Abstract
OBJECTIVE
Neurogenin 3 plays a pivotal role in pancreatic endocrine differentiation. Whereas mouse models expressing reporters such as eGFP or LacZ under the control of the Neurog3 gene enable us to label cells in the pancreatic endocrine lineage, the long half-life of most reporter proteins makes it difficult to distinguish cells actively expressing neurogenin 3 from differentiated cells that have stopped transcribing the gene.
RESEARCH DESIGN AND METHODS
In order to separate the transient neurogenin 3 –expressing endocrine progenitor cells from the differentiating endocrine cells, we developed a mouse model (Ngn3-Timer) in which DsRed-E5, a fluorescent protein that shifts its emission spectrum from green to red over time, was expressed transgenically from the NEUROG3 locus.
RESULTS
In the Ngn3-Timer embryos, green-dominant cells could be readily detected by microscopy or flow cytometry and distinguished from green/red double-positive cells. When fluorescent cells were sorted into three different populations by a fluorescence-activated cell sorter, placed in culture, and then reanalyzed by flow cytometry, green-dominant cells converted to green/red double-positive cells within 6 h. The sorted cell populations were then used to determine the temporal patterns of expression for 145 transcriptional regulators in the developing pancreas.
CONCLUSIONS
The precise temporal resolution of this model defines the narrow window of neurogenin 3 expression in islet progenitor cells and permits sequential analyses of sorted cells as well as the testing of gene regulatory models for the differentiation of pancreatic islet cells.
The mature pancreas is composed of exocrine (acinar and duct cells) and endocrine (α-, β-, δ-, ε-, and PP-cells) compartments. The differentiation of these distinct cell types is regulated by the coordinated expression of numerous transcription factors (1–3). Among these transcription factors, neurogenin 3 (Neurog3), a member of the basic helix-loop-helix transcription factor family, plays essential roles in initiating endocrine differentiation during embryonic development, regeneration, and transdifferentiation into functional insulin-producing cells (4–9). In addition, the transient nature of Neurog3 expression makes it a useful marker for uniquely identifying endocrine progenitor cells—cells that have committed to the endocrine lineage but have not yet differentiated into hormone-producing endocrine cells (10,11).
Mouse models expressing fluorescent reporter proteins have been used to sort specific cell populations. For example, cells sorted from Ngn3-eGFP mouse lines generated by different groups have been used to examine gene expression profiles during pancreatic endocrine differentiation (12,13). However, because of the long half-life (14), fluorescent reporter proteins persist after the Neurog3 gene itself has shut off; thus, the fluorescent cell population includes cells at different stages of differentiation. Destabilized fluorescent proteins have shorter half-lives but lower fluorescence (15). In addition, sorting cells at earlier time points may decrease the overlap with more differentiated cells as described previously (13); however, this approach cannot be used at later time points or for distinguishing more mature cells.
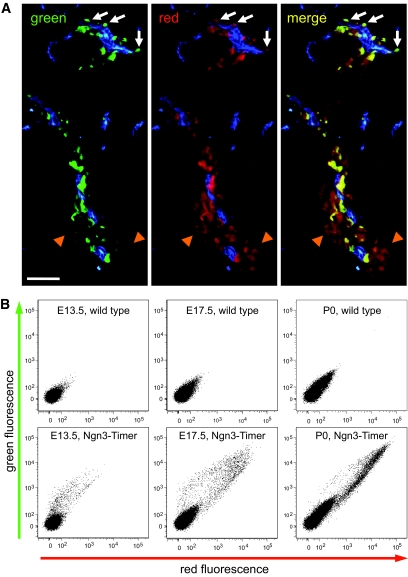
To solve this problem, we developed a novel transgenic mouse model (Ngn3-Timer) in which human NEUROG3 upstream and downstream sequences were used within a bacterial artificial chromosome (BAC) to drive expression of DsRed-E5, a variant of the Discosoma sp. red fluorescent protein that shifts its fluorescence emission peak from green to red in a time-dependent manner (16). Using fluorescence microscopy, green fluorescence could be detected in developing pancreata of Ngn3-Timer embryos as early as embryonic day 9.5 (E9.5) (data not shown). Both green and red fluorescent signals were readily detected in developing pancreata of Ngn3-Timer embryos from E12.5 to E18.5, whereas predominantly red fluorescence was observed at postnatal day 7 (P7) (supplemental Fig. 1, available in the online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0390/DC1), consistent with previous reports that few Neurog3-expressing cells persist after birth (5). At E17.5, histological analyses detected green-dominant and green/red double-positive fluorescent cells in close apposition with the ductal lumen, whereas red-dominant cells appeared in islet-like clusters (Fig. 1A), supporting a model whereby cells in the endocrine lineage emerge from ductal regions as Neurog3-positive cells and migrate away from the ductal region as differentiation progresses. Consistent with this model, staining for Neurog3 protein overlapped with predominantly green-dominant cells and some green/red double-positive cells (supplemental Fig. 2), whereas insulin staining overlapped with green/red double-positive cells and red-dominant cells (supplemental Fig. 3).
