Abstract
Defining the epitope specificity of CD8+ T cells is an important goal in autoimmune and immune-mediated disease research. We have developed a translational molecular approach to determine the epitope specificity of CD8+ T cells using the Theiler’s murine encephalomyelitis virus (TMEV) model of multiple sclerosis (MS). TMEV-specific CD8+ T cells were isolated from brains and spleens of 7-day TMEV-infected C57BL/6J mice and stimulated by Cos-7 cells that were co-transfected with expression vectors encoding the Db class I molecule along with overlapping segments of the TMEV genome. Both brain-infiltrating and spleen-derived CD8+ T cells expressed IFN-γ when Cos-7 cells were co-transfected with Db class I molecule and the TMEV genomic segment that encoded the immunodominant TMEV epitope. This demonstrated that peripheral and brain-infiltrating CD8+ T-cell responses were focused on peptide epitope(s) encoded by the same region of the TMEV genome. We propose that a similar molecular approach could also be used to determine the antigen specificity of suppressor CD8 T cells by the measurement of transforming growth factor–β (TGF-β) production. In addition, with a randomly generated library and peripheral blood or isolated CSF CD8+ T cells, this would be an effective method of predicting the epitope specificity of CD8+ T cells in human inflammatory CNS diseases, in animal models of MS or other organ-specific inflammatory diseases with a protective or pathogenic role of CD8 T cells.
Keywords: Theiler’s murine encephalomyelitis virus (TMEV), Effector CD8+ T cell, Suppressor CD8+ T cell, Epitope, Genomic library
1. Introduction
The role of central nervous system (CNS)–infiltrating CD8+ T cells is an active area of CNS inflammatory disease research. Studies have demonstrated that CD8+ T cells are prevalent in multiple sclerosis (MS) lesions, outnumbering other lymphocytes at all stages of lesion development [1,2]. CD8+ T cells have also been shown to be clonally expanded in lesions, cerebrospinal fluid (CSF), and peripheral blood, suggestive of a common epitope, or a restricted number of epitopes being recognized by this cell type [2,3]. In addition, it has been demonstrated that T-cell receptor sequences from clonally expanded CD8+ T cells can persist for years indicating ongoing activation of memory CD8+ T-cell clones. Whether these epitope specific CD8+ T cells act as suppressors of inflammation or effectors of pathology remains to be determined.
A major hypothesis generated through human tissue and animal model studies have provided evidence for a suppressor role of CD8+ T cells. Work in human samples has relied on characterizing functional aspects of cultured CD8+ T cells derived from the peripheral blood of MS patients. Findings from this work have suggested that a deficiency of suppressor function by CD8+ T cells may be linked to MS pathologic conditions [4]. The majority of suppressor CD8+ T-cell work has been performed in the experimental allergic encephalomyelitis (EAE) animal model of MS. In EAE, the absence of CD8+ T cells results in the worsening of disease or susceptibility to secondary induction of EAE [5–7]. Mechanistic studies have provided evidence that suppressor CD8+ T cells act via T-cell receptor interaction with the Qa-1 molecule presented by activated CD4+ T cells to cull inflammatory responses. Qa-1-independent mechanisms involving cytokine mediated suppression by CD8+ T cells have also been proposed [8,9].
Although not mutually exclusive from the suppressor CD8+ T-cell hypothesis, independent studies also support an effector role for this cell type in the pathology in MS patients. Studies in human tissue have demonstrated that all CNS cell types within an MS lesion have the capacity to express major histocompatability complex (MHC) class I molecules and can potentially be targeted by CD8+ T cell–mediated cytotoxicity [10]. Also supporting a role for cytotoxicity by activated CD8+ T cells is their close proximity to oligodendrocytes and demyelinated axons in brain tissue from MS patients. In EAE models, CD8+ T cells have been shown to be sufficient to adoptively transfer disease [11,12]. In the Theiler murine encephalomyelitis (TMEV) model, many aspects of clinical disease, including axonal dysfunction, demyelination, and motor dysfunction, have been linked to CD8+ T cells [13–16].
To determine the extent CD8+ T cells acquire suppressor and effector functions, the epitope specificity of CD8+ T cells in MS needs to be defined. Numerous approaches have been demonstrated to investigate the epitope specificity of CD8+ T cells in TMEV infection, including the use of overlapping peptide libraries, molecular expression of TMEV proteins, stable transfected cell lines, and recombinant vaccinia virus vectors [17–20]. These approaches have unequivocally demonstrated the utility in defining CD8 T-cell epitopes in TMEV infection. However, none of these previously developed systems are feasible approaches to define CD8 T-cell epitopes in human MS in which there are logarithmically higher numbers of potential antigenic targets and the genomic sequence encoding these antigenic determinants are unknown. We therefore have developed an unbiased translational molecular approach to identify CD8+ T-cell epitopes. In this proof-of-principle study, we successfully used this approach to identify the region of the TMEV genome that encodes the immunodominant epitope recognized by CNS-infiltrating CD8+ T cells in the C57BL/6 mouse strain. Although this mouse strain is resistant to chronic demyelination, as it effectively clears the virus in the acute meningo-encephalitis stage, it has been demonstrated that demyelination may occur in mice with C57BL/6 background by an intravenous injection of the immunodominant peptide on day 7, or by the use of mice deficient in interferon-γ receptor [21,22]. Furthermore, there is no theoretical limitation for this method to detect epitopes in demyelinating strains or to detect polyclonal CD8 T-cell responses. In this study, CD8+ T cells from the spleen were stimulated by the identical TMEV library segment as CD8+ T cells isolated from the brain. This interesting observation strongly suggests a specific focus among CD8+ T cells to a conserved immunodominant epitope, regardless of being isolated from brain or a peripheral lymphoid compartment. This molecular approach with a large organ-specific cDNA library derived from MS lesions, similarly to what has already been accomplished in autoimmune diabetes studies [23], has the potential to be adapted for clinical use with peripheral human CD8+ T cells derived from the blood or the CSF as a noninvasive method to predict the epitope specificity of CNS-infiltrating CD8+ T cells in MS patients.
