | 2.48 (3.26) | 1.94 (2.70) | 0.31 (0.27) |
| rsl075991 | 70.93 | 1.15 (3.15) | 1.32 (3.23) | −0.01 (−0.01) | |
| 17 | rs938350 | 121.21 | 2.10 (1.09) | 0.78 (0.99) | 1.22 (0.07) |
| X | rs548996 | 69.31 | 0.15 (1.80) | 1.71 (0.09) | 0.43 (0.09) |
Map distance is in Kosambi based on the deCODE genetic map.17
Dominant model LOD scores are given followed by recessive model LOD scores in parentheses.
LODs > 1.5 are highlighted in bold.
FIGURE 2.
Two-point and multipoint linkage results of chromosome 15 from all families. 2pt DOM, two-point linkage analysis dominant model; 2pt REC, two-point linkage analysis recessive model; Mpt LOD, multipoint linkage analysis LOD score.
Table 3 summarizes the peak markers with multipoint analysis LOD scores > 1. Chromosomes 1, 7, 15, 17, and X in multipoint analysis had LOD scores > 1. The finding on chromosome 1 is noteworthy, as this region is only 3 Mb away from the COL8A2 marker rs760594. Multipoint linkage analysis identified a significant LOD score of 1.97 on chromosome 7 and a linkage peak on 15q with LOD > 1. Similarly, we found potential regions of linkage on chromosomes 7 and X, when using the total dataset. Analysis of 21FAM indicates that these findings on chromosomes 7, 15, and X are derived from the small family aggregates.
TABLE 3.
Multipoint Analysis
| Analysis Models |
||||||
|---|---|---|---|---|---|---|
| Chromosome (Dataset) | SNP Marker | Map Position cM | LOD Exponential Pair | LOD Exponential All | LOD Linear Pair | Linear All |
| 1 (Faml) | rs270487 | 97.72 | 1.04 | 0.3 | 0.41 | 0.81 |
| rsl052238 | 194.99 | 1.91 | 1.91 | 0.33 | 0.92 | |
| 1 (21Fam) | rs315537 | 108.34 | 1.50 | 1.49 | 1.07 | 1.07 |
| rs716760 | 180.51 | 1.78 | 1.22 | 1.06 | 1.06 | |
| 7 (22Fam) | rs740295 | 38.65 | 1.97 | 1.85 | 1.44 | 1.37 |
| rs918980 | 58.86 | 1.62 | 0.94 | 1.84 | 1.76 | |
| 15 (22Fam) | rsl075456 | 65.73 | 1.04 | 0.84 | 1.15 | 1.22 |
| rsl445020 | 71.048 | 1.24 | 0.97 | 0.98 | 1.02 | |
| 17 (Faml) | rs938350 | 121.21 | 1.99 | 1.99 | 0.44 | 1.22 |
| 17 (22Fam) | rsl530348 | 120.69 | 1.57 | 2.53 | 1.12 | 1.45 |
| X (22Fam) | rs7881297 | 122.92 | 1.374 | 1.388 | 1.013 | 1.048 |
LODs > 1.5 are highlighted in bold.
The highest multipoint LOD score was from chromosome 17 which corresponds to LOD 2.53 using the overall dataset (22Fam). Chromosome 17, long arm (∼107–126-cM region), showed consistent results, with LOD > 2 by both two-point and multipoint analyses on the 22 Fam dataset, although the majority of linkage for 17q appeared to be contributed by family 1 (Table 2).
Identification of Reported COL8A2 Mutations
The maximum two-point HLOD score of 2.21 at rs760694 for the overall dataset is located only 3 Mb away from the COL8A2 gene on chromosome 1. We therefore screened all 92 samples in our dataset for the previously reported L450W and Q455K COL8A2 mutations,6–8 but we failed to identify these mutations in our FECD family cohort (data not shown). Although this finding does not exclude the possibility of COL8A2 as a candidate gene for the development of FECD, additional screening of the whole gene and putatively regulatory regions within additional samples is needed.
DISCUSSION
Our understanding of the genetic mechanisms associated with the development of FECD has lagged behind that of many other ocular disorders. To date, genome-wide linkage studies involving a large number of families with FECD have not been reported; indeed, most FECD genetic studies have involved only a few large multigenerational families. Although our study of 22 families is small compared with linkage studies for other disorders, it is, thus far, the largest study performed in FECD, using both multigenerational and complex families. Our finding of a linkage peak on chromosome 1 near the COL8A2 gene suggests that COL8A2 mutations within our current patient cohort may contribute to the peak; however, mutation screening of individuals from our families for the two previously reported COL8A2 mutations with the early-onset variant of FECD6 were not identified. These results raise the possibility that there is an alternative causal gene(s) within the linkage interval, additional mutations are contained within COL8A2, or the COL8A2 gene does not play a role in the late-onset form of FECD.
From both two-point and multipoint analyses, the linkage signals on chromosomes 7, 15, and X were from the overall dataset. Most of the multipoint LOD scores are lower than two-point LOD scores due to the lower LOD scores obtained on the markers near the two-point peak markers. The findings on chromosomes 7, 15, and X indicate that the linkage regions are derived from both small and large family aggregates, supporting a complex genetic cause in FECD. Our results also imply that the linkage signal on 17q was mainly contributed by family 1, a large multigenerational family, since excluding this family from the data set decreased the LOD score to as low as 0.78. In future studies, an increased sample size for family 1 may help to confirm or reject these findings.