FIG. 1.
Expression of the DsRed-E5 fluorescent protein in developing pancreas. A: The pancreas was dissected from an Ngn3-Timer embryo at E17.5 and stained with DBA lectin, a marker of pancreatic duct (blue). Green-dominant cells detected within ducts are labeled with white arrows; green/red double-positive and red-dominant cells (yellow or orange in color, right panel) are labeled with orange arrowheads. Scale bar: 50 μm. B: Pancreata from wild-type (A–C) and Ngn3-Timer mice (d–F) were dissociated at E13.5, E17.5, and P0 and analyzed using flow cytometry. Green fluorescence is shown on the vertical axis and red fluorescence on the horizontal axis. (A high-quality digital representation of this figure is available in the online issue.)
To quantify the shift in fluorescence during endocrine cell differentiation, flow cytometric analyses were performed with dissociated cells isolated from Ngn3-Timer mice. Labeled cells isolated from E13.5 pancreata emitted fluorescence predominantly in the green channel; however, a progressive increase in red fluorescence was observed in older embryos until P0, when the number of green-dominant cells dramatically declined compared with the earlier time points and most labeled cells fluoresced in the green/red double-positive area (Fig. 1B). These data support the hypothesis that the DsRed-E5 protein initially expressed in the Neurog3 lineage emits in the green spectrum and that as the cells turn off Neurog3 expression, age, and differentiate, it shifts to the red emission spectrum.
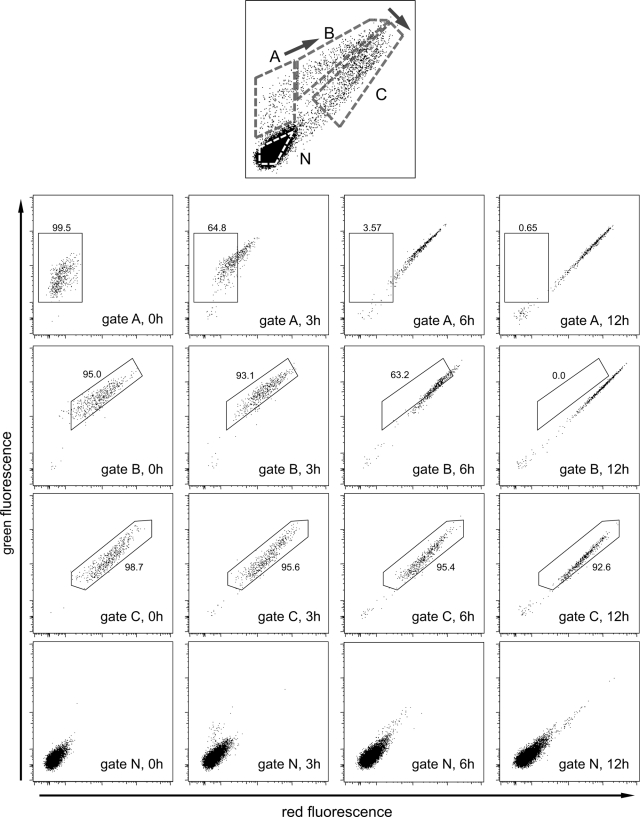
To verify this hypothesis and estimate the temporal resolution of this model, fluorescent cells were sorted by a fluorescence-activated cell sorter (FACS) into four different populations, placed in culture, and then reanalyzed by flow cytometry at various time points following culture. Green-dominant cells, sorted from gate A in Fig. 2, converted to green/red double-positive within 6 h, whereas green/red double-positive cells in gate B converted to the lower green/red ratio (red dominant) of gate C cells within 12 h. Therefore, the green-dominant cells were within a 6-h time window after initial DsRed-E5 expression and green/red double-positive cells within a 12-h time window. On the other hand, the fluorescent cells sorted using gate C changed little over 12 h, presumably because of the long half-life of DsRed-E5.
FIG. 2.