2. Subjects and methods
2.1. Animals
Five week old male C57BL/6 mice were obtained from Jackson laboratories (Bar Harbor, Maine). Mice were anesthetized with isoflurane and were intracranially infected with the Daniels strain of TMEV. Seven days postinfection the animals were euthanized, and tissue was harvested according to University of Cincinnati Institutional Animal Care and Use Committee standards.
2.2. Generation of TMEV library
Fifteen overlapping segments were amplified from the pDAFL plasmid containing the DA strain of TMEV using high-fidelity polymerase chain reaction (PCR), as presented in Table 1. Each segment was enzymatically digested then inserted into the pcDNA 3.1 His A vector (Invitrogen, Carlsbad, CA). The pcDNA 3.1 His A was digested with either EcoRI and XbaI to accommodate ligation of segments 1, 3, 5, 6, 7, 9, 10, 11, 12, 13, and 15 or BamHI and XbaI to accommodate ligation of segments 2, 4, and 8. In addition, the genomic sequence encoding the immunodominant TMEV peptide VP2121–130 was inserted into the pcDNA 3.1 His B vector.
Table 1.
Overlapping segments amplified from pDAFL plasmid containing Daniels strain of Theiler’s murine encephalomyelitis virus (TMEV).
| Segment | Location in TMEV genome | pcDNA3.1 His A cloning sites |
|---|---|---|
| TMEV 1 | 1038–1596 | EcoRI 5′; XbaI 3′ |
| TMEV 2 | 1536–2097 | BamHI 5′; XbaI 3′ |
| TMEV 3 | 2033–2596 | EcoRI 5′; XbaI 3′ |
| TMEV 4 | 2534–3099 | BamHI 5′; XbaI 3′ |
| TMEV 5 | 3031–3597 | EcoRI 5′; XbaI 3′ |
| TMEV 6 | 3532–4095 | EcoRI 5′; XbaI 3′ |
| TMEV 7 | 4030–4596 | EcoRI 5′; XbaI 3′ |
| TMEV 8 | 4531–5094 | BamHI 5′; XbaI 3′ |
| TMEV 9 | 5032–5596 | EcoRI 5′; XbaI 3′ |
| TMEV 10 | 5533–6096 | EcoRI 5′; XbaI 3′ |
| TMEV 11 | 6031–6597 | EcoRI 5′; XbaI 3′ |
| TMEV 12 | 6532–7095 | EcoRI 5′; XbaI 3′ |
| TMEV 13 | 7033–7597 | EcoRI 5′; XbaI 3′ |
| TMEV 14 | 7531–7971 | EcoRI 5′; XbaI 3′ |
| TMEV L* | 1064–1547 | EcoRI 5′; XbaI 3′ |
Fifteen overlapping segments were amplified from the pDAFL plasmid containing the TMEV genome using high-fidelity polymerase chain reaction. Segments were then digested with the specified restriction enzyme as defined by primer sequences and inserted into the pcDNA 3.1 His A vector to accommodate transfection into the Cos-7 cell line. The TMEV sequences overlap by 30 base pairs to ensure that all potential CD8+ T-cell epitopes are processed for antigen presentation.
Note that the L segment encodes a protein from an alternative reading frame important to TMEV infection.
2.3. Efficient co-transfection of Cos-7 cells with class I molecule and green fluorescent protein
Cos-7 cells (American Type Culture Collection number CRL-1651, African green monkey kidney cells) were plated into Costar 96-well culture plates (Corning Inc., Corning, NY) at approximately 70% confluency and incubated overnight. Cells were transfected using Fugene-6 (Roche Indianapolis, IN) according to the manufacturer’s protocol. Cos-7 cells were co-transfected with green fluorescent protein (GFP) MigR1vector and a vector containing either the Db or Kb MHC class I molecules (provided by Nilabh Shastri, University of California at Berkley, Berkeley, CA) which were expressed by the C57BL/6 strain of mouse. GFP MigR1 was co-transfected as a visual aid to distinguish wells that demonstrated efficient co-transfection. Peak GFP expression occurred in Cos-7 cells within 24–48 hours after transfection.
2.4. Isolation of brain mononuclear cells
Brains were pushed through 100-µm cell strainers into RPMI and 700 µg of collagenase type 4 (Worthington Lakewood, NJ) was added to each 5-ml quantity of slurry. Slurries were then incubated in a water bath at 42°C for 45 minutes. Each 5 ml of slurry was then added to Nalgene 50-ml ultra–high-speed centrifuge tubes (Nalge Nunc International, Rochester, NY) containing 1ml of 10X PBS, 9 ml of Percoll, and 35 ml of RPMI. Cell suspensions were then spun at 10,000 rpm (Sorvall SS-34 rotor) for 30 minutes. A lymphocyte layer was present in approximately the bottom 5 ml of media solution. Excess solution is aspirated off the lymphocyte layer. The lymphocyte layer was then resuspended into Falcon conical tubes, (Becton Dickinson, Franklin Lakes, NJ) and RPMI was added until 50 ml of total volume was reached. Cell suspensions were then spun at 1500 rpm for 10 minutes in a Sorvall Legend RT tabletop centrifuge (Thermo Scientific, Waltham, MA). Media was aspirated off and cell pellets were resuspended in RPMI media.