In this study we did not find potential linkage to chromosome 13, 18, and 20 as previously reported.12–14 Sundin et al.13,14 found linkage to these chromosomes in one and three large families. It is possible that our families, which consisted mostly of small pedigrees, reflect the complex inheritance pattern of FECD. Alternatively, family 1 illustrates the potential for an autosomal dominant mode of inheritance. Considering these results, one can hypothesize that FECD can be inherited both as autosomal dominant and in a complex fashion.
The search for genes involved in the late-onset form of FECD is ongoing. Our findings from a genetic linkage screen for FECD provide noteworthy leads for future studies. We have identified potential linkage regions on chromosomes 1, 7, 15, 17, and X. It should be noted that this study was based on only 22 families, which may be the reason that we did not find many linkage signals above the classic threshold of LOD score 3.0.18 However, the linkage regions reported in this study may provide guidance for a future search of susceptibility genes for FECD. Clearly, recruiting more families with FECD will help to verify this linkage evidence, as well as identify new linkage regions.
Acknowledgments
The authors thank all the families who generously participated in the study, John Hart for performing the linkage genotyping and sequencing of COL8A2, and Ling Zhang for statistical analysis of the linkage data.
Supported in part by a research grant from the National Eye Institute (R01 EY 016514). NAA has a research career development award from Research to Prevent Blindness.
Footnotes
Disclosure: N.A. Afshari, None; Y.-J. Li, None; M. Pericak-Vance, None; S. Gregory, None; G.K. Klintworth, None
References
- 1.Adamis AP, Filatov V, Tripathi BJ, Tripathi RC. Fuchs’ endothelial dystrophy of the cornea. Surv Ophthalmol. 1993;38:149–168. doi: 10.1016/0039-6257(93)90099-s. [DOI] [PubMed] [Google Scholar]
- 2.Kang PC, Klintworth GK, Kim T, Carlson AN, Afshari NA. Trends in the indications for penetrating keratoplasty 1980–2001. Cornea. 2005;24:801. doi: 10.1097/01.ico.0000157407.43699.22. [DOI] [PubMed] [Google Scholar]
- 3.Afshari NA, Pittard AB, Siddiqui A, Klintworth GK. Clinical study of Fuchs corneal endothelial dystrophy leading to penetrating keratoplasty. Arch Ophthalmol. 2006;124:777–780. doi: 10.1001/archopht.124.6.777. [DOI] [PubMed] [Google Scholar]
- 4.Joyce N. Cell cycle status in human corneal endothelium. Exp Eye Res. 2005;81:629–638. doi: 10.1016/j.exer.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Krachmer JH, Purcell JJ, Jr, Young CW, Bucher KD. Corneal endothelial dystrophy; a study of 64 families. Arch Ophthalmol. 1978;96:2036–2039. doi: 10.1001/archopht.1978.03910060424004. [DOI] [PubMed] [Google Scholar]
- 6.Biswas S, Munier FL, Yardley J, et al. Missense mutations in COL8A2, the gene encoding the a2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Gen. 2001;21:2415–2423. doi: 10.1093/hmg/10.21.2415. [DOI] [PubMed] [Google Scholar]
- 7.Gottsch JD, Zhang C, Sundin OH, Bell WR, Starck WJ, Green WR. Fuchs corneal dystrophy aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci. 2005;46:4504–4511. doi: 10.1167/iovs.05-0497. [DOI] [PubMed] [Google Scholar]
- 8.Gottsch JD, Sundin OH, Liu SH, et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–1939. doi: 10.1167/iovs.04-0937. [DOI] [PubMed] [Google Scholar]
- 9.Urquhart JE, Biswas S, Black GC, Munier FL, Sutphin J. Exclusion of COL8A1, the gene encoding the a2 (VIII) chain of type VIII collagen, as a candidate for Fuchs endothelial dystrophy and posterior polymorphous corneal dystrophy. Br J Ophthalmol. 2006;90:1430–1431. doi: 10.1136/bjo.2006.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aldave AJ, Rayner SA, Salem AK, et al. No pathologic mutations identified in the COL8A1 and COL8A2 genes in familial Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2006;47:3787–3790. doi: 10.1167/iovs.05-1635. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi A, Fujiki K, Murakami A, et al. Analysis of COL8A2 gene mutation in Japanese patients with Fuchs endothelial dystrophy ad posterior polymorphous dystrophy. Jpn J Ophthalmol. 2004;48:195–198. doi: 10.1007/s10384-003-0063-6. [DOI] [PubMed] [Google Scholar]
- 12.Vithana EN, Morgan PE, Ramprasad V, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy (FECD) Hum Mol Genet. 2008;17(5):656–666. doi: 10.1093/hmg/ddm337. [DOI] [PubMed] [Google Scholar]
- 13.Sundin OH, Jun AS, Broman KW, et al. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13p Tel-13q1213. Invest Ophthalmol Vis Sci. 2006;47:140–145. doi: 10.1167/iovs.05-0578. [DOI] [PubMed] [Google Scholar]
- 14.Sundin OH, Broman KW, Change HH, Vito EC, Starck WJ, Gottsch JD. A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2-q21.32. Invest Ophthalmol Vis Sci. 2006;47:3919–3926. doi: 10.1167/iovs.05-1619. [DOI] [PubMed] [Google Scholar]
- 15.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky H. Primer 3 on the WWW for general users and for biological programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 17.Kong A, Gudbjartsson DF, Sainz J, et al. A high resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 18.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]