Time-dependent shift of fluorescence in sorted cells after culture. Ngn3-Timer pancreata were dissociated at E17.5 and sorted by FACS. The sorted cells from the gates shown were analyzed immediately after FACS by flow cytometry (first column) or placed in culture and analyzed by flow cytometry after 3 h (second column), 6 h (third column), and 12 h (fourth column).
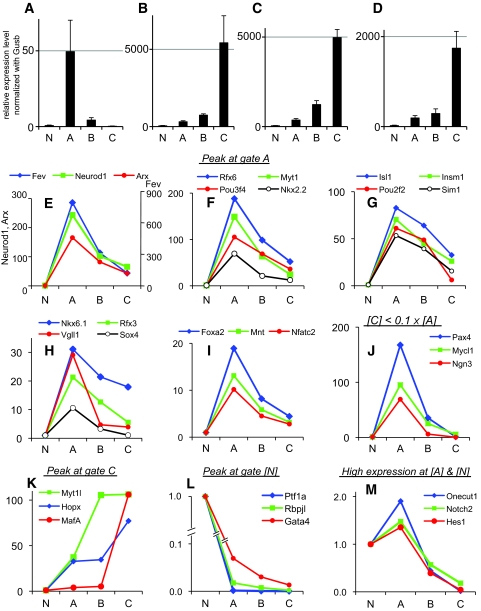
Real-time PCR analyses performed on RNA from cells sorted by FACS into the four populations defined in Fig. 2 confirmed the sequential maturation of these cells. The expression level of Neurog3 was highest in green-dominant cells and dramatically decreased in green/red double-positive cells (Fig. 3A), reflecting the rapid downregulation of Neurog3 once it has initiated endocrine differentiation. Expression of the islet hormones peaked later (Fig. 3B–D), with the mRNA encoding insulin and glucagon highest in cells from gate C, the most mature population.
FIG. 3.
Temporal transcriptome analysis in the pancreata of Ngn3-Timer embryos. A–D: Ngn3-Timer pancreata were dissected at E17.5 and sorted by FACS into four gates (gates A, B, C, and N). The sorted cell populations were analyzed by real-time RT-PCR for mRNA-encoding transcriptional regulators and endocrine hormones. All expression levels were normalized to β-glucuronidase. Neurog3 (A), insulin1 (B), insulin2 (C), glucagon (D). E–M: TaqMan array was performed for 145 transcription factors. Expression levels are shown relative to the level in gate N. Each data point represents the mean of three independent experiments.
To gauge the expression of known pancreatic transcriptional regulators (13,17) in these differentiating cell populations, TaqMan RT-PCR assays were performed with RNA from cells isolated by FACS from E17.5 Ngn3-Timer pancreata. The expression levels of 145 genes were normalized to β-glucuronidase, and the relative level of mRNA from each gate was determined with respect to the value in nonfluorescent cells (gate N). For 21 genes, expression in the earliest progenitor cells (gate A) exceeded expression in the nonfluorescent cells by more than 10-fold (gate [A]/gate [N] >10; Fig. 3E–J and supplemental Table).
Among these genes, three (Neurog3, Pax4, and Mycl1) demonstrated a sharp expression peak in gate A, with a subsequent decline of more than 10-fold from gate A to gate C (Fig. 3J)—which is consistent with the conclusion that these genes have expression restricted to the islet progenitor cells. These data confirm previous evidence that Neurog3 directly activates transient Pax4 gene expression followed by repression by Pax4 (18). The profile of Mycl1 paralleled those of Neurog3 and Pax4; therefore, it may also play a specific role in the transient endocrine progenitor cells.
Several of the other genes, including NeuroD, Nkx2. 2, Myt1, and Insm1, are also known targets of Neurog3 but persist in mature islet cells (1–3). In addition, this approach identified several factors with known expression in the pancreas (13,17) but without known roles in the Neurog3 pathway, including Mycl1, Mnt, Nfatc2, Pou2f2, Sim1, and Fev. Fev, which encodes an ETS transcription factor and showed the highest relative induction in the gate A cells (Fig. 3E), and we have recently confirmed its expression and function in the islet lineage (Y. Ohta and M.S.G., unpublished data). Finally, Rfx6, a member of the Rfx family of winged-helix transcription factors, has not been studied in the pancreas previously, but these data place it in the Neurog3 pathway and islet lineage.