2.5. Isolation of whole spleen cells
Spleens were pushed though 100-µm cell strainers into RPMI 1640. Cell suspensions were then spun at 1500 rpm for 10 minutes in a Sorvall Legend RT tabletop centrifuge. Media was aspirated off and cell pellets were resuspended in 2ml RPMI and 3 ml ACK (0.15 mol/l NH4Cl, 1.0 mol/l KHCO3, 0.1 mmol/l Na2-ethylenediaminetetraacetate). Cell suspensions were then spun at 1500 rpm for 5 minutes. Media was aspirated off and cell pellets were resuspended in RPMI 1640.
2.6. Purification of CD8+ cells
Isolated lymphocytes from the brain and spleen were resuspended in magnetic-activated cell separation (MACS) buffer (1×PBS and 0.5% bovine serum albumin) and incubated with MACS beads anti-CD8+ antibody (Miltenyi Biotec, Auburn, CA) at 4°C for 15 minutes. Cell suspensions were then diluted with 20 times additional MACS buffer and spun at 1500 rpm for 5 minutes on a tabletop centrifuge. Cell pellets were then resuspended in MACS buffer. Cell suspensions were then filtered through MACS LS separation columns (Miltenyi Biotec) according to the manufacturer’s protocol.
2.7. Intracellular staining of splenocytes
Splenocytes were isolated from 7-day TMEV-infected animals as discussed above. Cells were stimulated with either VP2121–130 peptide or E7 control peptide. Cells were treated according to the manufacturer’s protocol using the BD Cytofix/Cytoperm Plus Fixation/Permeabilization kit (with BD GolgiPlug protein transport inhibitor containing brefeldin A; BD Bioscience, San Diego, CA). Cells treated with E7 control peptide yielded no cells positive for interferon (IFN)–γ(data not shown).
2.8. Staining cells for flow-cytometric analysis
Cells were resuspended in fluorescence activated cell sorter (FACS) buffer (1X phosphate-buffered saline solution, 0.025% sodium azide and 1% fetal calf serum). Brain-derived lymphocytes were stained with Db:VP2121–130 tetramer for 40 minutes followed by 20 minutes of staining with anti-CD8 antibody. Cells were then washed twice with FACS buffer and resuspended in 1% paraformaldehyde. Cos-7 cells were stained with biotinalated anti-Kb,Db primary antibody (BD Pharmingen clone # 28-8-6) for 20 minutes. Cells were washed with FACS buffer and incubated with streptavidin phycoerithrin for 20 minutes. Cells were then washed twice with FACS buffer and fixed with 1% paraformaldehyde. All cell samples were analyzed on a BD LSR II flow cytometer.
2.9. Enzyme-linked immunoabsorbent assay
The eBiosciences Mouse IFN-γ Femto-HS enzyme-linked immunoabsorbent assay (ELISA) kit (cat# 88–8314) (eBiosciences, San Diego, CA) was used to detect the presence of IFN-γ in the media of wells containing virus-specific CD8+T cells plated onto Cos-7 cells expressing GFP and an MHC class I/TMEV peptide complex. IFN-γ detection was performed according to the manufacturer’s protocol.
3. Results
3.1. Generation of the TMEV library
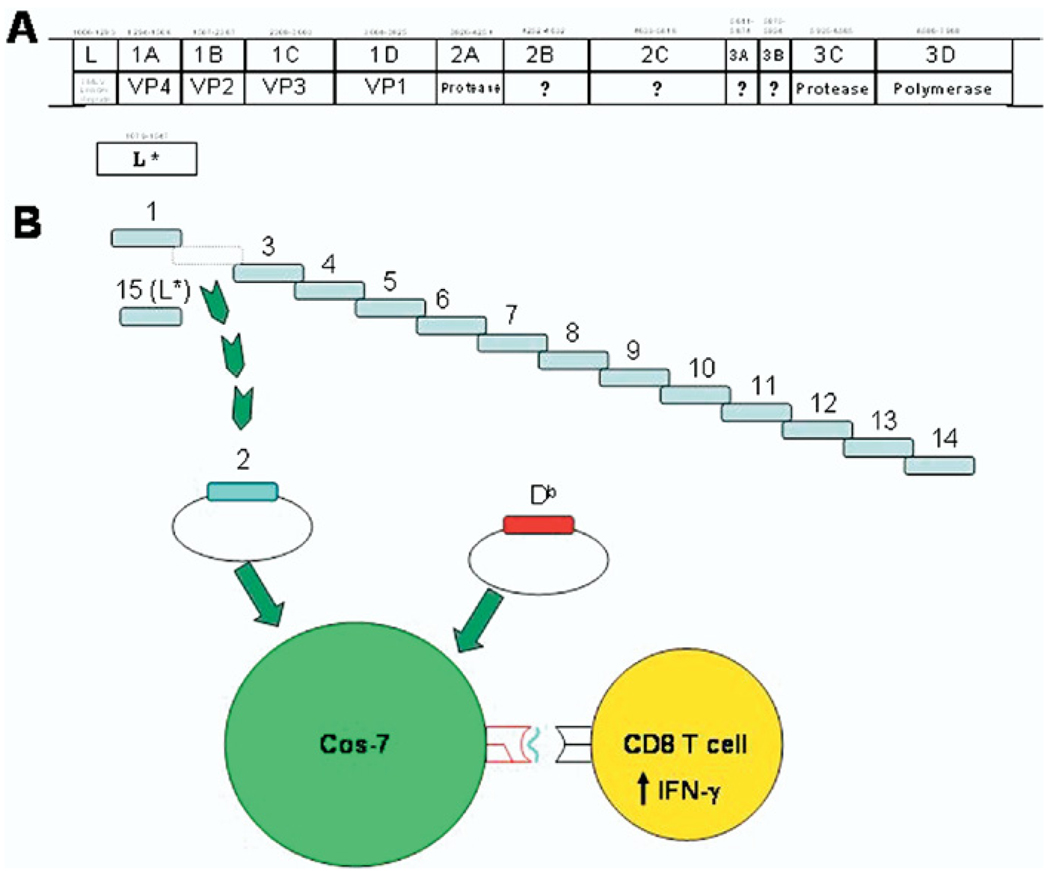
To define the TMEV epitopes recognized by CD8+ T cells directly isolated ex vivo from brains and spleen, 14 segments approximately 560 base pairs in length and overlapping by approximately 30 base pairs were PCR amplified from the TMEV genome (Figure 1A). The overlap in this library enables a comprehensive analysis of all potential class I binding peptides encoded by the TMEV genome. Segment 15 encoded the L* protein, an alternate reading frame that has been noted to produce protein critical to TMEV infection [24]. Each segment of the genome was inserted into the pcDNA 3.1 His A vector and sequenced (Figure 1B). The pcDNA 3.1 His A vector expresses well in mammalian cell lines ideally matched for transfecting Cos-7 cells. Individual wells of Cos-7 cells were then co-transfected with the Db or Kb MHC class I molecule and a TMEV genomic segment. CD8+ T cells isolated from the brain or spleen of a 7-day TMEV-infected mouse were cultured with co-transfected Cos-7 cells. Upon epitope recognition CD8+ T cells expressed IFN-γ into culture media later detected by ELISA. Therefore, IFN-γ was expressed only in wells co-transfected with an MHC class I molecule/TMEV epitope recognized by an ex vivo isolated CD8+ T cell.
Fig. 1.
Molecular approach to detect CD8+ T-cell epitope specificity directly ex vivo. Overlapping segments were amplified from the Theiler’s murine encephalomyelitis virus (TMEV) genome as noted in (A) and Table 1. In (B), overlapping TMEV genomic segments were cloned into pcDNA 3.1 His A. Individual wells of Cos-7 cells are co-transfected with either major histocompatibility (MHC) class I molecule Kb or Db and a specific TMEV genomic segment. At 24–48 hours after transfection, Cos-7 cells express TMEV peptide presenting MHC class I molecules. Upon epitope recognition, central nervous system (CNS)–infiltrating and spleen-derived CD8+ T cells express detectible levels of interferon (IFN)–γ. This molecular approach is designed to enable recognition of all TMEV restricted CD8+ T-cell responses.
3.2. Efficient co-transfection of class I and GFP expressing plasmid vectors in Cos-7 cells
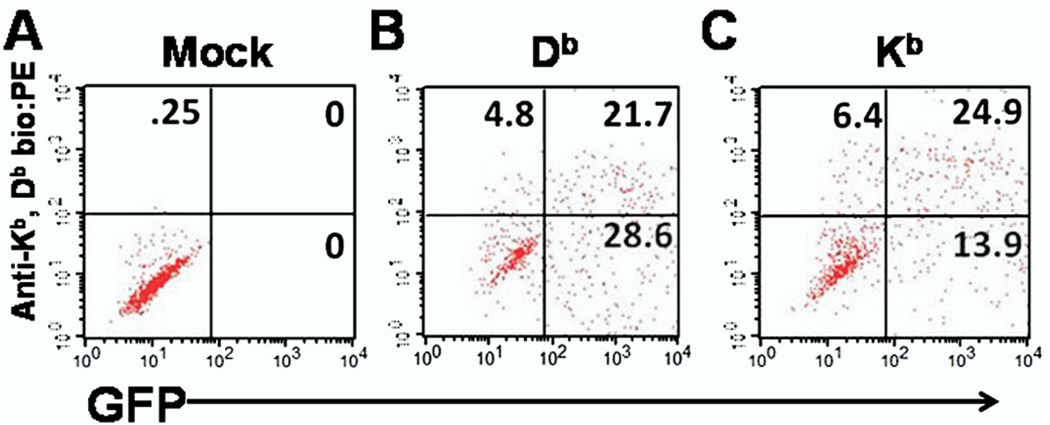
Next we addressed the efficacy of co-transfection of class I molecules and GFP in Cos-7 cells. To accomplish this, Cos-7 cells were sham transfected or co-transfected with Kb or Db and with GFP and analyzed by flow cytometry (Figure 2). Wells sham transfected with empty pcDNA 3.1 His A vector or co-transfected with Kb or Db class I molecule plus GFP were incubated for 48 hours. Cells were then resuspended from wells using cold EDTA PBS and stained with antibody specific for both Kb and Db. No expression of GFP, Kb, or Db was observed in sham-transfected Cos-7 cells (Figure 2A). In contrast, both Kb and Db had marked expression simultaneously with GFP. Approximately 21.7% of co-transfected Cos-7 cells expressed both Db and GFP (Figure 2B). Meanwhile, 24.9% of co-transfected Cos-7 cells expressed both Kb and GFP (Figure 2C). This experiment showed that a significant portion of Cos-7 cells could be co-transfected with class I molecule and a second plasmid encoding GFP, demonstrating the feasibility of this approach to stimulate ex vivo isolated CD8+ T cells with TMEV genome segments.