Of the 145 genes only three, Mafa, Hopx, and Myt1l, showed a robust increase from gate A to gate C (Fig. 3K) that paralleled the expression profiles of Ins1 and Ins2 (Fig. 3B and C). MafA appears late in pancreatic development specifically in mature β-cells (19) and directly transactivates the insulin gene in conjunction with NeuroD1 and Pdx1, which are expressed earlier. Therefore, MafA may play a critical role in the final maturation of β-cells. It follows that Hopx and Myt1l potentially also contribute to the maturation of the endocrine cells.
As expected, Ptf1a and Rbpjl, known exocrine genes, were confined to gate N (Fig. 3L). On the other hand, mRNA encoding Hes1, Notch2, and Onecut1 (Hnf6), which regulate the expression of Neurog3 (4,20,21), persisted in gate A but rapidly declined as cells matured (Fig. 3M).
In summary, the Ngn3-Timer mouse provides a useful in vivo tool for temporal dissection of the differentiation processes. The model allowed us to accurately isolate cells from distinct narrow time points along the differentiation pathway and also to study the characteristics of those cells. We have used this tool to identify genes that likely play unique roles at these different steps of differentiation. The availability of this tool also permits relatively facile methods for studying signals that impact the initiation or completion of differentiation, the rate of differentiation, or the proliferation or death of differentiating cells. Likewise, similar transgenic models could be used to study the temporal features of differentiation in other cell populations.
RESEARCH DESIGN AND METHODS
Generation of Ngn3-Timer transgenic mouse.
Starting with a BAC containing 134 kb upstream and 30 kb downstream of the human NEUROG3 gene (clone RP11-343J3 [Sanger Institute, U.K.]), the NEUROG3 coding sequence was replaced by the DsRed-E5 coding region (pTimer; Clontech, Palo Alto, CA) via homologous recombination. The modified BAC was purified using cesium chloride gradient ultracentrifugation, dialyzed, and microinjected into the pronuclei of B6SJL/F1 oocytes to generate the Ngn3-Timer transgenic mice. The mice were genotyped by PCR using the forward primer 5′-cgctgctcatcgctctcta-3′ in the 5′-flanking sequence of NEUROG3 and the reverse primer 5′-GGTGTTGTGGCCCTCGTAG-3′ in the coding region of DsRed-E5. A total of six lines of Ngn3-Timer mice were generated and analyzed by means of flow cytometry and microscopy, and two highly fluorescent lines were maintained for further analyses. Both high-expressing lines gave the same expression pattern. One of these two lines was used for all of the studies reported here.
Mice were housed on a 12-h light/dark cycle in a controlled climate. Timed matings were carried out with E0.5 being set as midday of the day of discovery of a vaginal plug. All studies involving mice were approved by the University of California San Francisco Institutional Animal Care and Use Committee.
Whole-mount observation and histological analyses.
Transgenic Ngn3-Timer embryos were killed from E12.5 to P7, and macroscopic appearance and fluorescence of the Ngn3-Timer mice were examined using the fluorescent dissecting microscope. For histological analyses, tissues were fixed in 4% paraformaldehyde in PBS at 4°C, washed in PBS alone, and then immersed into sucrose solution in PBS overnight at 4°C. The next day, the tissues were embedded and frozen in Tissue-Tek (OCT Compound, Sakura). Tissues were sectioned at 6 μm, permeabilized with 0.1% Triton X-100, blocked with 5% goat serum (Invitrogen) and incubated with biotinylated lectin Dolichos biflorus agglutinin (DBA; Vector Laboratories) diluted 1:200 in PBS, and then visualized using Alexa Fluor 350–conjugated streptavidin (Molecular Probes, Eugene, OR).
Pancreatic cell dispersion and flow cytometry.
The Ngn3-Timer transgenic mice were distinguished from control littermates using the fluorescent dissecting microscope, and the whole pancreata were manually dissected from other organs. Pancreata were treated with 0.05% trypsin/0.53 mmol/l EDTA (Invitrogen, Carlsbad, CA) at 37°C for 5 min, and the digestion was inactivated by addition of FBS. The dissociated cells were resuspended in FACS buffer (2% FBS in PBS) and then analyzed using an LSRII flow cytometer (PerkinElmer) or sorted using a MoFlo cell sorter (Dako Cytomation, Carpinteria, CA). Dead cells were excluded with DNA dye TO-PRO-3 (Molecular Probes, Eugene, OR).
Cell culture.