Fig. 2.
Flow cytometry analysis of efficient class I and green fluorescent protein (GFP) co-transfection in Cos-7 cells. Co-transfected Cos-7 cells were stained with antibody that binds both the Db and Kb class I molecule. Shown are a representative well of (A) cells transfected with empty pcDNA 3.1 His A vector, (B) Db plus GFP expression vectors, and (C) Kb plus GFP expression vectors. Note that cells within the upper right quadrant denotes successful co-expression of GFP and class I molecules.
3.3. Recognition of TMEV specific CD8 T cells derived from brain and spleen is restricted to the library fragment encoding the immunodominant epitope
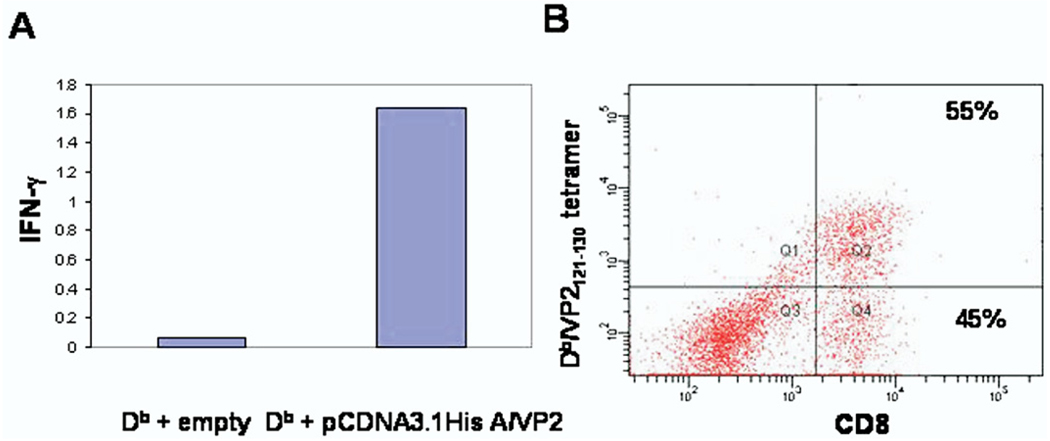
After establishing that Kb and Db class I molecules were expressed on the surface of transfected Cos-7 cells, the efficiency of antigen processing and presentation was tested. Cos-7 cells were co-transfected with Db and pcDNA 3.1 His A/VP2, which encodes the immunodominant VP2121–130 peptide (genomic base pairs 1867–1896) or Db plus an empty pcDNA 3.1 His A vector. The Db:VP2121–130 complex has been established as the immunodominant epitope detected by brain-infiltrating CD8+ T cells in C57BL/6 mouse stain when infected with TMEV [25]. CD8+ T cells isolated from the brains of 7 day TMEV infected C57BL/6 mice were plated onto these transfected Cos-7 cells. A higher signal for IFN-γ was detected by ELISA in wells co-transfected with Db and pcDNA3.1 His A/VP2 than in wells transfected with Db plus empty pcDNA 3.1 His A vector (Figure 3A). CNS-infiltrating CD8+ T cells were also analyzed by flow-cytometric analysis. Consistent with previous reports, approximately 55% epitope dominance for Db:VP2121–130 among CD8+ T cells was observed [15] (Figure 3B). This demonstrated that CD8+ T cells isolated from the CNS could directly recognize the immunodominant Db:VP2121–130 epitope presented by Cos-7 cells co-transfected with Db plus pcDNA 3.1 His A/VP2.
Fig. 3.
Central nervous system (CNS)–infiltrating CD8+ T cells recognize the immunodominant Db:VP2121–130 epitope as determined by our Cos-7 co-transfection system. (A) Enzyme-linked immunoabsorbent assay was used to detect interferon (IFN)–γ expression by central nervous systesm (CNS)–infiltrating CD8+ T cells co-cultured with Cos-7 cells co-transfected with Db plus pcDNA 3.1 His A/VP2. Db plus empty pcDNA 3.1 His A vector resulted in no IFN-γ expression. In (B), Db:VP2121–130 tetramer staining demonstrated that at 7 days postinfection, approximately 55% of all brain-infiltrating CD8+ T cells were specific for the VP2121–130 epitope confirming the results in (A). These data demonstrate that VP2121–130 peptide was successfully expressed, processed, and presented by Db class I molecule demonstrating the feasability of our co-transfection molecular approach to stimulate the response of ex vivo isolated CD8+ T cells. Values presented in (A) are the mean of two wells.
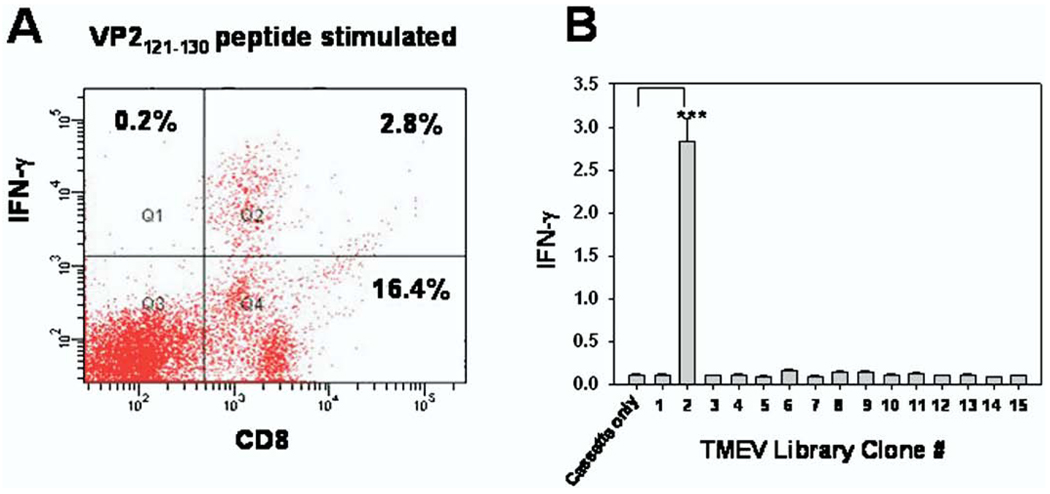
We next wanted to determine the extent that CD8+ T cells isolated from peripheral lympoid compartments were focused on specific TMEV epitopes 1 week post–intracranial infection. Previous work involving the use of Db:VP2121–130 tetramers determined that low numbers of Db:VP2121–130 epitope-specific CD8+ T cells were observed in the spleen of 7-day TMEV-infected C57BL/6 mice [25]. We therefore used the TMEV library approach to determine how focused the CD8+ T-cell response was towards the immunodominant VP2121–130 peptide. Our molecular approach, unlike tetramer technology, would also determine whether there were epitopes other than Db:VP2121–130 recognized by spleen-derived CD8+ T cells. To analyze the epitope recognition of spleen-derived CD8+ T cells, each of the 15 segments of the TMEV library was co-transfected with the Db class I molecule in Cos-7 cells. Magnetically sorted CD8+ T cells isolated from the spleens of 7 day TMEV-infected C57BL/6 mice were cultured with Cos-7 cells co-transfected with each of the 15 library segments and Db. Twenty-four hours after the addition of CD8+ T cells, a sample of culture media was taken from each well to be analyzed for IFN-γ expression. Despite low numbers of detectable Db:VP2121–130 epitope CD8+ T cells isolated from the spleen (Figure 4A), there was significant response towards the TMEV genomic segment 2 encoding the immunodominant VP2121–130 peptide, as confirmed by IFN-γ expression (Figure 4B). These experiments demonstrated that despite the low numbers of Db:VP2121–130–specific spleen-derived CD8+ T cells, epitope recognition remains exclusively focused towards this immunodominant epitope. In addition, no other TMEV genome segments encoded additional epitopes detectable among spleen-derived CD8+T cells with our library system (Figure 4B). Using this system, we have determined that the central and peripheral CD8+ T-cell compartments appear have similar epitope specificity. Clonal expansion and identical T-cell receptor use among CD8 T cells in cerebral spinal fluid and peripheral blood has also been reported in human studies [3].
Fig. 4.
Spleen derived CD8+ T cells are restricted to the epitope derived from the same Theiler’s murine encephalomyelitis virus (TMEV) genomic segment as central nervous system (CNS)–infiltrating CD8+ T cells. (A) Lymphocytes isolated from the spleen of 7-day TMEV-infected mice were cultured for 4 hours with VP2121–130 peptide and assessed for interferon (IFN)–γ expression with intracellular cytokine staining. No IFN-γ–positive cells were found in wells stimulated with the E7 control peptide (data not shown). In (B), CD8+ T cells isolated from the spleen were co-cultured with Cos-7 cells co-transfected with Db and all 15 individual TMEV genomic segments. CD8+ T cells demonstrated specificity for segment 2 of the TMEV genome, which encodes the immunodominant VP2121–130 peptide, when compared with stimulation with Cos-7 cells co-transfected with Db and empty pcDNA 3.1 His A vector as predicted by enzyme-linked immunoabsorbent assay IFN-γ detection (***p = 0.0004). All wells in (B) were performed in quadruplicate.
4. Discussion
In this study we have developed an unbiased molecular approach to determine CD8+ T-cell epitopes without the use of culture systems. Unlike the use of peptide MHC tetramer technology, this system has the potential to analyze the full inflammatory CD8+ T-cell response towards multiple epitopes simultaneously. Using this system in TMEV-infected C57BL/6 mice, we have determined that the CD8+ T cells isolated from both the brain and the spleen peripheral lymphoid compartment are specific for the same region of TMEV. This is the first direct comparison of spleen and brain CD8+ T-cell responses using a library approach to analyze all potential class I epitopes. With the knowledge that peripheral CD8 T cells are specific for the same MHC class I target in the CNS emerges the possibility that a similar approach could be designed for human work using a much more complex, randomly generated library, as has been performed by others [26,27]. Such a system could incorporate HLA-E and the detection of suppressor cytokines in addition to classical HLA molecules and detection of Th1 cytokines. A humanized system would be a marked advantage over current systems that require the culture of CD8+ T cells from peripheral blood or CSF cells which are subject to activation induced cell death and promiscuity of the T-cell receptor [27,28]. In addition, our system could also aid in the characterization of “driver clones,” as recently described in the EAE model of MS by Menezes et al. [29]. Whether such clones exist in other murine models of MS or in human MS is unknown at this time. A potential pitfall in the proposed future use of this technology in suppressor CD8 T cells is that there may be non–epitope-specific suppressor CD8 T cells, as recently demonstrated [30,31]. In addition, there may be unknown effects of antigen processing related to the interspecies mismatch between monkey cells (Cos7) and murine or human cells.