The Ngn3-Timer pancreata were dissociated at E17.5 and sorted by FACS into the different populations shown. The sorted cells were incubated in growth medium (DME H-16 50%/F-12 50% with 10% FBS, antibiotics, and insulin-transferrin-selenium) at 37°C in 5% CO2. After 2–12 h of culture, the cells were dissociated with 0.05% trypsin/0.53 mmol/l EDTA, transferred into FACS tubes, and then analyzed using an LSRII flow cytometer (PerkinElmer).
Real-time quantitative PCR.
Total RNA was extracted from cells sorted by FACS from three independent groups of E17.5 Ngn3-Timer embryos using the RNeasy Plus micro kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The quality and quantity of extracted RNA were assessed with the Agilent 2100 Bioanalyzer using the RNA 6000 Pico Assay kit (Agilent Technologies, Santa Clara, CA), and 10 nanograms of the total RNA were linearly amplified and converted into cDNA with the NuGEN WT-Ovation RNA amplification system (NuGen, San Carlos, CA). Individual cDNAs were quantified by real-time PCR using the TaqMan low density array system (Applied Biosystems, Foster City, CA), designed for pancreas-specific genes including endocrine hormones and transcription factors (supplemental Table). Gene expression levels of the assayed genes were normalized to the expression levels of β-glucuronidase. The means ± SE are shown in the supplemental Table.
Supplementary Material
Acknowledgments
This work was supported by Larry L. Hillblom Foundation Grant 2007/1B, Juvenile Diabetes Research Foundation Fellowship Award 3-2007-721, National Institutes of Health Grant R01 DK021344, a grant from the Nora Eccles Treadwell Foundation, and core laboratories supported by National Institutes of Health Grant P30 DK63720.
No potential conflicts of interest relevant to this article were reported.
We thank Gerold Grodsky, Francis Lynn, and all members of the German laboratory for helpful advice and criticism and Xuyu Zhou, Shuwei Jiang, and Shigeru Oshima for help with experiments.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Zaret KS: Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet 2008; 9: 329– 340 [DOI] [PubMed] [Google Scholar]
- 2.Oliver-Krasinski JM, Stoffers DA: On the origin of the beta cell. Genes Dev 2008; 22: 1998– 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gittes GK: Developmental biology of the pancreas: a comprehensive review. Dev Biol 2009; 326: 4– 35 [DOI] [PubMed] [Google Scholar]
- 4.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H: Notch signalling controls pancreatic cell differentiation. Nature 1999; 400: 877– 881 [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl G, Dierich A, LeMeur M, Guillemot F: neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 2000; 97: 1607– 1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu G, Dubauskaite J, Melton DA: Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 2002; 129: 2447– 2457 [DOI] [PubMed] [Google Scholar]
- 7.Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M: Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci U S A 2004; 101: 13245– 13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, D'Hoker J, Stange G, Bonne S, De Leu N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, Bouwens L, Scharfmann R, Gradwohl G, Heimberg H: Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 2008; 132: 197– 207 [DOI] [PubMed] [Google Scholar]
- 9.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA: In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008; 455: 627– 632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD, Serup P: Independent development of pancreatic alpha- and β-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes 2000; 49: 163– 176 [DOI] [PubMed] [Google Scholar]
- 11.Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS: Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 2000; 127: 3533– 3542 [DOI] [PubMed] [Google Scholar]
- 12.Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA: Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development 2004; 131: 165– 179 [DOI] [PubMed] [Google Scholar]
- 13.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH: Defining pancreatic endocrine precursors and their descendants. Diabetes 2008; 57: 654– 668 [DOI] [PubMed] [Google Scholar]
- 14.Corish P, Tyler-Smith C: Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 1999; 12: 1035– 1040 [DOI] [PubMed] [Google Scholar]
- 15.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR: Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 1998; 273: 34970– 34975 [DOI] [PubMed] [Google Scholar]
- 16.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P: “Fluorescent timer”: protein that changes color with time. Science 2000; 290: 1585– 1588 [DOI] [PubMed] [Google Scholar]
- 17.Kong YM, Macdonald RJ, Wen X, Yang P, Barbera VM, Swift GH: A comprehensive survey of DNA-binding transcription factor gene expression in human fetal and adult organs. Gene Expr Patterns 2006; 6: 678– 686 [DOI] [PubMed] [Google Scholar]
- 18.Smith SB, Gasa R, Watada H, Wang J, Griffen SC, German MS: Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J Biol Chem 2003; 278: 38254– 38259 [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R: The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A 2004; 101: 2930– 2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP: Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol 2000; 20: 4445– 4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, German MS: Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 2001; 50: 928– 936 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.