Randomly generated cDNA expression libraries using the Cos7 system have been used to define epitopes in complex diabetes models as well as large herpes viruses [23,32]. In general, these approaches use random cDNA libraries generated through incomplete Sau3AI 4–base pair cutting enzymatic digestion. The resulting digested cDNA fragments are cloned into pCDNA3.1 expression vectors in three separate reading frames. Pools of these randomly generated clones are then transiently expressed along with a class I molecule of interest. This approach uses a high throughput method to transiently express all potential CD8 T-cell epitopes along with a class I of interest. cDNA clones are identified only after demonstration of successful stimulation of CD8 T cells. In this manner, all potential epitopes have the capacity to be presented by a chosen HLA molecule also co-transfected into the Cos7 cell. The resulting library therefore is interchangeable with other class I molecules. Such a system could incorporate HLA-E and the detection of suppressor cytokines in addition to classical HLA molecules and detection of the Th1 cytokines.
The two prevailing hypotheses that clonally expanded CD8+ T-cell subsets are acting as suppressors or effectors in MS demonstrates the importance of ultimately defining class I:peptide epitopes. Defining epitopes in CNS inflammatory diseases will be critical to both understand their role in disease as well as manipulate their function to positively impact disease outcome. We also demonstrated that nearly undetectable CD8 T-cell populations in the periphery of the mouse are focusing on the same response as the brain, a novel finding of our study. The observation that peripheral CD8+ T cells are focused on the same target epitope as CNS-infiltrating CD8+ T cells also demonstrates that a similar more complex organ specific cDNA library approach could potentially be applied to clinical studies in MS.
Acknowledgments
We acknowledge Nilabh Shastri for providing the Kb- and Db-expressing plasmids, and likewise thank Feng Gao for technical assistance. This work was supported by National Institutes of Health grant RO1 NS058698, a National MS Society Pilot Grant, Waddell Center for MS, and intramural funds.
Footnotes
The authors have no conflicting financial interests.
References
- 1.Neumann H, Medana IM, Bauer J, Lassmann H. Cytotoxic T lymphocytes in autoimmune and degenerative CNS diseases. Trends Neurosci. 2002;25:313. doi: 10.1016/s0166-2236(02)02154-9. [DOI] [PubMed] [Google Scholar]
- 2.Babbe H, Roers A, Waisman A, Lassmann H, Goebels N, Hohlfeld R, Friese M, et al. Clonal expansions of CD8(+) T cells dominate the T-cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med. 2000;192:393. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skulina C, Schmidt S, Dornmair K, Babbe H, Roers A, Rajewsky K, et al. Multiple sclerosis: Brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc Natl Acad Sci U S A. 2004;101:2428. doi: 10.1073/pnas.0308689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennakoon DK, Mehta RS, Ortega SB, Bhoj V, Racke MK, Karandikar NJ. Therapeutic induction of regulatory, cytotoxic CD8+ T cells in multiple sclerosis. J Immunol. 2006;176:7119. doi: 10.4049/jimmunol.176.11.7119. [DOI] [PubMed] [Google Scholar]
- 5.Hu D, Ikizawa K, Lu L, Sanchirico ME, Shinohara ML, Cantor H. Analysis of regulatory CD8 T cells in Qa-1-deficient mice. Nat Immunol. 2004;5:516. doi: 10.1038/ni1063. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, Zhang SI, Pernis B. Role of CD8+ T cells in murine experimental allergic encephalomyelitis. Science. 1992;256:1213. doi: 10.1126/science.256.5060.1213. [DOI] [PubMed] [Google Scholar]
- 7.Koh DR, Fung-Leung WP, Ho A, Gray D, Acha-Orbea H, Mak TW. Less mortality but more relapses in experimental allergic encephalomyelitis in CD8−/−mice. Science. 1992;256(5060):1210. doi: 10.1126/science.256.5060.1210. [DOI] [PubMed] [Google Scholar]
- 8.Najafian N, Chitnis T, Salama AD, Zhu B, Benou C, Yuan X, et al. Regulatory functions of CD8+CD28- T cells in an autoimmune disease model. J Clin Invest. 2003;112:1037. doi: 10.1172/JCI17935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilliet M, Soumelis V, Watanabe N, Hanabuchi S, Antonenko S, de Waal-Malefyt R, Liu YJ. Human dendritic cells activated by TSLP and CD40L induce proallergic cytotoxic T cells. J Exp Med. 2003;197:1059. doi: 10.1084/jem.20030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoftberger R, Aboul-Enein F, Brueck W, Lucchinetti C, Rodriguez M, Schmidbauer M, et al. Expression of major histocompatibility complex class I molecules on the differenT-cell types in multiple sclerosis lesions. Brain Pathol. 2004;14:43. doi: 10.1111/j.1750-3639.2004.tb00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huseby ES, Liggitt D, Brabb T, Schnabel B, Ohlen C, Goverman J. A pathogenic role for myelin-specific CD8(+) T cells in a model for multiple sclerosis. J Exp Med. 2001;194:669. doi: 10.1084/jem.194.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D, Whitaker JN, Huang Z, Liu D, Coleclough C, Wekerle H, Raine CS. Myelin antigen-specific CD8+ T cells are encephalitogenic and produce severe disease in C57BL/6 mice. J Immunol. 2001;166:7579. doi: 10.4049/jimmunol.166.12.7579. [DOI] [PubMed] [Google Scholar]
- 13.Ure DR, Rodriguez M. Preservation of neurologic function during inflammatory demyelination correlates with axon sparing in a mouse model of multiple sclerosis. Neuroscience. 2002;111:399. doi: 10.1016/s0306-4522(02)00012-x. [DOI] [PubMed] [Google Scholar]
- 14.Murray PD, McGavern DB, Lin X, Njenga MK, Leibowitz J, Pease LR, Rodriguez M. Perforin-dependent neurologic injury in a viral model of multiple sclerosis. J Neurosci. 1998;18:7306. doi: 10.1523/JNEUROSCI.18-18-07306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AJ, Upshaw J, Pavelko KD, Rodriguez M, Pease LR. Preservation of motor function by inhibition of CD8+ virus peptide-specific T cells in Theiler’s virus infection. FASEB. 2001;15:2760. doi: 10.1096/fj.01-0373fje. [DOI] [PubMed] [Google Scholar]
- 16.Tsunoda I, Kuang LQ, Kobayashi-Warren M, Fujinami RS. Central nervous system pathology caused by autoreactive CD8+ T-cell clones after virus infection. J Virol. 2005;79:14640. doi: 10.1128/JVI.79.23.14640-14646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyman MA, Lee HG, Kang BS, Kang HK, Kim BS. Capsid-specific cytotoxic T lymphocytes recognize three distinct H-2D(b)-restricted regions of the BeAn strain of Theiler’s virus and exhibit different cytokine profiles. J Virol. 2002;76:3125. doi: 10.1128/JVI.76.7.3125-3134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borson ND, Paul C, Lin X, Nevala WK, Strausbauch MA, Rodriguez M, Wettstein PJ. Brain-infiltrating cytolytic T lymphocytes specific for Theiler’s virus recognize H2Db molecules complexed with a viral VP2 peptide lacking a consensus anchor residue. J Virol. 1997;71:5244. doi: 10.1128/jvi.71.7.5244-5250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsunoda I, Kuang LQ, Fujinami RS. Induction of autoreactive CD8+ cytotoxic T cells during Theiler’s murine encephalomyelitis virus infection: Implications for autoimmunity. J Virol. 2002;76:12834. doi: 10.1128/JVI.76.24.12834-12844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsunoda I, Libbey JE, Kobayashi-Warren M, Fujinami RS. IFN-gamma production and astrocyte recognition by autoreactive T cells induced by Theiler’s virus infection: Role of viral strains and capsid proteins. J Neuroimmunol. 2006;172:85. doi: 10.1016/j.jneuroim.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Pirko I, Gamez J, Johnson AJ, Macura SI, Rodriguez M. Dynamics of MRI lesion development in an animal model of viral-induced acute progressive CNS demyelination. Neuroimage. 2004;21:576. doi: 10.1016/j.neuroimage.2003.09.037. [DOI] [PubMed] [Google Scholar]
- 22.Pirko I, Suidan GL, Rodriguez M, Johnson AJ. Acute hemorrhagic demyelination in a murine model of multiple sclerosis. J Neuroinflammation. 2008;5:31. doi: 10.1186/1742-2094-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong FS, Karttunen J, Dumont C, Wen L, Visintin I, Pilip IM, et al. Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat Med. 1999;5:1026. doi: 10.1038/12465. [DOI] [PubMed] [Google Scholar]
- 24.Ghadge GD, Ma L, Sato S, Kim J, Roos RP. A protein critical for a Theiler’s virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J Virol. 1998;72:8605. doi: 10.1128/jvi.72.11.8605-8612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AJ, Njenga MK, Hansen MJ, Kuhns ST, Chen L, Rodriguez M, Pease LR. Prevalent class I-restricted T-cell response to the Theiler’s virus epitope Db: VP2121-130 in the absence of endogenous CD4 help, tumor necrosis factor alpha, gamma interferon, perforin, or costimulation through CD28. J Virol. 1999;73:3702. doi: 10.1128/jvi.73.5.3702-3708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judkowski V, Pinilla C, Schroder K, Tucker L, Sarvetnick N, Wilson DB. Identification of MHC class II-restricted peptide ligands, including a glutamic acid decarboxylase 65 sequence, that stimulate diabetogenic T cells from transgenic BDC2.5 nonobese diabetic mice. J Immunol. 2001;166:908. doi: 10.4049/jimmunol.166.2.908. [DOI] [PubMed] [Google Scholar]
- 27.Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, et al. How the T-cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Tallquist MD, Yun TJ, Pease LR. A single T-cell receptor recognizes structurally distinct MHC/peptide complexes with high specificity. J Exp Med. 1996;184:1017. doi: 10.1084/jem.184.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menezes JS, van den Elzen P, Thornes J, Huffman D, Droin NM, Maverakis E, Sercarz EE. A public T-cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE. J Clin Invest. 2007;117:2176. doi: 10.1172/JCI28277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filaci G, Rizzi M, Setti M, Fenoglio D, Fravega M, Basso M, et al. Non-antigen-specific CD8(+) T suppressor lymphocytes in diseases characterized by chronic immune responses and inflammation. Ann N Y Acad Sci. 2005;1050:115. doi: 10.1196/annals.1313.013. [DOI] [PubMed] [Google Scholar]
- 31.Filaci G, Fravega M, Fenoglio D, Rizzi M, Negrini S, Viggiani R, Indiveri F. Non-antigen specific CD8+ T suppressor lymphocytes. Clin Exp Med. 2004;4:86. doi: 10.1007/s10238-004-0042-3. [DOI] [PubMed] [Google Scholar]
- 32.Koelle DM. Expression cloning for the discovery of viral antigens and epitopes recognized by T cells. Methods. 2003;29:213. doi: 10.1016/s1046-2023(02)00344-4. [DOI] [PubMed] [Google Scholar]